Abstract
Work carried out over the last 30 years unveiled the role of structural dynamics in controlling protein function. Cavity networks modulate structural dynamics trajectories and are functionally relevant; in globins they have been assigned a role in ligand migration and docking. These findings raised renewed interest for time-resolved structural investigations of myoglobin (Mb), a simple heme protein displaying a photosensitive iron-ligand bond. Photodissociation of MbCO generates a nonequilibrium population of protein structures relaxing over a time range extending from picoseconds to milliseconds. This process triggers ligand migration to matrix cavities with clear-cut effects on the rate and yield of geminate rebinding. Here, we report subnanosecond time-resolved Laue diffraction data on the triple mutant YQR-Mb [Leu-29(B10)Tyr, His-64(E7)Gln, Thr-67(E10)Arg] that depict the sequence of structural events associated with heme and protein relaxation from 100 ps to 316 ns and above. The photodissociated ligand rapidly (<0.1 ns) populates the Xe-binding cavity distal to the heme. Moreover, the heme relaxation toward the deoxy configuration is heterogeneous, with a slower phase (≈ns) evident in these experiments. Damping of the heme response appears to result from a strain exerted by the E-helix via the CD-turn; Phe-43(CD1), in close contact with heme, opposes tilt until the strain is relieved. A comparison with crystallographic data on wild-type Mb and mutants Leu(29)Phe or Leu(29)Trp suggests that the internal structure controls the rate and amplitude of the relaxation events. A correlation between structural dynamics as unveiled by Laue crystallography and functional properties of Mb is presented.
Keywords: heme proteins, conformational landscape, packing defects, functional control
Since the seminal work on myoglobin (Mb) published >30 years ago by Austin et al. (1), the role of protein dynamics in controlling function has become a very active and challenging theme of research. The concept of conformational substates of a protein and the description of its energetics in terms of a complex landscape (2, 3) are fundamental steps forward in our understanding of the relationships between structure and function in proteins. This perspective is based on the notion that proteins are dynamic structures, selected to optimize thermodynamic stability and internal flexibility. Analysis of the structure of globular proteins revealed that internal packing defects result in the formation of cavities (4), which may correspond to real, thermodynamically costly structural voids; thereby it was assumed that they have been conserved during evolution to achieve functional tasks.
In Mb, the four cavities shown to bind xenon (5) were assigned a role in ligand migration and docking (6–9). This fact raised the interest for in-depth investigations of the structural dynamics of Mb, a “paradigm of complexity” (10), using protein engineering, transient spectroscopy, molecular dynamics (MD) simulations, or kinetic crystallography. To answer the general question of “how does the structure of Mb control its function?” an efficient strategy has been to assess how point mutations affect the dynamics of the conformational changes leading from the ligand bound to the deoxy state. With such data at hand, several aspects of the question may be addressed: can a common pattern of events after ligand photodissociation be identified in different Mb variants? Are structural relaxations specific to each variant in either kind or extent? Is the ligand migration pathway through the web of internal cavities affected by mutations? Is there a correlation between the directly observed conformational changes and the functional properties of the different variants?
Since the pioneering work on wild-type Mb (wt-Mb) by K. Moffat and his collaborators (11, 12), time-resolved Laue crystallography has emerged as a major approach to answering these questions. Recently, molecular movies could be produced with high spatial resolution and wide temporal sampling (100 ps to ms) (13–17). In a study of a triple mutant of sperm whale Mb called YQR-Mb [Leu-29(B10)Tyr, His-64(E7)Gln, Thr-67(E10)Arg] (18) it was seen that migration of the photodissociated ligand from the Xe4 cavity on the distal side of the heme to the Xe1 on the proximal side correlated with larger motions of the protein and lagged ≈100 ns in time after photolysis (13, 17). Schotte et al. (14), in a study on the Leu-29(B10)Phe Mb mutant (F-Mb), identified several docking sites visited by CO in <1 ns, whose access is largely controlled by conformational fluctuations of Phe-29. Very recently, time-resolved crystallographic and spectroscopic data (16, 19) on the Leu-29(B10)Trp Mb mutant (W-Mb) have revealed that Trp-29 acts as a dynamic plug that closes distal cavities shortly after photolysis; a quantitative analysis of the transient structural kinetics was successfully attempted in the ns-to-s time range (16, 19). The transient structures of YQR-Mb and F-Mb obtained by Laue crystallography (13, 14) proved to be largely consistent with MD simulations (20, 21), cross-validating the results by both techniques when available.
In this article, we extend our investigation by Laue crystallography on YQR-Mb to the ps regime. Our data show that photodissociated CO populates the distal Xe4 site by 100 ps, at variance with F-Mb and W-Mb, and that heme tilting displays heterogeneous kinetics, with a slower contribution extending to ns. These observations reinforce the concept of extended structural relaxation in Mb and allow an evaluation of the role of the internal structure on the dynamics and functional control.
