Abstract
Comprehensive identification of cis-regulatory elements is necessary for accurately reconstructing gene regulatory networks. We studied proximal promoters of human and mouse genes with differential expression across 56 terminally differentiated tissues. Using in silico techniques to discover, evaluate, and model interactions among sequence elements, we systematically identified regulatory modules that distinguish elevated from inhibited expression in the corresponding transcripts. We used these putative regulatory modules to construct a single predictive model for each of the 56 tissues. These predictors distinguish tissue-specific elevated from inhibited expression with statistical significance in 80% of the tissues (45 of 56). The predictors also reveal synergy between cis-regulatory modules and explain large-scale tissue-specific differential expression. For testis and liver, the predictors include computationally predicted motifs. For most other tissues, the predictors reveal synergy between experimentally verified motifs and indicate genes that are regulated by similar tissue-specific machinery. The identification in proximal promoters of cis-regulatory modules with tissue-specific activity lays the groundwork for complete characterization and deciphering of cis-regulatory DNA code in mammalian genomes.
Keywords: cis-regulatory modules, transcription factor binding
A major first step toward comprehensively understanding the differential control of gene expression in specific tissues and developmental stages is mapping the functional cis-regulatory modules (CRMs) (1) responsible for transcription regulation. CRMs are autonomous units of transcription programs encoded in DNA (2–4) and are largely composed of transcription factor-binding sites (TFBSs). Identification and categorization of the entire repertoire of TFBSs are among the greatest challenges in systems biology (5–7). Although CRM identification in lower eukaryotes, such as various yeast species, has been progressing rapidly (8, 9), similar efforts in vertebrates have proven to be especially difficult due to the genomic and regulatory complexity of the higher organisms (10). CRMs in vertebrate regulatory regions not only control transcription during the life of individual cells but also must orchestrate cellular communication during tissue differentiation, morphogenesis, and body-part formation (11), as well as maintain specific patterns of transcription in terminally differentiated tissues (12).
Recent work to predict expression using cis-regulatory elements includes studies on yeast (6, 9, 13, 14), identification of target genes and binding sites for specific transcription factors (15, 16), and an evaluation of the effect of TFBS quality (17) on target regulation. Bussemaker et al. (13) and Conlon et al. (14) used linear regression to fit the count of predicted cis-regulatory elements (13) or the sum of the likelihood ratios of all potential cis-regulatory elements (14) to expression intensity. Beer and Tavazoie (6) used cis-regulatory elements in promoters to successfully predict gene expression patterns in yeast. Das et al. (9) showed that CRMs in yeast promoters can be used to predict gene expression during the cell cycle; they demonstrated that nonlinear synergistic TFBS models significantly improve fit with expression. Smith et al. (16) showed that CRMs composed of synergistic TFBS pairs in human promoters predict chromatin localization intensity of certain hepatocyte nuclear factors in human liver and pancreatic islets. Papatsenko and Levine (17) experimented with several enhancer evaluation methods, identifying the score of the highest-scoring site and the average score across high-scoring sites as the best predictors for Dorsal binding in the Drosophila embryo.
Here we show that CRMs in proximal promoters can be used to predict differential expression of downstream target transcripts in terminally differentiated tissues from human and mouse. For each tissue, we identify a single CRM-based predictor that distinguishes elevated from inhibited tissue-specific expression. We analyze predictors for a selected set of human tissues. These include predictors based on experimentally validated cis-regulatory elements and predictors that are composed of cis-regulatory elements identified in silico and provide direction for targeted experiments to follow.
Results
We constructed predictors for 28 tissues with data in both human and mouse. We show that 80% of these predictors distinguish elevated from inhibited tissue-specific expression with statistical significance. Full descriptions of predictors, including CRM composition and interaction models, for each of the 56 tissues are available from the authors. Here, we describe the most significant components of the CD4 and CD8 T cell predictors and demonstrate a relationship between correct prediction in these tissues and location of highest-likelihood ETS-1 binding sites. We highlight properties of the liver predictors, which are composed of CRMs that were identified in silico. We discuss similarities between predictors for trigeminal ganglion, dorsal root ganglia, skeletal muscle, ovary, and salivary gland; predictors for these tissues suggest large-scale cell-cycle arrest phenotype, and the inhibited genes with common predictor signatures in these tissues are likely to be regulated by similar transcription-factor complexes. We conclude with a study of the predictive ability of tissue-specific predictors on other human tissues, arguing that functional similarity exists between tissues with common prediction patterns.
