Abstract
Vernalization is the acceleration of flowering by prolonged cold that aligns the onset of reproductive development with spring conditions. A key step of vernalization in Arabidopsis is the epigenetic silencing of FLOWERING LOCUS C (FLC), which encodes a repressor of flowering. The vernalization-induced epigenetic silencing of FLC is associated with histone deacetylation and H3K27me2 and H3K9me2 methylation mediated by VRN/VIN proteins. We have analyzed whether different histone methyltransferases and the chromodomain protein LIKE HETEROCHROMATIN PROTEIN (LHP)1 might play a role in vernalization. No single loss-of-function mutation in the histone methyltransferases studied disrupted the vernalization response; however, lhp1 mutants revealed a role for LHP1 in maintaining epigenetic silencing of FLC. Like LHP1, VRN1 functions in both flowering-time control and vernalization. We explored the localization of VRN1 and found it to be associated generally with Arabidopsis chromosomes but not the heterochromatic chromocenters. This association did not depend on vernalization or VRN2 function and was maintained during mitosis but was lost in meiotic chromosomes, suggesting that VRN1 may contribute to chromatin silencing that is not meiotically stable.
Keywords: chromatin, mitosis, vernalization, flowering, meiosis
Many plants require prolonged cold before they will flower, as a mechanism to ensure that they flower in spring. The acceleration of flowering by cold is a process called vernalization, and it has been dissected by using a molecular genetic approach in Arabidopsis (1–3). A central player in vernalization is a MADS box protein FLOWERING LOCUS C (FLC) (3–6). FLC represses flowering by delaying the activation of a set of genes called floral pathway integrators, required to switch the meristem from a vegetative to floral fate (7). FLC expression is down-regulated by prolonged cold, and this repression is epigenetically maintained during the subsequent development of the plant. This epigenetic control of FLC by vernalization is mediated by VIN3, a PHD finger protein; VRN2, a homologue of the Polycomb protein Su(z)12; and VRN1, a plant-specific protein containing DNA-binding domains (8–10).
The VRN/VIN proteins are required for vernalization-dependent histone modifications at FLC, which include reduction of H3 acetylation and increased H3K9 dimethylation (me2) and H3K27me2 (10, 11). A possible sequence of events may be: VIN3 activity initiates histone modifications (deacetylation) at FLC, enabling H3K27 methylation by a VRN2-containing Polycomb complex equivalent to Polycomb repressive complex (PRC)2 (12) and/or H3K9 methylation by a VRN1-containing complex. This model predicts the involvement of a number of factors that have not yet been identified in the genetic analysis of vernalization. In the PRC2 complex, Su(z)12 is thought to confer nucleosome binding, whereas the H3K27 histone methyltransferase activity depends predominantly on ENHANCER OF ZESTE [E(z)] (13). An E(z) function may therefore be associated with the VRN2 complex. The methylation of H3K9 may also involve a second histone methyltransferase activity associated with the plant equivalent of PRC1 (14). Unlike the components of PRC2, those of PRC1 do not appear to be evolutionarily conserved and cannot be identified in the Arabidopsis genome (15). An interesting possibility is that a VRN1-containing complex may undertake a similar function to PRC1 in plants. If this is the case, it is likely to be associated with a K9 methyltransferase activity. A major contributor to H3K9me2 in Arabidopsis is KRYPTONITE (KYP), a member of the Su(var)3–9 class of histone methyltransferases first isolated from Drosophila (16) and identified in Arabidopsis through its role in transcriptional silencing of both endogenous genes and transgenes (17, 18). Recently, a related protein SUVH2 was shown to have in vitro histone methyltransferase activity and to be required in vivo for H3K9, H3K27, and H4K20 methylation (19). KYP and SUVH2 are 2 of 14 members constituting the Arabidopsis SUVH/SUVR class of proteins that contain SET domains predicted to methylate histone 3 (20).
Another key component in epigenetic silencing in Drosophila and vertebrates is HETEROCHROMATIN PROTEIN (HP)1. HP1 was shown to bind specifically to methylated H3K9 and to be involved in the formation of heterochromatin (21), and it has been shown that tethering HP1 to chromatin is sufficient to induce silencing of surrounding genes (22). Usually, HP1 is associated with heterochromatic regions but has also been shown to associate with euchromatin (23, 24). The Arabidopsis genome encodes only one HP1-related protein (LHP1) which is much larger than mammalian or Drosophila HP1, with a moderately conserved chromo domain and weakly conserved chromo shadow domain (25). Analysis of plants carrying a complementing LHP1/GFP fusion, driven by either the LHP1 or 35S promoters, has indicated that LHP1 is predominantly localized outside the heterochromatic chromocenters (26, 27). In contrast, transient expression assays have shown that LHP1 localizes to chromocenters and that this localization depends on sequences that recognize methylated H3 lysine 9 (28). lhp1 mutants, also referred to as tfl2 (29) and tu8 (30) do not appear to cause misexpression of heterochromatic regions (17, 27, 31, 32) but are early flowering because of ectopic expression of genes, including FT, AP3, PI, and SEP. The one HP1 homologue in the Arabidopsis genome thus appears to function rather differently from its animal counterparts.
The histone modifications at FLC induced by vernalization are epigenetically stable throughout vegetative development, but, at some stage, FLC expression must be reset to ensure a vernalization requirement in each generation of seedlings. Unlike animals, the germ line in plants is not laid down during embryo development. In flowering plants, specialized reproductive cells differentiate from somatic tissue within the flower. These cells undergo meiosis to produce haploid megaspores that then undergo further rounds of mitotic division to produce the pollen grain and embryo sac. The gametes are formed from these multicellular haploid structures. Self-fertilization is the norm in Arabidopsis, after which the embryo develops through characteristic stages until it desiccates, and the mature seed is formed. Erasure of the vernalization-induced histone modifications at FLC must occur before final embryo development to give the high FLC levels at germination and thus prevent precocious flowering (33).