Results and Discussion
Overall Summary of Experiments.
We have obtained four x-ray diffraction data sets from photo-excited YQR-MbCO single crystals, at 100 ps and six more delay times (316 ps, 512 ps, 1 ns, 3.16 ns, 316 ns, or 3.2 ms) (data not shown) and −10 ns, equivalent to “dark.” The snapshots acquired at delays >3.16 ns facilitated comparison with our previous results (13). Structural changes after CO photodissociation were assessed from the analysis of experimental difference electron density maps (Fig. 1). Despite relatively low levels of photolysis (from ≈10% to ≈17%, Table 1), the time evolution of these changes is clearly detected (as detailed in Materials and Methods). Importantly, similar trends were observed in analyzing either the positive or the negative difference electron density (Fig. 4, which is published as supporting information on the PNAS web site), indicating that reversible photolysis-induced disorder in the crystal, which would slow down the onset of positive density, has no significant influence.
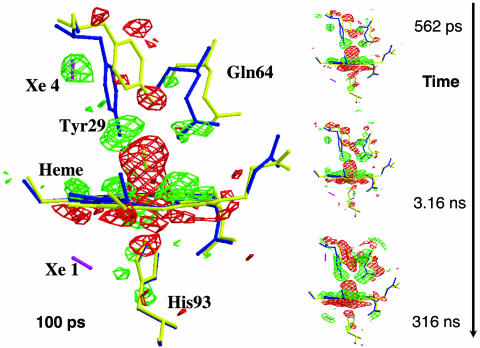
Fig. 1.
Structural changes in the heme vicinity of YQR-Mb detected at 0.1–316 ns after CO photolysis; the time frames at 316 ps and 1 ns are not shown. (Flight − Fdark) difference electron density maps (red: negative; green: positive; contoured at 3.0 σ, where σ is the standard deviation of the electron density difference) are overlaid on models of YQR-MbCO (yellow, Protein Data Bank code 1MYZ) and YQR-Mb (blue, Protein Data Bank code 1MZ0). For clarity, a magenta bar indicates the position of CO in both the Xe1 and Xe4 pockets.
Table 1.
Diffraction data
| Parameter | Data set |
|||
|---|---|---|---|---|
| YQR19b | YQR19c | YQR20 | YQR20b | |
| Resolution, Å | 30.00–1.65 | 30.00–1.70 | 30.00–1.75 | 30.00–1.75 |
| Outer shell | 1.74–1.65 | 1.79–1.70 | 1.84–1.75 | 1.84–1.75 |
| No. of unique reflections | 23,062 (336) | 21,319 (282) | 19,670 (240) | 19,620 (164) |
| Redundancy | 4.1 (0.2) | 4.0 (0.3) | 3.7 (0.2) | 4.0 (0.3) |
| Outer shell | 1.6 (0.1) | 1.8 (0.2) | 1.8 (0.1) | 1.7 (0.1) |
| Completeness, % | 88.1 (1.2) | 89.1 (1.2) | 89.8 (1.1) | 89.5 (0.8) |
| Outer shell | 71.2 (3.9) | 78.9 (2.2) | 74.4 (2.7) | 72.8 (2.8) |
| Rsym, %* | 7.2 (0.6) | 8.3 (0.7) | 6.9 (0.3) | 7.0 (0.6) |
| Outer shell | 16.2 (1.0) | 17.0 (0.8) | 15.3 (1.2) | 14.1 (1.4) |
| Mean I/σ(I) | 14.6 (1.7) | 14.1 (1.4) | 14.2 (0.7) | 15.2 (0.9) |
| Outer shell | 2.7 (0.6) | 3.1 (1.0) | 3.8 (0.6) | 4.2 (0.7) |
| Photolysis level, % | 10.2 (2.9)† | 15.1 (2.8) | 17.2 (2.6) | 12.4 (3.9)† |
The average of eight data sets [dark, 100 ps, 316 ps, 562 ps, 1 ns, 3 ns, 316 ns (YQR19c and YQR20) or 3,200 ns (YQR19b and YQR20b), −10 ms] is shown. Values in parenthesis correspond to the “stretch” along the series of data sets, ie the (maximum − minimum) values encountered in the series.
*Rsym = ΣjΣh|Ih,j − 〈Ih〉|/ΣjΣh Ih,j.
†The stretch for five data sets (100 ps, 316 ps, 562 ps, 1 ns, 3 ns) is shown, as the yield at 3,200 ns for YQR19b and YQR20b was found to be significantly above average.
Description of Structural Changes.