Tissue-Specific Expression Predictors.
Each expression predictor describes combinatorial and synergistic relations between experimentally verified or in silico-identified cis-regulatory motifs and CRMs; cis-regulatory motifs are modeled by using position weight matrices (18, 19), and CRMs are sets of motifs hypothesized to act synergistically. Prediction errors, their statistical significance, and corresponding classification errors are given in Table 1 for each of the 28 human tissues. Results for mouse tissues are given in Table 2, which is published as supporting information on the PNAS web site. Our results indicate that information in proximal promoters can predict differential expression of downstream target transcripts in terminally differentiated tissues from human and mouse with statistically significant accuracy.
Table 1.
Prediction accuracy for 28 human tissues
| Tissue | Terms | ClassErr | PredctErr | P value |
|---|---|---|---|---|
| Skeletal muscle | 7 | 0.313 | 0.343 | 2.9E−23 |
| Liver | 4 | 0.281 | 0.344 | 5.5E−23 |
| Dorsal root ganglia | 4 | 0.340 | 0.347 | 3.9E−22 |
| Salivary gland | 3 | 0.321 | 0.351 | 4.8E−21 |
| Trigeminal ganglion | 2 | 0.344 | 0.357 | 1.9E−19 |
| Lung | 3 | 0.329 | 0.359 | 6.2E−19 |
| Ovary | 3 | 0.372 | 0.385 | 6.5E−13 |
| Testis | 3 | 0.357 | 0.397 | 1.5E−10 |
| Olfactory bulb | 5 | 0.365 | 0.400 | 5.4E−10 |
| Heart | 5 | 0.346 | 0.402 | 1.2E−09 |
| Kidney | 5 | 0.360 | 0.402 | 1.2E−09 |
| CD8 T cells | 3 | 0.381 | 0.404 | 2.8E−09 |
| Placenta | 6 | 0.359 | 0.408 | 1.3E−08 |
| CD4 T cells | 6 | 0.370 | 0.415 | 1.8E−07 |
| Prostate | 7 | 0.378 | 0.425 | 5.2E−06 |
| Cerebellum | 6 | 0.352 | 0.430 | 2.4E−05 |
| Adrenal gland | 5 | 0.408 | 0.431 | 3.2E−05 |
| Amygdala | 6 | 0.365 | 0.436 | 1.3E−04 |
| Thymus | 4 | 0.379 | 0.436 | 1.3E−04 |
| Pituitary | 2 | 0.418 | 0.444 | 1.0E−03 |
| Thyroid | 3 | 0.420 | 0.445 | 1.7E−03 |
| Pancreas | 3 | 0.371 | 0.448 | 2.7E−03 |
| Lymph node | 3 | 0.392 | 0.453 | 9.8E−03 |
| Bone marrow | 2 | 0.402 | 0.456 | 1.5E−02 |
| Uterus | 2 | 0.423 | 0.456 | 1.5E−02 |
| Trachea | 4 | 0.393 | 0.467 | 1.2E−01 |
| Adipose tissue | 4 | 0.399 | 0.472 | 2.1E−01 |
| Hypothalamus | 3 | 0.398 | 0.487 | 1.0E+00 |
For each human tissue, we present the number of MARS terms selected for building the predictor to minimize prediction error (PredctErr), the classification error (ClassErr) using this number of MARS terms, the prediction error according to 10-fold crossvalidation, and the corresponding Bonferroni corrected P value (corrected for MARS term selection). After correction, predictors for bone marrow, uterus, trachea, adipose tissue, and hypothalamus fail to predict significantly (P > 0.01).
Detailed Treatment of Predictors.