To further investigate the components involved in Polycomb silencing in plants, we have investigated whether Arabidopsis mutants defective in either histone methyltransferase activities or LHP1 accumulation are impaired in vernalization. The results enabled further comparative analysis between the Polycomb-silencing systems of plants and animals. In addition, we have explored the role of VRN1 in chromatin silencing and discovered its widespread association with Arabidopsis chromosomes but not the heterochromatic chromocenters. This association was mitotically stable but was lost in meiotic chromosomes, suggesting VRN1’s involvement in mitotic but not meiotic epigenetic inheritance.
Results
Single suvh and suvr Mutants Are Still Responsive to Vernalization.
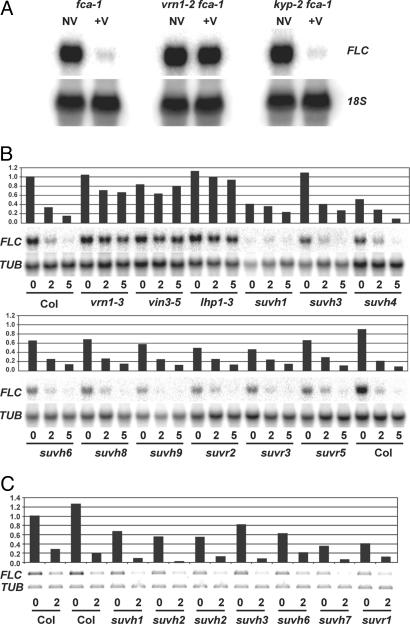
The KYP histone methyltransferase (also known as SUVH4) has been shown to be essential for most of the H3K9me2 in Arabidopsis (32). To analyze whether KYP was involved in the vernalization-induced H3K9 methylation at FLC, the kyp-2 mutation [in Landsberg erecta (Ler)] was introduced into fca-1 (also in Ler). fca-1 causes a vernalization-responsive late-flowering phenotype that can provide the background to test the role of various factors in vernalization. kyp-2 fca-1 flowered at approximately the same time as fca-1 and showed a similar suppression of FLC expression and accelerated flowering after vernalization; thus, we can conclude that KYP is not necessary for FLC regulation (Fig. 1A and Table 1). To expand the search for a histone methyltransferase involved in vernalization, we examined FLC levels in seven additional suvh mutants and four suvr mutants (see Table 2, which is published as supporting information on the PNAS web site). The suvh and suvr alleles (in Columbia) contain mutations in genes related to KYP that contain SET domains predicted to methylate histone 3 (20). The mutants include a T-DNA insertion in SUVH2, a gene recently described as being essential for an array of repressive histone methylation marks (19). These mutants were examined by a combination of Northern blots and RT-PCR of nonvernalized and vernalized tissue (Fig. 1 B and C). Although many of the suv mutants displayed moderately reduced levels of FLC in nonvernalized seedlings, none was impaired in the ability to repress FLC in response to vernalization.
Fig. 1.
FLC expression in different histone methyltransferase mutants. (A) Northern blot analyzing FLC expression in fca-1, vrn1-2 fca-1, and kyp-2 fca-1. Seedlings were grown for 19 days (NV) or vernalized 4 weeks as imbibed seeds and harvested after 16 days growth (+V). The blot was subsequently stripped and probed with 18S rDNA as a loading control. (B) Northern blot analyzing FLC expression in Columbia, suvh, suvr, vrn1-3, and vin3-5 single mutants in Columbia either nonvernalized (0) or vernalized as seeds for 2 or 5 weeks on soil and harvested after 15 days of growth. The blot was subsequently stripped and probed with TUBULIN (TUB) as a loading control. (C) RT-PCR performed on total RNA extracted from plants that were nonvernalized or vernalized for 2 weeks as seeds and harvested after 12 days of growth. Plants from two different seed lots of suvh2 were included.
Table 1.
Flowering time of different genotypes
| Genotype | ESD NV | ESD +V |
|---|---|---|
| fca-1 | 27.3 ± 0.3 | 9.1 ± 0.2* |
| vrn1-2 fca-1 | 35.6 ± 0.4 | 24.3 ± 0.4* |
| VRN1 GFP vrn1-2 fca-1 | 35.5 ± 0.6 | 11.9 ± 0.2* |
| fca-1 | 45.8 ± 1.6 | 12.8 ± 0.3† |
| vrn1-2 fca-1 | 46.2 ± 1.7 | 41.3 ± 2.9† |
| kyp-2 fca-1 | 57.2 ± 3.7 | 12.3 ± 0.8† |
| lhp1-3 FRI FLC/luciferase | 24.8 ± 1.3 | 13.5 ± 0.6* |
| Col FRI FLC/luciferase | 86.0 ± 2.2 | 15.8 ± 0.7* |
ESD, extended short day; NV, nonvernalized; +V, vernalized; Col, Columbia.
∗, +V, 6 weeks.
†, +V, 4 weeks.
LHP1 Is Required for Maintenance of Vernalization-Induced Repression of FLC.