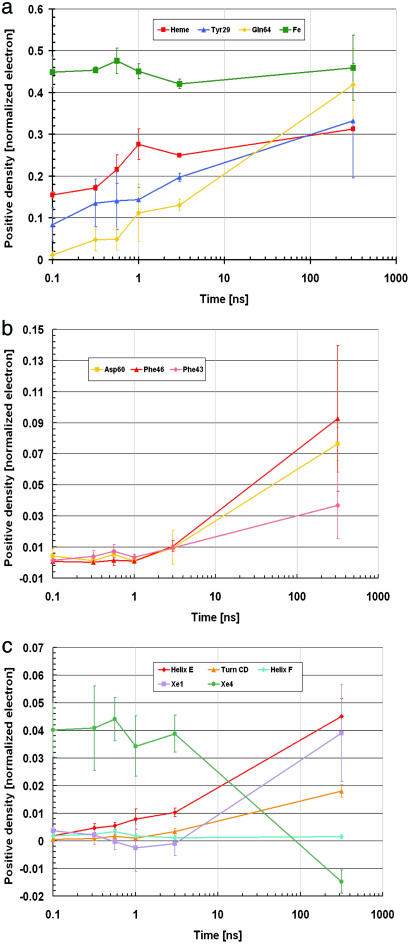
Figs. 1 and 2 depict the photolysis-induced time-dependent electron density changes in the neighborhood of the heme. Essential structural features are: (i) The photolysis of CO and the iron motion (≈0.3 Å) toward the proximal His-93(F8) are fully developed at 100 ps and remain constant up to 316 ns. CO recombination starts from ≈100 ms and leads back to the dark state at ≥3 ms (13). (ii) The heme tilt toward the distal side, already observable at 100 ps, is only partial (≈ 50% of total) and continues to develop for ≈1 ns. (iii) The photolyzed CO, found in the distal Xe4 cavity with maximum density at 100 ps, stays put up to 3.16 ns, and thereafter migrates to the proximal Xe1 site. (iv) The descent of Tyr-29(B10) toward the Fe is already seen at 100 ps but continuously progresses up to 316 ns. Relative to the 100-ps map, the corresponding difference electron density is about three times stronger at 3 ns and five times stronger at 316 ns; this moderate signal increase at long delays is at variance with our previous observation (13). (v) The motions experienced by Gln-64(E7) and the whole E-helix (not shown in Fig. 1), weak <1 ns, gain much more amplitude after ≈3 ns and reach their maximum occupancy at >316 ns. Relative to the 100-ps map, the signal of Gln-64(E7) is 10 times stronger at 3 ns and 40 times stronger at 316 ns. (vi) The motions experienced by the CD corner, especially Phe-46(CD4), and Asp-60(E3) (not shown in Fig. 1), only appear from 3.16 ns onward. (vii) Phe-43(CD1) (not shown in Fig. 1), in close contact with heme pyrrole C, displays a slowly developing motion along the whole time range; <1 ns this motion is judged significant, although at the limit of detection. (viii) The motions v and vi correlate in time with the migration of the photodissociated ligand from the distal Xe4 to the proximal Xe1 docking sites (see Fig. 2).
Fig. 2.
Time dependence of difference electron densities for key structural features. The numerical values reflect the integral of the positive electron density beyond 3.0 σ and are corrected for variations in photolysis yield. They are normalized so that the negative bound-CO feature is assigned a value of 1. Average values weighted by photolysis yield over the four independent data sets are shown. Error bars correspond to twice the weighted standard deviations between data sets. (a) Key features that appear promptly. (b) Residues involved in the strain of the CD turn, lagging behind. (c) Population of CO in the Xe1 and Xe4 sites and changes in secondary structures; in the latter, the average integrated density per residue is shown.
Questions Raised by the Early Time Points.
These experimental data reveal the extended character of protein relaxation as its structure evolves toward the deoxy state. Two features deserve special attention and will be discussed in light of the data reported by Schotte et al. (14, 15) on wt-Mb and F-Mb and by Schmidt et al. (16) on W-Mb. First, the heme tilt seen at 100 ps in wt-Mb, F-Mb, and YQR-Mb is similar in sign and direction. However, our data show unambiguously that in the case of YQR-Mb the (ensemble averaged) tilt has not reached its full extent by 100 ps, but rather continues to develop up to ns (Fig. 2a). Although this observation agrees with the stretched relaxation time course seen by time-resolved spectroscopy (22), it was not made explicitly in earlier studies. A first question is therefore to identify the structural origins of the extended heme relaxation. Second, in YQR-Mb the photolyzed CO migrates to the Xe4 cavity, 8 Å from the heme iron, within 100 ps, whereas in F-Mb it remains resident in the primary docking site at that time (14, 21) and requires several hundred ps to migrate to the Xe4 site. In wt-Mb photodissociated CO remains resident in the primary docking site up to 100 ns or just above (12, 15), whereas in W-Mb (16) CO is never detected distal to the heme and is seen only when located in the proximal Xe1 cavity. A second question is to identify the structural elements that permit the very rapid migration of CO to the Xe4 site in YQR-Mb.