Predictors that include known tissue-specific DNA motifs and previously observed synergy include ETS, YY1, and CREB-binding motifs in human and mouse T cells and lymph node; serum response factor and basic helix–loop–helix-domain motifs in human heart; myocyte enhancer factor 2 (MEF-2)- and hepatocyte nuclear factor 4 (HNF)-4-binding motifs in mouse heart; myogenin, ETS- and FoxJ-binding motifs in mouse heart; and HNF-4- and activating protein 2 (AP-2)-binding motifs in mouse kidney. Human liver, human testis, and mouse testis are the only tissues in which predictors using a combination of novel and experimentally verified motifs significantly outperformed predictors based only on experimentally verified motifs. Our emphasis on large-scale expression prediction implies that tissue-specific transcription regulators that act on a relatively small set of target promoters may not be included in final predictors. Motifs and modules (constructed from both known and novel motifs) that best predict elevated or inhibited tissue-specific expression on their own are available from the authors. Tables 3 and 4, which are published as supporting information on the PNAS web site, include top experimentally verified binding-site motifs for human CD4 T cells and liver. Tables 5 and 6, which are published as supporting information on the PNAS web site, include modules constructed from experimentally verified binding-site motifs for the same two tissues. An analysis of the testis predictor is given in Fig. 4, which is published as supporting information on the PNAS web site. Experimentally verified binding site motifs that are not included in predictors but are good predictors of elevated tissue-specific expression on their own include MEF-2 and AP-2 motifs in mouse skeletal muscle, HNF-1 and -4 motifs in mouse kidney, and an HNF-4 motif in human liver. Finally, to measure tissue specificity of predictors, we apply each statistically significant human predictor to other human tissues, showing that most human predictors are specific to a single related tissue or a small set of related tissues. The resulting prediction errors are highly correlated (P < 1.0E-16) with common expression patterns calculated using the Eisen similarity score (20). A full description of the human CD8 T cell predictor is given in Table 7 and Fig. 5, which are published as supporting information on the PNAS web site.
T Cells.
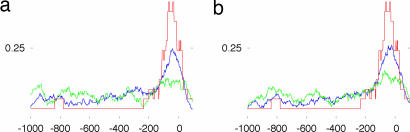
Positive (and negative) sets for CD4 and CD8 human T cells have large intersections (see Table 5), and their predictors include common modules. Motifs associated with factors from the ETS family play a central role in CD4 and CD8 T cell expression prediction in both human and mouse. Fig. 1 shows that the distribution of highest-likelihood ETS-binding sites in correctly predicted positive-set promoters (true positives) is in agreement with the distribution of experimentally verified ETS-binding sites in human and mouse promoters, even though position preference is not a feature used by the predictor. Scores of highest-likelihood ETS-binding sites for false-positive predictions are comparable to scores in true-positive promoters but are less likely to occur near the transcription start site. Binding sites for ETS-1 in human and ELK-1 in mouse are the best individual expression predictors (see Table 3). CD4 and CD8 human T cell predictors rely on synergistic relations between experimentally verified DNA-binding motifs for ETS, YY1, and factors from the CREB family. ETS-family factors are ubiquitously expressed (21) but are well known to have an important role in regulation of transcription in T cells (22, 23). Members of the ETS and CREB families are known to interact (24–26). YY1 is a zinc-finger regulator that acts as an activator or repressor in different contexts (27–29), and it has been observed to repress transcription of genes outside of T cells and enhance their transcription in T cells (30).
Fig. 1.
Distributions of highest-likelihood ETS-binding site positions (relative to the TSS) in true-positive promoters in CD4 (a) and CD8 (b) T cells agree with the distribution of relative positions of experimentally verified ETS-binding sites. The distribution of 29 experimentally verified human and mouse ETS-binding sites is given in red; 6 of the 29 occur outside of the [−1,000, 100] region and are counted but not presented. The distributions of highest-likelihood ETS-binding site positions in the 299 and 325 correctly predicted positive-set promoters (true positives) are given in blue, and the distributions of the 169 and 206 incorrectly predicted negative-set promoters (false positives) are given in green. Each graph point gives the proportion of (verified or putative) binding sites in a window of length 100 that is centered at that point. False-positive predictions may include true ETS-binding sites and show enrichment of sites near the transcription start site, but this enrichment is considerably less pronounced.
Liver.