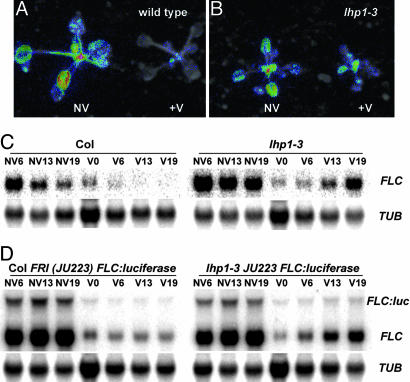
To discover whether LHP1 plays a role in FLC silencing, the vernalization response of plants deficient in LHP1 was analyzed. The lhp1-3 allele was crossed into a line containing an active FRIGIDA allele and an FLC/luciferase translational fusion. The FLC/luciferase fusion mirrors the regulation of the endogenous FLC gene in response to both FRIGIDA, fca mutations and vernalization and, so, provides an effective assay to follow FLC expression (3). The lhp1-3 mutation did not alter FLC/luciferase expression in nonvernalized seedlings but resulted in increased expression in vernalized seedlings (Fig. 2A and B). Detailed time-course analysis of FLC expression in Columbia and FRIGIDA + backgrounds showed that FLC expression was repressed in lhp1-3 mutants like wild type during the cold but then increased during subsequent growth in warm temperatures (Fig. 2 C and D). To ensure that this was not an effect of a mutation linked to lhp1-3, we tested the vernalization response of lhp1-4 and lhp1-5 and found that all lhp alleles were equally compromised in their ability to suppress FLC by vernalization (data not shown). A reduction of FLC levels in the cold, followed by increased expression later is similar to that observed in the vrn mutants (8, 9) and suggests that LHP1 is required for maintenance of FLC repression postvernalization. Analysis of flowering time in lhp1-3 FRI plants was complicated by ectopic expression of FT in the lhp1-3 background, a floral promoter that functions downstream of FLC, demonstrating the importance of assaying FLC expression rather than flowering time (Table 1).
Fig. 2.
Effect of lhp1 mutations on FLC repression by vernalization. Merged FLC/luciferase (pseudocolor) and photographic (black/white) images taken of Col FRI FLC/luciferase grown for 3 weeks either without vernalization (NV) or after 6 weeks vernalization (+V) (A) and lhp1-3 FRI FLC/luciferase nonvernalized (NV) or vernalized (+V) (B). (C and D) Northern blot time-course analysis of FLC expression in lhp1-3 single mutants and Columbia (C) or lhp1-3 FRI FLC/luciferase and Col FRI FLC/luciferase (D). Plants were harvested from plates at three stages of nonvernalized growth (NV6–NV19 days), immediately after 6 weeks cold (V0) and three stages of growth after vernalization (V6–V19). Blots were subsequently stripped and probed with TUBULIN (TUB) as a loading control.
VRN1 Is Associated with Many Arabidopsis Chromosomal Regions Throughout Mitosis.
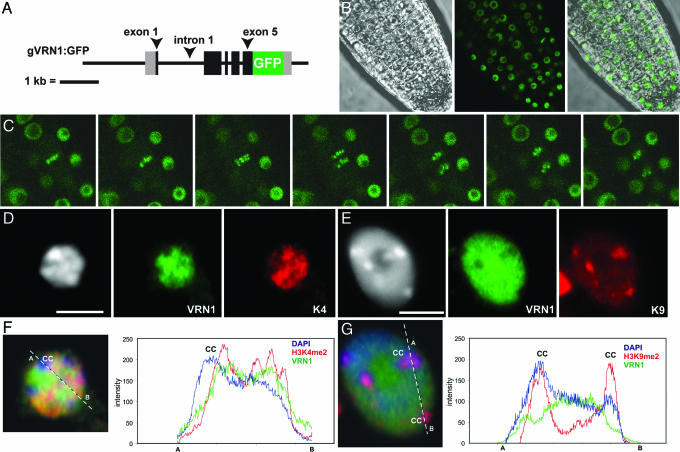
VRN1 is a protein with two plant-specific B3 domains and binds dsDNA non-sequence-specifically in vitro (9). Chromatin immunoprecipitation experiments have shown that VRN1 is associated with FLC chromatin (10). To further analyze its in vivo DNA-binding properties, Arabidopsis transformants carrying a VRN1/GFP translational fusion (GFP at the translation termination site in a VRN1 genomic fragment) were generated (Fig. 3A). A single locus line was chosen and found to be fully functional, because it could restore the vernalization response of the late flowering vrn1-2 fca-1 double mutant to that of fca-1 (Table 1). In vivo, VRN1/GFP localized strongly and evenly throughout the nucleoplasm, with very little fluorescence in the nucleolus and cytoplasm (Fig. 3 B and C). Examination of root tips revealed that VRN1/GFP appeared to localize to all Arabidopsis chromosomes (only four are clearly seen in the figure), remaining associated throughout mitosis (Fig. 3C). The VRN1/GFP pattern in interphase cells also indicated a specific subnuclear location. To examine this finding further, we performed immunodetection experiments using an antibody raised against full-length VRN1 in parallel with H3K4me2 and H3K9me2 antibodies. The latter two are cytological marks for euchromatin and visible heterochromatin, respectively. Whereas H3K9me2 labels the chromocenters, the VRN1 and H3K4me2 signals were both located outside the chromocenters (Fig. 3 D–G). However, VRN1 and H3K4me2 showed different labeling intensities at particular locations, suggesting localization at different targets within the chromosomal arms. This localization is illustrated by the green-yellow-orange-red pattern caused by overlay of DAPI, K4, and VRN1 immunofluorescence signals (Fig. 3F).
Fig. 3.