A Mechanism for the Delayed Heme Relaxation.
To unveil the structural constraints that may damp the heme response in YQR-Mb, it is useful to recapitulate the sequential conformational changes occurring during the transition from the carboxyl to the deoxy conformation (Fig. 3). The deoxy conformation is characterized by a H bond between Tyr-29(B10) and Gln-64(E7). The transition to this conformation initiates with the displacement of Tyr-29(B10), which descends toward the vacated CO binding site and inhibits fast geminate recombination; the adjacent side chain, Gln-64(E7), slides into a position where it can form a H bond with Tyr-29(B10). The motion of Gln-64(E7) tends to drag the whole E-helix toward the heme, exerting a downward force onto the CD-turn, via Asp-60(E3). This force is not relieved until the main-chain carbonyl oxygen of Asp-60(E3) forces Phe-46(CD4) to undergo a substantial tilt in its ring orientation. As seen in Fig. 2, the latter conformational change starts at several ns and takes >100 ns to complete, thus lagging behind the heme relaxation. On the other hand, as CO dissociates, the CD-turn experiences an upward force caused by Phe-43(CD1) sensing the heme doming. This invariant residue, lying flat and in close contact with the heme (Fig. 3), is therefore strained from both directions and, as a result, tends to oppose the heme tilt. Confirmation of the strain experienced by Phe-43(CD1) during the first ns can be visualized in our data: the small upward displacement of this residue is one of the rare features displaying negative electron density more prominent than positive at the early time points (Fig. 5, which is published as supporting information on the PNAS web site). This stronger negative density is consistent with transient disorder being induced on Phe-43(CD1), which we assign to conflicting strain on the phenyl ring. Analysis of the time course (Fig. 2) of these complex structural changes relative to the extended heme relaxation suggests a distribution of conformational substates characterized by different kinetics and probably variable levels of strain on the nearby side chains [such as Phe-43(CD1) and Phe-46(CD4)].
Fig. 3.
A schematic view of a possible mechanism to account for the structural constraints involved in damping the heme response after CO photolysis (see text). Displacements caused by the transition from the CO-bound state (dark gray) to the deoxy state (light gray), together with relevant residues and the heme, are shown. Arrows highlight stereochemical clashes.
A point of interest is whether the sequential structural changes observed here are common to all Mbs. Is there a similar pattern of motions among the variants with differences limited to amplitude and time constants, or does each variant behave differently? Here, it is interesting to note that the conformational change of Phe-46(CD4) seen in YQR-Mb is present in only 3 of 10 deoxy mutants investigated so far by crystallography. Analysis of the structures of these Mb variants (plus wt-Mb) in the deoxy state highlights a correlation between the orientation of Phe-46(CD4) on one hand and the distance from the Cα of residue E7 to the heme iron on the other (Table 2, which is published as supporting information on the PNAS web site). When this distance is shorter than ≈8.3 Å (as in YQR-Mb, H64T-Mb, and H64Q-Mb), indicating a more pronounced displacement of the E-helix toward the heme, Phe-46(CD4) is forced to flip by Asp-60(E3). Nevertheless, time-resolved Laue data suggest that a slow phase of the heme relaxation is also seen in wt-Mb and F-Mb, with varying amplitudes relative to YQR-Mb (see the movies published in refs. 14 and 15). Therefore, we propose that the extended heme relaxation dynamics is a general feature in Mb, with the amplitude and time range modulated by a varying degree of strain imposed by the specific nature of a few distal residues.
The Early Migration of CO to the Xe4 Site.
In YQR-Mb, the occupancy of the Xe4 cavity by CO is already maximal at 100 ps (Figs. 1 and 2). This finding is consistent with the observation in a cryotrapping experiment (18) that, on photolysis at 20 K, CO populates this cavity, suggesting that migration to the Xe4 site must be fast and involves little protein motion. By comparison, ligand migration from the primary docking site to Xe4 in F-Mb is delayed by a few hundred ps but is still faster than in wt-Mb. This early difference in the migration of CO in F-Mb appears to be caused mostly by Phe-29(B10), His-64(E7), and the water molecule near the latter, as indicated by MD simulations (21). Whereas the presence of His-64(E7) in wt-Mb and F-Mb keeps CO in the primary docking site, Gln-64(E7) in YQR-Mb, by staying put immediately after photolysis, allows CO to jump to the Xe4 cavity (Fig. 1). In addition, Tyr-29(B10), by reducing the volume available in the primary docking site and providing a nonfavorable electrostatic environment, promotes early migration of photodissociated CO in the Xe4 cavity without blocking the passage. In the extreme case of the mutant W-Mb (16), the passage to Xe4 is blocked by the bulky Trp residue, explaining why CO is not detected distal to the heme.