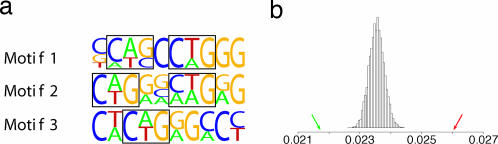
We predict differential expression in human liver with an error rate of 0.344 (P < 5.5E-23). Binding motifs for HNF-4 and -1 are among the best experimentally verified motifs at predicting elevated expression in human and mouse liver; these factors have been repeatedly shown to regulate transcription in liver (15, 31). However, our predictor does not include binding motifs for these factors and instead is composed of computationally predicted motifs that provide significantly better prediction of differential expression. The liver predictor includes three modules. One is overrepresented in the positive set, and the remaining two are overrepresented in the negative set. The positive module has the greatest contribution to prediction; it includes three compensatory C/G-rich motifs (modeled as additive) and is described in Fig. 2. Motifs 1 and 2 include a pair of CAG/CTG patterns with different spacing, and motif 3 includes one CAG/CTG pattern. The CAG/CTG pattern is an excellent expression predictor in its own right, with classification error rate of 0.323 compared with the predictor’s 0.281.
Fig. 2.
Transcripts for which the sum of the scores of the best matches for the three novel motifs is >13.5 are predicted to have elevated expression in human liver. (a) The three motifs are enriched with CAG/CTG (emphasized), and CAG/CTG frequency is a significant predictor of differential expression with classification error of 0.323 compared with classification error of 0.281 of the liver predictor. (b) CAG/CTG frequency in promoters of transcripts with inhibited (0.0217 pointed in green) and elevated (0.0260 pointed in red) expression in human liver are compared with CAG/CTG frequency distribution in 10,000 randomly selected human promoter sets (of the same size) from CSHLmpd. CAG/CTG frequency in the randomly selected human promoter sets is Gaussian with (μ, σ) = (0.0235, 0.00024).
Tissues with Cell-Cycle-Related Inhibition.
Predictors of differential expression in human trigeminal ganglion, skeletal muscle, ovary, and salivary gland have a prediction error <0.36 and include binding-site motifs for cell-cycle-related factors E2F¶ , NRF-1, and ETF. These binding motifs are found in combination with motifs for members of the CREB family and NF-μE1 in trigeminal ganglion, YY1 in ovary, and C/EBP in salivary gland. The human dorsal root ganglia predictor has a prediction error <0.35, and it includes a C/G-rich binding motif for E2F and binding motifs for ETF, CREB, and YY1. Negative promoter sets in the five tissues have an unusually large intersection (see Table 8, which is published as supporting information on the PNAS web site), and Fig. 3 shows that predictors for any one of these tissues do well on the other tissues even when this intersection is excluded. In addition, cell-cycle activation and inhibition through interactions among E2F-1 and CREB, NRF-1, YY1, DP, or ETF have been previously described (32, 33). These cell-cycle-related motifs are enriched in the negative sets, suggesting that the five tissues have unusually strong cell-cycle-related inhibition. This finding is consistent with physiological properties shared by the five tissues. Adult trigeminal ganglion, dorsal root ganglia, and skeletal muscle are in cell-cycle arrest, which is mediated by E2F factors in synergistic activity with Rb, Myc, and cyclin factors (34–36). In Drosophila, which is often used as a model organism for studying the mammalian cell cycle, endocycle (37), ovary, and salivary gland cells as they enter the endocycle and become polyploid. In this state, many E2F-regulated and mitosis-associated transcripts are inhibited (38). Genes correctly predicted in ovary and salivary gland negative sets include nuclear mitotic apparatus protein 1 (NUMA1), HLA-B associated transcript 3 (BAT3), kelch domain containing 3 (KLHDC3), and DEAH box polypeptide 30 (DHX30). Genes correctly predicted in negative sets common to the five tissues are implicated in protein transport and membrane trafficking and include adaptor-related protein complex 3 (AP3D1), pleckstrin homology Sec7 coiled-coil domains 2 (PSCD2), and arsA arsenite transporter ATP-binding homolog 1 (ASNA1). Complete sets of transcripts from positive and negative sets are available from the authors, along with the predicted class of each transcript.