VRN1/GFP associates with mitotic chromosomes and not heterochromatin. (A) A genomic VRN1/GFP construct was produced by mutating the VRN1 stop codon into a BamHI site for in-frame fusion with GFP coding sequence. (B) GFP expression in root tissue of VRN1/GFP vrn1-2 fca-1. (C) Time series showing association of VRN1/GFP with chromosomes during mitosis, images taken at 30-second intervals. (D) Immunodetection in wild-type Ler using VRN1 (green) and H3K4me2 (red) antibodies. (E) Immunodetection in wild-type Ler using VRN1 (green) and H3K9me2 (red) antibodies. (F) Quantitative line profiles of DAPI (blue), VRN1 (green), and H3K4me2 (red). (G) Quantitative line profiles of DAPI (blue), VRN1 (green), and H3K9me2 (red). (Scale bars, 5 μm.)
VRN1 Is Not Associated with Chromosomes in Male Meiosis.
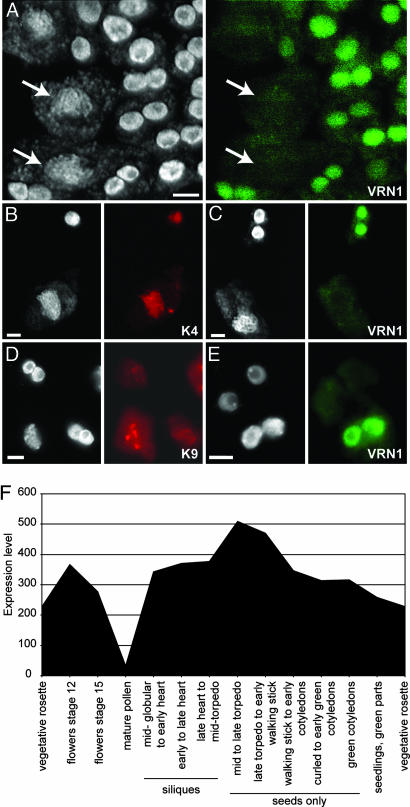
Because VRN1 associates with mitotic chromosomes, we investigated whether it was also associated with meiotic chromosomes by immunolocalization on chromosome-spread preparations from anthers. In somatic nuclei, chromocenters and nucleoli can be observed, whereas, in meiotic nuclei, individual chromosome fibers become visible. We found that somatic cells stained strongly for VRN1, whereas no signal was detectable in meiotic nuclei at midprophase I (Fig. 4A and C). This condition persisted to a later stage of meiosis, where the VRN1 signal was detected in somatic cells but not in microspores (Fig. 4E). This result is not due to an inability to label meiotic cells, because an H3K4me2 signal was detectable in midprophase nuclei (Fig. 4B). Somatic cells, such as the binucleate tapetum cells, also labeled well with anti-H3K4me2. The H3K9me2 antibody also labeled the chromocenters of both tapetum and midprophase nuclei (Fig. 4D). Analysis of subsequent pollen stages was prevented by the formation of impenetrable cell walls. The loss of VRN1 association in meiotic chromosomes could be caused by a changed localization of the protein or reduced expression of VRN1. Examination of the publicly available microarray data (34) shows that VRN1 expression can be detected throughout vegetative development in flowers and developing embryos but is ≈10-fold lower in mature pollen (Fig. 4F). Thus, it seems likely that a reduced expression causes the loss of VRN1 association to meiotic chromosomes.
Fig. 4.
Immunodetection of anther preparations. (A) DAPI staining (Left, white) and VRN1 immunodetection (Right, green). The larger meiotic cells, which are weakly stained with VRN1 antibody, are marked by arrows. (B) The H3K4me2 antibody (red) is able to penetrate and label the somatic binucleate tapetum nuclei (above, binucleate and stronger DAPI) and mid-prophase meiotic nuclei (below, mononucleate and weaker DAPI stain). (C) The VRN1 antibody (green) labels somatic (above) but not midprophase meiotic nuclei (below). (D) The H3K9me2 antibody (red) labels the chromocenters of somatic (above and Right) and mid-prophase meiotic nuclei (below, Left). (E) The VRN1 antibody (green) labels the tapetum nuclei (below) but not the nuclei of microspores (above). (F) Selected AtGenExpress data (34) showing drop in VRN1 expression in mature pollen. (Scale bars, 5 μm.)
VRN1 Localization Does Not Change with Vernalization or in Polycomb Mutants.
The requirement for prolonged cold for VRN1 action on FLC led us to analyze whether the VRN1/GFP localization was altered by vernalization. No obvious changes were detected at the level of resolution provided by confocal microscopy (data not shown); however, this assay would not detect changes occurring at specific loci. We also addressed whether VRN1 may be recruited to its site of action by a function associated with VRN2 activity. The general chromosome association of VRN1 supports the idea that it is not an FLC-specific regulator (9), which also appears to be the case for VRN2 (35). Localization of VRN1/GFP was therefore analyzed in a vrn2 mutant background, but no obvious difference to wild-type plants was observed (see Fig. 5, which is published as supporting information on the PNAS web site). VRN2 is therefore not required for the general chromosomal association of VRN1; however, this assay would not detect changes occurring at specific loci. VRN2 is part of a three-member gene family in the Arabidopsis genome (8) with EMF2 (36) and FIS2 (37). VRN2 and EMF2 are expressed in vegetative tissue (8, 36), whereas FIS2 function is thought to be restricted to seed development (37, 38). The similar expression pattern and documented functional redundancy between VRN2 and EMF2 (35) led us to test whether VRN1/GFP chromosome association in seedlings was altered in a vrn2-1 emf2-3 double mutant. Given the likely functional association of VRN2 with an ENHANCER OF ZESTE [E(z)] protein, we also tested VRN1/GFP chromosomal association in a double mutant defective in the E(z) homologues CURLY LEAF (CLF) and SWINGER (SWN) clf-50 swn-3 (35, 39). The general chromosomal association of VRN1/GFP, however, appeared to be wild type in both vrn2-1 emf2-3 and clf-50 swn-3 (Fig. 5).