The subsequent migration of CO along the web of cavities displays a common pattern of motions, because in all variants (12–14, 16) it populates the Xe1 cavity on the proximal side within a few tens (F-Mb) or hundreds of ns.
Dynamic Control of Function?
A point emerging from the time-resolved Laue studies on wt-Mb and three different Mb mutants (12–17) is that the structure of the heterogeneous protein matrix controls the rate and amplitude of the conformational changes, from ps to ms. We now present some speculation on the functional significance of these structural changes in relation to ligand binding. The notion that protein dynamics is a crucial component of functional control was established by extensive solution studies on the geminate recombination of O2, NO, and CO (1, 2, 23–31). Early geminate recombination of the dissociated ligand was shown to be heterogeneous and was proposed to be controlled by prosthetic group motions and the specific nature of the pocket residues. The extended relaxation of the heme (up to ns) demonstrated in this work is consistent with and supports a time dependence of the inner barrier. This important aspect for the interpretation of the early geminate rebinding in wt-Mb has been extensively investigated by time-resolved spectroscopy at low to room temperature, by studying the effect of viscosity, and by modeling the dependence of the barrier height on the protein coordinate (22–25, 28, 29, 32). In addition, it may be recalled that the probability of reaction has been correlated to the number of hits between the metal and the dissociated ligand while trapped in the protein. MD simulations (26) indicated that the average number of ligand/iron collisions per ps is much increased when the dissociated ligand is “confined” close to the heme. In the same article (26), the size of the amino acid side chain at position 29(B10) was found to correlate with diffusion of the ligand to the primary docking site, leading to an increase in geminate rates for bulkier residues. The immediate NO geminate recombination (ps) was significantly faster when Leu-29(B10) in wt-Mb was replaced by Phe, whereas it was grossly decreased when replaced by Ala or Val. These findings are consistent with the ps crystallographic data on F-Mb (14), which show that the photodissociated CO is confined in the primary docking site for a shorter time than in the case of wt-Mb. On the other hand, late geminate rebinding was invoked to depend on ligand migration away from the distal pocket, based on the effect of filling the cavities by mutations or Xe (30, 31). In fact, for the same Mb variants the rates of delayed (ns) geminate recombination of NO, reflecting back migration from the protein matrix toward the heme iron, follow an opposite order, suggesting that once NO has diffused in the protein, Phe-29(B10) provides a larger steric barrier to back diffusion. Consistent with these results, the overall NO dissociation rate constant for F29-Mb was found to be five times slower than for wt-Mb and ≈30 times slower than for A29-Mb (26), affecting overall ligand affinity by confinement of the (thermally) dissociated NO. Although geminate NO recombination for YQR-Mb was measured only in the ns range (33), data showed that: (i) the ns photolysis quantum yield for NO is ≈4%, i.e., much higher than for any other Mb variant, indicating that ps recombination must be negligible, and (ii) the delayed ns geminate recombination is remarkably slow (>100 ns) and is affected by Xe filling the cavities. These findings, which suggested immediate migration of the photolyzed ligand away from the distal pocket, are consistent with the very rapid partial movement of the bulky Tyr-29(B10) toward the heme and the immediate migration of the ligand into Xe4 seen at 100 ps, as described here.
Is the Extended Mb Relaxation a General Phenomenon?
Available time-resolved Laue data on photolyzed MbCO (11–16) prove that relaxation to the deoxy state extends over a large dynamic range in time. These studies provide the structural basis for interpreting the extensive investigations obtained by transient spectroscopy in solution, showing events extending from ps to ms. The huge time range, covering ≈9 orders of magnitude, has been described by a stretched exponential function reflecting the relaxation of the conformational substates of the protein within its complex energy landscape (3, 22, 29). The novelty unveiled by the Laue diffraction data is the direct visualization of the corresponding structural relaxation events with unprecedented time resolution. Here, we have provided evidence that relaxation of residues near the active site [such as Tyr-29(B10), Gln-64(E7), or Phe-43(CD1)], the heme tilting, the collapse of the E-helix, and the coupled strain on the CD corner all are stretched, covering a very large time range. Comparing the structural relaxations of the different Mb variants, we observe that the conformational changes are similar in kind but modulated in magnitude and speed by specific internal mutations, with some reasonable correlation to functional control (see above). Likewise, the internal migration of photolysed CO is in all variants (including wt-Mb) leading to maximal occupancy of the Xe1 cavity by a few hundred ns, but the population of the other docking sites en route is heavily modulated by a few distal residues.