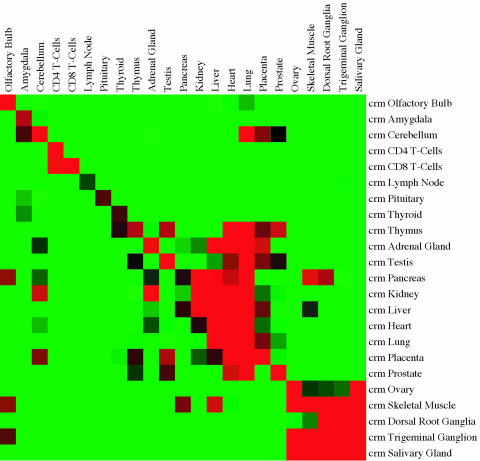
Fig. 3.
Prediction error of CRM-based predictors (on the right) trained on specific tissues and tested on all tissues (at the top). Errors below, at, and above 45% are displayed in red, black, and green, respectively. The diagonal, corresponding to predictors trained and tested on the same tissue, gives prediction error <10-fold crossvalidation. When applying a predictor to a tissue other than that on which it was trained, promoters common to both tissues are excluded. Tissues and corresponding predictors with crossvalidation error P value above the 0.01 significance cutoff are omitted. Tissue order corresponds to physiological properties: brain tissues, T cells, endocrine glands, internal organs, and tissues in cell-cycle arrest. Complete predictors for all tissues, promoter sets, and predictor calls for each promoter are available from the authors.
Tissue-Specific Predictor Evaluation on Other Tissues.
We wanted to determine whether the predictors trained on one tissue could provide statistically significant predictions in other tissues. Fig. 3 summarizes the prediction quality of each human predictor on each human tissue, excluding tissues and corresponding predictors that fail the statistical significance test. Fig. 6, which is published as supporting information on the PNAS web site, has the analogous data for mouse. We are interested in prediction quality and not classification quality and do not consider promoters that were included in the training set: a promoter that is common to tissues T1 and T2 is not included when using the predictor trained on T1 to predict expression in T2. Tables 8 and 9, which are published as supporting information on the PNAS web site, describe the common promoter composition in the positive and negative sets of each tissue pair. Tissues with common predictor patterns tend to be functionally related. Predictors include common patterns in amygdala, cerebellum, and, to a lesser extent, olfactory bulb; CD4 and CD8 T cells; internal organs; and tissues in cell-cycle arrest. Lung, and to a lesser degree heart, can be predicted with statistical significance using C/G content; predictors that reward high C/G content in the positive set and high A/T content in the negative set are more likely to predict these tissues with significance.
Discussion
We demonstrate that CRMs in human and mouse proximal promoters contain sufficient information to predict with statistically significant accuracy whether target genes have elevated or inhibited expression in many terminally differentiated tissues. Our work proves that CRM-based in silico expression prediction is possible for mammals even without the use of distal regions. The majority of our predictors are statistically significant, but our best predictors have a prediction error of 35%, which is high relative to previously observed expression prediction errors in yeast (6). Sequence-based prediction of expression in mammals will significantly improve as cell-based (instead of tissue-based) data become available, with transcript- instead of gene-centric expression microarrays, with improved transcription start site (TSS) annotation that includes alternative TSS data, with better characterization of general and specific TFBSs, and with computational methods that account for chromatin remodeling and distal CRMs further upstream and in intronic regions (39, 40).
We present a detailed in silico study of predictors for human CD4 and CD8 T cells and a predictor for human liver that is composed of novel motifs. We show that promoters of tissue-specific inhibited transcripts in human trigeminal ganglion, dorsal root ganglia, skeletal muscle, ovary, and salivary gland include binding-site patterns for cell-cycle regulators; these terminally differentiated tissues are known to be in cell-cycle arrest. Predictors trained to recognize tissue-specific expression in one tissue recognize tissue-specific expression in that tissue and a few related tissues with common function or lineage. Promoter sets corresponding to transcripts with tissue-specific enhanced and inhibited expression together with calls by tissue-specific predictors are available from the authors. We also provide known and computationally predicted motifs and modules that best predict tissue-specific enhanced and inhibited expression. Predictors, motifs, modules, and transcript sets present opportunity for further study, including identification of cooperative core, proximal and distal elements, verification of predicted synergistic relations and computationally predicted motifs, and construction of better-informed gene networks. Tools developed in this work are freely available and can be reapplied to study transcription regulation of other differentially expressed gene sets.