Discussion
The regulation of the gene encoding the Arabidopsis floral repressor FLC by vernalization provides an excellent system to dissect the sequence of events involved in epigenetic silencing induced by environmental change. Previous analyses had demonstrated the role of VIN3, VRN2, and VRN1 in establishing histone modifications, H3 deacetylation, and H3K27me2 and H3K9me2 (10, 11), typical of silenced chromatin states in yeast and animals. The predicted sequence of events suggests the involvement of histone methyltransferases and proteins that bind to the methylated histone marks necessary to establish the stable epigenetically silenced state. In this study, we investigated the involvement of SUVH and SUVR histone methyltransferases and an Arabidopsis HP1 homologue, LHP1, in the maintenance of FLC repression. We also investigated the role of VRN1, a plant-specific protein, in the epigenetic silencing mechanism.
Increased H3K9me2 at FLC induced by vernalization suggested the involvement of a histone methyltransferase activity in vernalization (11). A strong candidate was KYP (18), which has been shown to regulate H3K9me2 levels of the transposons Ta2 and Ta3, the silenced floral repressor FWA, the zinc finger floral regulator SUPERMAN (40), and chromocenters (32). Mutants impaired in the H3K9 histone methyltransferase activity at FLC would share properties of vrn mutants. Our finding that kyp and all tested suvh and suvr single mutants do not show this phenotype indicates that none of the SUV proteins tested are absolutely required for the histone methyltransferase activity required for modification at FLC but does not rule out that they function redundantly. Thus, many components required for FLC silencing remain to be identified, and they may differ from other plant silencing machinery, because, unlike many other silenced loci, repression of FLC by vernalization is not associated with changes in DNA methylation (41).
LHP1 is the only HP1-like homologue encoded by the Arabidopsis genome, and analysis of the misexpression of genes in lhp1 mutants suggests that its function is predominantly to repress expression of a range of euchromatic genes, including many floral genes normally repressed during vegetative development (17, 25, 27, 31, 32). lhp1 mutants showed a defect specifically in the maintenance of FLC silencing in plants that had experienced prolonged cold. Thus, it is likely that LHP1 is a component of the epigenetic silencing machinery at FLC, perhaps binding to the methylated H3K9, causing heterochromatin formation. However, recent data has shown that mammalian HP1 can influence histone modifications as well as bind to modified histones, so LHP1 could also contribute to both steps (42).
Perhaps consistent with the non-sequence-specific DNA binding and multiple roles predicted for VRN1 function (9), the VRN1/GFP fusion was found to generally associate with Arabidopsis chromosomes. However, the continued association of VRN1 with chromosomes all the way through mitosis was unexpected. Most Polycomb, Polycomb-associated proteins, and transcription factors have been found to be displaced from their recognition sequences during mitosis (43–47); however, some do remain, including Drosophila GAGA factor and Pipsqueak, which function as sequence-specific binding proteins and are involved in recruitment of Polycomb complexes to the Polycomb response element and high-mobility-group proteins, which bind DNA non-sequence-specifically (48–52). Knowledge of VRN1 interactors may help elucidate why it remains associated during mitosis and the significance of this finding for epigenetic stability of FLC silencing.
Despite the lack of gross microscopic changes observed in VRN1/GFP localization after vernalization, chromatin immunoprecipitation studies have suggested that subtle conformational changes of VRN1 association with FLC chromatin do occur (11). Immunoprecipitation with an H3K4me2 antibody of a region of the first intron of FLC known to be required for maintenance of FLC repression (53) was reduced in vernalized seedlings. However, this reduction was not observed in vrn1 mutants. This result was interpreted as vernalization inducing the tight association of a complex dependent on VRN1 activity or containing VRN1 to a cis-element in intron 1. This conformational change might then occlude the specific H3K4me2 epitope (11). Alternatively, VRN1 activity might result in nucleosome repositioning, leaving that region of intron 1 devoid of histones. Because VRN1 is required for the H3K9me2 mark at FLC, its activity may be required for LHP1 association with FLC. The common targets of FLC and FT and the coordinate drop in expression of both genes in pollen (LHP1 expression drops ≈3-fold in pollen) (34) might indicate that VRN1 and LHP1 function closely together. Preliminary yeast two-hybrid assays, however, have shown that VRN1 and LHP1 do not appear to interact (L.B., unpublished data).
At some stage during gamete or seed development, the epigenetic repression of FLC is removed, and expression is reset, because, in all species studied, the vernalization requirement is reestablished each generation. The lack of VRN1/GFP association with the chromosomes during meiosis I and II is, thus, particularly intriguing. This regulation appears to be at the level of expression, based on the drop of VRN1 expression detected in pollen (34). Analysis of the same microarray data shows that other VRN proteins and known chromatin regulators remain expressed in pollen; however, as discussed earlier, LHP1 expression drops, as does another Arabidopsis protein CRYPTOCHROME2 (CRY2). CRY2 is a blue-light photoreceptor that functions in the long-day promotion of flowering and whose expression is repressed by FLC (54). Interestingly, CRY2-GFP has also been found to associate generally with Arabidopsis chromosomes during mitosis (55). These microarrays may provide a good screen to identify proteins that function in mitotically stable silencing that is reset at meiosis. VRN1 cannot play a global role in maintaining K9 methylation, because the H3K9me2 at pericentric heterochomatin was still detectable during the meiotic stages where VRN1 is likely to be absent. However, the immunolocalization technique is not sufficiently sensitive to detect K9 methylation in genes dispersed throughout the chromosome arms, so we cannot address the role of VRN1 in maintaining K9 methylation of cryptic heterochromatin.