A crucial point to be asked is to what extent is this nonexponential progression of conformational change a general feature of all proteins, and how can we address this question, especially for the faster time regime. In this context, the value of MD simulations, now extending to tens of ns and applicable to all proteins, is self-evident. Two studies recently published (20, 21) show satisfactory agreement with experiments, which carries more advantages, such as (i) features at the limit of reliability in the diffraction maps may be more confidently assigned to real protein motions, and (ii) the question related to the properties of single molecules can be compared with the average behavior detected by crystallography. Thus a combined approach may help to solve the fundamental problem of how the extended conformational relaxation documented by Laue crystallography relates to trajectories of individual protein molecules.
The conclusions reached from this Laue time-resolved crystallography study strongly support the concepts of conformational substates and energy landscapes in proteins (3, 10) and affirm the relevance of very fast structural dynamics to biology. The new challenge is to expand on this framework and explore the role of fast dynamics for other proteins, including those displaying allosteric control.
Materials and Methods
Protein Crystals.
YQR-Mb was produced in Escherichia coli from a synthetic gene with the substitution Asp-122–Asn, a conservative mutation with negligible functional and structural effects, but one that coaxes the protein to crystallize in space group P6 (34). Crystallization conditions under CO were as reported by Bourgeois et al. (13).
Laser Photolysis and Data Collection Strategy.
YQR-MbCO crystals were photolysed with light pulses (spectral range 520–560 nm) of ≈40 μJ that had been stretched to several tens of picoseconds by passage through 3 m of multimode optical fiber. A detailed description of the laser sources and optical setup is given in ref. 15. To extract the excess heat caused by visible light absorption by the sample, the capillary was bathed in a ≈10°C gas stream (Oxford Cryosystem 600). The achieved level of photolysis did not exceed ≈20% and in some instances was <10%, presumably because of the presence of a minor amount of oxidized met-Mb (estimated by optical spectroscopy to be <5%) or a residual mismatch between the pumped and probed volumes of the sample.
Laue diffraction data were acquired on the ID09B beamline at the European Synchrotron Radiation Facility. The synchrotron was operated in the so-called four-bunch mode, where four equally spaced electron bunches circulate in the storage ring. X-ray pulses of ≈150 ps (full width at half maximum) containing ≈1010 photons per pulse were generated in a relative bandwidth of ≈3.5% (full width at half maximum) peaked at 0.79 Å (15).
Four complete time-resolved diffraction data sets were collected, each from separate regions of two different large crystals. Each data set was comprised of 31 images spanning 60°. To improve the signal-to-noise ratio, each image represented the integration of 16 pump-probe measurements accumulated at 3 Hz on a MarCCD detector (Mar USA, Evanston, IL). To minimize systematic errors between time points, time was used as the “fast variable.” At a given crystal orientation, all of the diffraction images at the investigated pump-probe delays were collected, and then the crystal was rotated and the process was repeated. With this approach, fluctuations in photolysis yield and radiation damage are distributed equally over all time points, thereby minimizing their effects on the observed time-dependent structural changes (14, 15, 35).
Data Analysis.
Diffraction data were reduced in an automated way by using lauegen, prow, and lscale (Table 1) (36). Experimental electron density difference maps (Flight − Fdark) were generated with ccp4 (37) using phases from a dark-state model (Protein Data Bank code 1MYZ, refined from earlier data on similar crystals), after optimization of the difference amplitudes with a Bayesian statistics-based method (38). To estimate the photolysis yield, the negative difference electron density feature at the bound CO was integrated within a mask excluding the contribution of the iron motion. This density was then compared with the one obtained from simulated data of equal completeness and corresponding to complete photolysis of CO. For all data set series, the photolysis yield was found to be constant (± ≈1%) over the time range 100 ps to 3.16 ns. To evaluate the progress of the structural changes with time at different locations of the structure, the difference (Flight − Fdark) maps were integrated above/below ±3.0 σ with homemade software based on a 3D connectivity search algorithm. For each Mb atom, difference electron density above/below ±3.0 σ was searched at a maximum distance of 1.5 Å from the atom. If density was found, integration was carried out with the connectivity search algorithm, within a limiting cube of 8 Å3. Voxels above/below the σ threshold could be selected several times (by several atoms), but were considered only once. Results were then normalized to the negative density feature at the CO for each time delay, which was assigned an arbitrary value of 1.