The general distribution of VRN1 with Arabidopsis chromosomes makes it surprising that vrn1 does not have a more pleiotropic phenotype. Overexpression of VRN1 led to a range of developmental defects, only some of which were caused by ectopic expression of the floral regulator FT (see Fig. 6, which is published as supporting information on the PNAS web site), so VRN1 can clearly target a range of genes. Functionally, redundancy could account for the relative lack of phenotypes in the loss-of-function mutant, because there are many B3 domain proteins encoded in the Arabidopsis genome (56), including a relatively close homologue of VRN1 (At1g49480) called RTV1 (RELATED TO VERNALIZATION1). Mechanistic redundancy may also cover loss of VRN1 function for some targets, and, in this respect, we have found that vrn1 cry2 double mutants flower much later than either parent (Y. Y. Levy and C.D., unpublished results). CRY2 may, therefore, substitute for loss of VRN1 for some targets, so the similar chromosome localization and expression drop in pollen of CRY2 and VRN1 may be relevant. The particular questions for VRN1 are how prolonged cold specifies FLC as a target, why other proteins cannot cover for this function, and whether loss of VRN1 association is a prerequisite for FLC resetting.
Materials and Methods
Plant Materials and Growth Conditions.
For details of growth conditions, refer to figure legends; and see Supporting Materials and Methods, which is published as supporting information on the PNAS web site. Details of mutants included in this study and methods for genotyping are included in Table 3, which is published as supporting information on the PNAS web site. To analyze the FLC expression pattern in lhp1-3, a parent line in Ler containing FLC/luciferase and functional FRIGIDA (3) was introgressed five times into Columbia before crossing to lhp1-3, also referred to as tfl2-1 (29).
VRN1 Constructs and Confocal Microscopy.
The VRN1/GFP translational fusion contains a Ler VRN1 genomic fragment region from 1,879-bp upstream of the ATG to 630-bp downstream of the stop codon. The VRN1 stop codon was modified into a BamHI site by using the QuikChange Site-Directed Mutagenesis kit (Stratagene), which was used to insert in-frame coding sequences for GFP from pAVA120 (57). The VRN1/GFP fusion was cloned into the binary vector pSLJ75516 (58) and transformed into the vrn1-2 fca-1 double mutant (9). VRN1/GFP plants were imaged by using a Leica SP1 confocal system. GFP was excited by using 488-nm light from an Argon Ion laser and imaged by using emission filter 500–50 nm.
Chromosome Preparation and Immunolabeling.
For immunodetection of VRN1 during meiosis, VRN1/GFP vrn1-2 fca-1 anthers were fixed and prepared as described in Jasencakova et al. (59), with minor modifications. Samples were immunolabeled essentially as described in Soppe et al. (60). The primary antibodies used were rabbit anti-dimethyl-lysine 4 of histone H3 (07-030 Lot#22672, 1:100; Upstate Biotechnology, Lake Placid, NY), rabbit anti-dimethyl-lysine 9 of histone H3 (07-212 Lot#22704, 1:50; Upstate Biotechnology), and a polyclonal VRN1 antibody raised in rat against His 10-tagged full-length VRN1 protein expressed in pET19b (9). For details of detection antibodies, refer to Supporting Materials and Methods.
Supplementary Material
Acknowledgments
We thank Paul Boss for confirming vrn1-3 (SAIL_1247_D06); Koji Goto for tfl2-2 and tfl2-3 (referred to as lhp1-4 and lhp1-5); Fuquan Liu for kyp-2 fca-1; Justin Goodrich for vrn2-1 emf2-3/+ and clf-50 swn-3/+; Grant Calder for assistance with microscopy; Mervyn Smith for excellent care of Arabidopsis plants; and Gordon Simpson, Justin Goodrich, and Dean laboratory members for commenting on the manuscript. This work was supported by a Biotechnology and Biological Sciences Research Council (BBSRC) core strategic grant to the John Innes Centre, European Commission Grant QLK5-CT-2001-01412 and BBSRC Grant BB/C517633/1 (both to J.S.M.), a BBSRC CASE studentship with Plant Bioscience, Ltd. (to L.B.), and a European Molecular Biology Organization long-term fellowship (to S.M.).
Abbreviations
- Ler
Landsberg erecta
- PRC
Polycomb repressive complex.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Boss P. K., Bastow R. M., Mylne J. S., Dean C. Plant Cell. 2004;16:S18–S31. doi: 10.1105/tpc.015958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chandler J., Wilson A., Dean C. Plant J. 1996;10:637–644. doi: 10.1046/j.1365-313x.1996.10040637.x. [DOI] [PubMed] [Google Scholar]
- 3.Mylne J., Greb T., Lister C., & Dean C. In: Epigenetics. Stillman B., Stewart D., editors. Vol. LXIX. Cold Spring Harbor, New York: Cold Spring Harbor Lab. Press; 2004. pp. 457–464. [DOI] [PubMed] [Google Scholar]
- 4.Michaels S. D., Amasino R. M. Plant Cell. 1999;11:949–956. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sheldon C. C., Burn J. E., Perez P. P., Metzger J., Edwards J. A., Peacock W. J., Dennis E. S. Plant Cell. 1999;11:445–458. doi: 10.1105/tpc.11.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henderson I. R., Shindo C., Dean C. Annu. Rev. Genet. 2003;37:371–392. doi: 10.1146/annurev.genet.37.110801.142640. [DOI] [PubMed] [Google Scholar]
- 7.Simpson G. G., Dean C. Science. 2002;296:285–289. doi: 10.1126/science.296.5566.285. [DOI] [PubMed] [Google Scholar]
- 8.Gendall A. R., Levy Y. Y., Wilson A., Dean C. Cell. 2001;107:525–535. doi: 10.1016/s0092-8674(01)00573-6. [DOI] [PubMed] [Google Scholar]
- 9.Levy Y. Y., Mesnage S., Mylne J. S., Gendall A. R., Dean C. Science. 2002;297:243–246. doi: 10.1126/science.1072147. [DOI] [PubMed] [Google Scholar]
- 10.Sung S., Amasino R. M. Nature. 2004;427:159–164. doi: 10.1038/nature02195. [DOI] [PubMed] [Google Scholar]
- 11.Bastow R., Mylne J. S., Lister C., Lippman Z., Martienssen R. A., Dean C. Nature. 2004;427:164–167. doi: 10.1038/nature02269. [DOI] [PubMed] [Google Scholar]
- 12.Müller J., Hart C. M., Francis N. J., Vargas M. L., Sengupta A., Wild B., Miller E. L., O’Connor M. B., Kingston R. E., Simon J. A. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 13.Nekrasov M., Wild B., Müller J. EMBO Rep. 2005;6:348–353. doi: 10.1038/sj.embor.7400376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R. S., Zhang Y. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 15.Goodrich J., Tweedie S. Annu. Rev. Cell Dev. Biol. 2002;18:707–746. doi: 10.1146/annurev.cellbio.18.040202.114836. [DOI] [PubMed] [Google Scholar]
- 16.Tschiersch B., Hofmann A., Krauss V., Dorn R., Korge G., Reuter G. EMBO J. 1994;13:3822–3831. doi: 10.1002/j.1460-2075.1994.tb06693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malagnac F., Bartee L., Bender J. EMBO J. 2002;21:6842–6852. doi: 10.1093/emboj/cdf687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson J. P., Lindroth A. M., Cao X., Jacobsen S. E. Nature. 2002;416:556–560. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- 19.Naumann K., Fischer A., Hofmann I., Krauss V., Phalke S., Irmler K., Hause G., Aurich A. C., Dorn R., Jenuwein T., Reuter G. EMBO J. 2005;24:1418–1429. doi: 10.1038/sj.emboj.7600604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baumbusch L. O., Thorstensen T., Krauss V., Fischer A., Naumann K., Assalkhou R., Schulz I., Reuter G., Aalen R. B. Nucleic Acids Res. 2001;29:4319–4333. doi: 10.1093/nar/29.21.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bannister A. J., Zegerman P., Partridge J. F., Miska E. A., Thomas J. O., Allshire R. C., Kouzarides T. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 22.Danzer J. R., Wallrath L. L. Development (Cambridge, U.K.) 2004;131:3571–3580. doi: 10.1242/dev.01223. [DOI] [PubMed] [Google Scholar]
- 23.Ayyanathan K., Lechner M. S., Bell P., Maul G. G., Schultz D. C., Yamada Y., Tanaka K., Torigoe K., Rauscher F. J., III Genes. Dev. 2003;17:1855–1869. doi: 10.1101/gad.1102803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cryderman D. E., Grade S. K., Li Y., Fanti L., Pimpinelli S., Wallrath L. L. Dev. Dyn. 2005;232:767–774. doi: 10.1002/dvdy.20310. [DOI] [PubMed] [Google Scholar]
- 25.Gaudin V., Libault M., Pouteau S., Juul T., Zhao G., Lefebvre D., Grandjean O. Development (Cambridge, U.K.) 2001;128:4847–4858. doi: 10.1242/dev.128.23.4847. [DOI] [PubMed] [Google Scholar]
- 26.Libault M., Tessadori F., Germann S., Snijder B., Fransz P., Gaudin V. Planta. 2005;222:910–925. doi: 10.1007/s00425-005-0129-4. [DOI] [PubMed] [Google Scholar]
- 27.Nakahigashi K., Jasencakova Z., Schubert I., Goto K. Plant Cell Physiol. 2005;46:1747–1756. doi: 10.1093/pcp/pci195. [DOI] [PubMed] [Google Scholar]
- 28.Zemach A., Li Y., Ben-Meir H., Oliva M., Mosquna A., Kiss V., Avivi Y., Ohad N., Grafi G. Plant Cell. 2006;18:133–145. doi: 10.1105/tpc.105.036855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larsson A. S., Landberg K., Meeks-Wagner D. R. Genetics. 1998;149:597–605. doi: 10.1093/genetics/149.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim J. H., Durrett T. P., Last R. L., Jander G. Plant Mol. Biol. 2004;54:671–682. doi: 10.1023/B:PLAN.0000040897.49151.98. [DOI] [PubMed] [Google Scholar]
- 31.Kotake T., Takada S., Nakahigashi K., Ohto M., Goto K. Plant Cell Physiol. 2003;44:555–564. doi: 10.1093/pcp/pcg091. [DOI] [PubMed] [Google Scholar]