Figs. 1 and 3 were drawn by using bobscript (39) and raster3d (40). In Fig. 2, the data shown refer to the weighted average behavior of the structural features as assessed from analysis of all four data sets. Weights w were taken as w = MAX(0, |photolysis yield − 10|), so that individual data sets with a photolysis yield <10%, judged to have a too high level of noise, were discarded. Error bars corresponding to twice the weighted standard deviations between data sets at every time point are also shown in Fig. 2. However, it must be kept in mind that these error estimates include all kinds of systematic deviations from crystal to crystal, and therefore give an overly pessimistic outlook of the data. Because time was used as the fast variable, the time evolution of the electron density for a single crystal is believed to be much more reliable than suggested by these error bars, although no other statistical evaluation is available.
Supplementary Material
Acknowledgments
We thank Dr. M. Wolff, ID09, European Synchrotron Radiation Facility. This research was supported in part by Ministero dell’Istruzione, dell Università e della Ricerca Grants RBLA03B3KC_004 2003 and RBNE01KXC9 2005 (to M.B.) and the Intramural Research Program of the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases (P.A.A.).
Abbreviations
- Mb
myoglobin
- wt-Mb
wild-type Mb
- YQR-Mb
Mb triple mutant [Leu-29(B10)Tyr, His-64(E7)Gln, Thr-67(E10)Arg]
- F-Mb
Leu-29(B10)Phe Mb mutant
- W-Mb
Leu-29(B10)Trp Mb mutant
- MD
molecular dynamics.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Austin R. H., Beeson K. W., Eisenstein L., Frauenfelder H., Gunsalus I. C. Biochemistry. 1975;14:5355–5373. doi: 10.1021/bi00695a021. [DOI] [PubMed] [Google Scholar]
- 2.Frauenfelder H., Parak F., Young R. D. Annu. Rev. Biophys. Chem. 1988;17:451–479. doi: 10.1146/annurev.bb.17.060188.002315. [DOI] [PubMed] [Google Scholar]
- 3.Frauenfelder H., Sligar S. G., Wolynes P. G. Science. 1991;254:1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 4.Richards F. M. Annu. Rev. Biophys. Bioeng. 1977;6:151–176. doi: 10.1146/annurev.bb.06.060177.001055. [DOI] [PubMed] [Google Scholar]
- 5.Tilton R. F., Jr., Kuntz I. D., Jr., Petsko G. A. Biochemistry. 1984;23:2849–2857. doi: 10.1021/bi00308a002. [DOI] [PubMed] [Google Scholar]
- 6.Elber R., Karplus M. J. Am. Chem. Soc. 1990;112:9161–9175. [Google Scholar]
- 7.Carlson M. L., Regan R. M., Gibson Q. H. Biochemistry. 1996;35:1125–1136. doi: 10.1021/bi951767k. [DOI] [PubMed] [Google Scholar]
- 8.Decatur S. M., DePillis G. D., Boxer S. G. Biochemistry. 1996;35:3925–3932. doi: 10.1021/bi952625t. [DOI] [PubMed] [Google Scholar]
- 9.Brunori M., Gibson Q. H. EMBO Rep. 2001;2:674–679. doi: 10.1093/embo-reports/kve159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frauenfelder H., McMahon B. H., Fenimore P. W. Proc. Natl. Acad. Sci. USA. 2003;100:8615–8617. doi: 10.1073/pnas.1633688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Srajer V., Teng T., Ursby T., Pradervand C., Ren Z., Adachi S., Schildkamp W., Bourgeois D., Wulff M., Moffat K. Science. 1996;274:1726–1729. doi: 10.1126/science.274.5293.1726. [DOI] [PubMed] [Google Scholar]
- 12.Srajer V., Ren Z., Teng T. Y., Schmidt M., Ursby T., Bourgeois D., Pradervand C., Schildkamp W., Wulff M., Moffat K. Biochemistry. 2001;40:13802–13815. doi: 10.1021/bi010715u. [DOI] [PubMed] [Google Scholar]
- 13.Bourgeois D., Vallone B., Schotte F., Arcovito A., Miele A. E., Sciara G., Wulff M., Anfinrud P., Brunori M. Proc. Natl. Acad. Sci. USA. 2003;100:8704–8709. doi: 10.1073/pnas.1430900100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schotte F., Lim M., Jackson T. A., Smirnov A. V., Soman J., Olson J. S., Phillips G. N., Jr., Wulff M., Anfinrud P. A. Science. 2003;300:1944–1947. doi: 10.1126/science.1078797. [DOI] [PubMed] [Google Scholar]