- 32.Lindroth A. M., Shultis D., Jasencakova Z., Fuchs J., Johnson L., Schubert D., Patnaik D., Pradhan S., Goodrich J., Schubert I., et al. EMBO J. 2004;23:4286–4296. doi: 10.1038/sj.emboj.7600430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quesada V., Macknight R., Dean C., Simpson G. G. EMBO J. 2003;22:3142–3152. doi: 10.1093/emboj/cdg305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmid M., Davison T. S., Henz S. R., Pape U. J., Demar M., Vingron M., Scholkopf B., Weigel D., Lohmann J. U. Nat. Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- 35.Schubert D., Clarenz O., Goodrich J. Curr. Opin. Plant Biol. 2005;8:553–561. doi: 10.1016/j.pbi.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 36.Yoshida N., Yanai Y., Chen L., Kato Y., Hiratsuka J., Miwa T., Sung Z. R., Takahashi S. Plant Cell. 2001;13:2471–2481. doi: 10.1105/tpc.010227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo M., Bilodeau P., Koltunow A., Dennis E. S., Peacock W. J., Chaudhury A. M. Proc. Natl. Acad. Sci. USA. 1999;96:296–301. doi: 10.1073/pnas.96.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo M., Bilodeau P., Dennis E. S., Peacock W. J., Chaudhury A. Proc. Natl. Acad. Sci. USA. 2000;97:10637–10642. doi: 10.1073/pnas.170292997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chanvivattana Y., Bishopp A., Schubert D., Stock C., Moon Y.-H., Sung Z. R., Goodrich J. Development (Cambridge, U.K.) 2004;131:5263–5276. doi: 10.1242/dev.01400. [DOI] [PubMed] [Google Scholar]
- 40.Johnson L. M., Cao X., Jacobsen S. E. Curr. Biol. 2002;12:1360–1367. doi: 10.1016/s0960-9822(02)00976-4. [DOI] [PubMed] [Google Scholar]
- 41.Finnegan E. J., Kovac K. A., Jaligot E., Sheldon C. C., Peacock W. J., Dennis E. S. Plant J. 2005;44:420–432. doi: 10.1111/j.1365-313X.2005.02541.x. [DOI] [PubMed] [Google Scholar]
- 42.Kourmouli N., Sun Y.-m., van der Sar S., Singh P. B., Brown J. P. Biochem. Biophys. Res. Comm. 2005;337:901–907. doi: 10.1016/j.bbrc.2005.09.132. [DOI] [PubMed] [Google Scholar]
- 43.Michelotti E. F., Sanford S., Levens D. Nature. 1997;388:895–899. doi: 10.1038/42282. [DOI] [PubMed] [Google Scholar]
- 44.Buchenau P., Hodgson J., Strutt H., Arndt-Jovin D. J. J. Cell Biol. 1998;141:469–481. doi: 10.1083/jcb.141.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miyagishima H., Isono K., Fujimura Y., Iyo M., Takihara Y., Masumoto H., Vidal M., Koseki H. Histochem. Cell Biol. 2003;120:111–119. doi: 10.1007/s00418-003-0551-2. [DOI] [PubMed] [Google Scholar]
- 46.Furuyama T., Tie F., Harte P. J. Genesis. 2003;35:114–124. doi: 10.1002/gene.10173. [DOI] [PubMed] [Google Scholar]
- 47.Voncken J., Schweizer D., Aagaard L., Sattler L., Jantsch M., van Lohuizen M. J. Cell Sci. 1999;112:4627–4639. doi: 10.1242/jcs.112.24.4627. [DOI] [PubMed] [Google Scholar]
- 48.Koga H., Matsui S., Hirota T., Takebayashi S., Okumura K., Saya H. Oncogene. 1999;18:3799–3809. doi: 10.1038/sj.onc.1202732. [DOI] [PubMed] [Google Scholar]
- 49.Plath K., Talbot D., Hamer K. M., Otte A. P., Yang T. P., Jaenisch R., Panning B. J. Cell Biol. 2004;167:1025–1035. doi: 10.1083/jcb.200409026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mak W., Baxter J., Silva J., Newall A. E., Otte A. P., Brockdorff N. Curr. Biol. 2002;12:1016–1020. doi: 10.1016/s0960-9822(02)00892-8. [DOI] [PubMed] [Google Scholar]
- 51.Schwendemann A., Lehmann M. Proc. Natl. Acad. Sci. USA. 2002;99:12883–12888. doi: 10.1073/pnas.202341499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harrer M., Lührs H., Bustin M., Scheer U., Hock R. J. Cell Sci. 2004;117:3459–3471. doi: 10.1242/jcs.01160. [DOI] [PubMed] [Google Scholar]
- 53.Sheldon C. C., Conn A. B., Dennis E. S., Peacock W. J. Plant Cell. 2002;14:2527–2537. doi: 10.1105/tpc.004564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.El-Assal S.-E., Alonso-Blanco C., Peeters A. J., Wagemaker C., Weller J. L., Koornneef M. Plant Physiol. 2003;133:1504–1516. doi: 10.1104/pp.103.029819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cutler S. R., Ehrhardt D. W., Griffitts J. S., Somerville C. R. Proc. Natl. Acad. Sci. USA. 2000;97:3718–3723. doi: 10.1073/pnas.97.7.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsukagoshi H., Saijo T., Shibata D., Morikami A., Nakamura K. Plant Physiol. 2005;138:675–685. doi: 10.1104/pp.104.057752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.von Arnim A. G., Deng X.-W., Stacey M. G. Gene. 1998;221:35–43. doi: 10.1016/s0378-1119(98)00433-8. [DOI] [PubMed] [Google Scholar]
- 58.Jones J. D. G., Shlumukov L., Carland F., English J., Scofield S. R., Bishop G. J., Harrison K. Transgenic Res. 1992;1:285–297. doi: 10.1007/BF02525170. [DOI] [PubMed] [Google Scholar]
- 59.Jasencakova Z., Soppe W. J. J., Meister A., Gernand D., Turner B. M., Schubert I. Plant J. 2003;33:471–480. doi: 10.1046/j.1365-313x.2003.01638.x. [DOI] [PubMed] [Google Scholar]
- 60.Soppe W. J. J., Jasencakova Z., Houben A., Kakutani T., Meister A., Huang M. S., Jacobsen S. E., Schubert I., Fransz P. F. EMBO J. 2002;21:6549–6559. doi: 10.1093/emboj/cdf657. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.