- 15.Schotte F., Soman J., Olson J. S., Wulff M., Anfinrud P. A. J. Struct. Biol. 2004;147:235–246. doi: 10.1016/j.jsb.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 16.Schmidt M., Nienhaus K., Pahl R., Krasselt A., Anderson S., Parak F., Nienhaus G. U., Srajer V. Proc. Natl. Acad. Sci. USA. 2005;102:11704–11709. doi: 10.1073/pnas.0504932102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brunori M., Bourgeois D., Vallone B. J. Struct. Biol. 2004;147:223–234. doi: 10.1016/j.jsb.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 18.Brunori M., Vallone B., Cutruzzola F., Travaglini-Allocatelli C., Berendzen J., Chu K., Sweet R. M., Schlichting I. Proc. Natl. Acad. Sci. USA. 2000;97:2058–2063. doi: 10.1073/pnas.040459697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nienhaus K., Ostermann A., Nienhaus G. U., Parak F. G., Schmidt M. Biochemistry. 2005;44:5095–5105. doi: 10.1021/bi047513t. [DOI] [PubMed] [Google Scholar]
- 20.Bossa C., Anselmi M., Roccatano D., Amadei A., Vallone B., Brunori M., Di Nola A. Biophys. J. 2004;86:3855–3862. doi: 10.1529/biophysj.103.037432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hummer G., Schotte F., Anfinrud P. A. Proc. Natl. Acad. Sci. USA. 2004;101:15330–15334. doi: 10.1073/pnas.0405295101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson T. A., Lim M., Anfinrud P. Chem. Phys. 1994;180:131–140. [Google Scholar]
- 23.Henry E. R., Sommer J. H., Hofrichter J., Eaton W. A. J. Mol. Biol. 1983;166:443–451. doi: 10.1016/s0022-2836(83)80094-1. [DOI] [PubMed] [Google Scholar]
- 24.Petrich J. W., Lambry J. C., Kuczera K., Karplus M., Poyart C., Martin J. L. Biochemistry. 1991;30:3975–3987. doi: 10.1021/bi00230a025. [DOI] [PubMed] [Google Scholar]
- 25.Steinbach P. J., Ansari A., Berendzen J., Braunstein D., Chu K., Cowen B. R., Ehrenstein D., Frauenfelder H., Johnson J. B., Lamb D. C., et al. Biochemistry. 1991;30:3988–4001. doi: 10.1021/bi00230a026. [DOI] [PubMed] [Google Scholar]
- 26.Gibson Q. H., Regan R., Elber R., Olson J. S., Carver T. E. J. Biol. Chem. 1992;267:22022–22034. [PubMed] [Google Scholar]
- 27.Ikeda-Saito M., Dou Y., Yonetani T., Olson J. S., Li T., Regan R., Gibson Q. H. J. Biol. Chem. 1993;268:6855–6857. [PubMed] [Google Scholar]
- 28.Ansari A., Jones C. M., Henry E. R., Hofrichter J., Eaton W. A. Biochemistry. 1994;33:5128–5145. doi: 10.1021/bi00183a017. [DOI] [PubMed] [Google Scholar]
- 29.Hagen S. J., Eaton W. A. J. Chem. Phys. 1996;104:3395–3398. [Google Scholar]
- 30.Scott E. E., Gibson Q. H. Biochemistry. 1997;36:11909–11917. doi: 10.1021/bi970719s. [DOI] [PubMed] [Google Scholar]
- 31.Scott E. E., Gibson Q. H., Olson J. S. J. Biol. Chem. 2001;276:5177–5188. doi: 10.1074/jbc.M008282200. [DOI] [PubMed] [Google Scholar]
- 32.Agmon N., Hopefield J. J. J. Phys. Chem. 1983;79:2042–2053. [Google Scholar]
- 33.Brunori M., Cutruzzola F., Savino C., Travaglini-Allocatelli C., Vallone B., Gibson Q. H. Biophys. J. 1999;76:1259–1269. doi: 10.1016/S0006-3495(99)77289-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phillips G. N., Jr., Arduini R. M., Springer B. A., Sligar S. G. Proteins. 1990;7:358–365. doi: 10.1002/prot.340070407. [DOI] [PubMed] [Google Scholar]
- 35.Ihee H., Rajagopal S., Srajer V., Pahl R., Anderson S., Schmidt M., Schotte F., Anfinrud P. A., Wulff M., Moffat K. Proc. Natl. Acad. Sci. USA. 2005;102:7145–7150. doi: 10.1073/pnas.0409035102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bourgeois D., Wagner U., Wulff M. Acta Crystallogr. D. 2000;56:973–985. doi: 10.1107/s0907444900006958. [DOI] [PubMed] [Google Scholar]
- 37.Collaborative Computational Project. Acta Crystallogr. D. 1994;50:760–763. [Google Scholar]
- 38.Ursby T., Bourgeois D. Acta Crystallogr. A. 1997;53:564–575. [Google Scholar]
- 39.Esnouf R. M. Acta Crystallogr. D. 1999;55:938–940. doi: 10.1107/s0907444998017363. [DOI] [PubMed] [Google Scholar]
- 40.Merritt E. A., Bacon D. J. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.