We use proximal promoter elements to predict expression and do not consider contribution from distal regulatory elements. Distal regulatory elements are rare in yeast but are likely to be major contributors to the diversity seen in higher eukaryotes (3, 41) and are important for understanding the dynamics of metazoan gene regulation. They are known to regulate chromatin modification and often include motifs that are also present in the proximal or core promoter (10). We identify a significant role for proximal promoter elements in determining tissue specificity. Our results complement, rather than conflict with, theories that describe distal mediation of cell type- or tissue-specific regulation. Observed synergistic activity between distal, proximal, and core elements (3, 10, 42) and proximal element competition for distal CRMs (43, 44) suggests that recruitment of distal CRMs may be the responsibility of elements located near transcription start sites. In this paper, we focus on demonstrating that tissue-specific patterns of expression could be predicted by proximal promoter motifs and give only a limited description of the predictive models in a few tissues. To accurately evaluate and compare prediction errors, we use large and equal-size transcript sets. One of the drawbacks of such inquiry is that tissue-specific cis-regulatory motifs and CRMs with relatively few target genes do not appear significant, even when they play a central role in tissue-specific functions. An investigation of transcripts with higher tissue specificity reveals overrepresentation of binding sites for factors that are known to regulate tissue-specific expression but are not included among our top motif and module predictors. A comprehensive study that aims to better catalog all tissue-specific cis-regulatory motifs and modules in human and mouse proximal promoters will be published separately.
Materials and Methods
We focus on large-scale tissue-specific differential expression and identify patterns of regulation that are common to promoters of hundreds of transcripts with tissue-specific behavior. For each tissue, we construct a positive and negative promoter set, each of size 500. This size is a compromise between tissue specificity and statistical power. These uniform-size sets include promoters of transcripts that are not tissue-specific in some tissues and exclude promoters of transcripts that are tissue-specific in other tissues. Benefits of using large and uniform-size sets include consistent prediction estimates that are comparable across tissues and robustness to outliers and features that are shared by few promoters. Drawbacks include higher estimates of prediction error and increased noise. The positive set for each tissue includes Cold Spring Harbor Laboratory Mammalian Promoter Database (CSHLmpd) (45) promoters of transcripts with the most elevated expression, measured in terms of standard deviations from their mean intensity over all tissues studied by Su et al. (46). The negative set includes CSHLmpd promoters of transcripts with inhibited expression under the same criteria. Each promoter sequence is taken 1,000 bp upstream to 100 bp downstream from the CSHLmpd-annotated transcription start site.
Predictors are constructed sequentially, identifying potential master-regulator binding-site motifs (47) and potential synergistic motifs, combining them into small CRMs, and modeling CRM interactions to predict elevated or inhibited expression. Master-regulator binding motifs and potential synergistic motifs are selected from experimentally verified motifs in the TRANSFAC (48) database (called known motifs) and supplemented with motifs identified in silico using dme (49) (called novel motifs). Motifs are combined into pairs and triples to form putative CRMs based on their combined ability to differentiate positive from negative promoters. Predictors are constructed from CRMs using multivariate adaptive regression splines (MARS) (9, 16, 50). MARS can predict the linear and nonlinear relation between CRMs, capturing a multitude of interactive behaviors.
We outline our methods below. Motif identification software is available by request from the authors. Motif analysis, CRM construction, and CRM evaluation programs are a part of the open source package cread, which is available from the authors.
Data Preparation.
The microarray expression experiments of Su et al. (46) include 28 tissues with experiments on human and mouse (see Table 1). We concentrate on these tissues and assign probes to transcripts through mapping probe location back to National Center for Biotechnology Information human genome assembly Hs33 and mouse genome assembly v3C (dating to February 2003). Each transcript was assigned mean intensity over experiments and probes and mapped to a transcription start site in CSHLmpd (45), when possible. Transcripts with no transcription start site annotation in CSHLmpd were discarded. Each transcript with known transcription start site was assigned a proximal promoter consisting of the DNA sequence from −1,000 to 100 relative to its transcription start site. Positive and negative promoter sets for each tissue, each of size 500, were assembled by using unique promoters of transcripts with the most elevated and inhibited (respectively) tissue-specific expression, measured in terms of the number of standard deviations from the transcript’s mean intensity over all tissues. Summary information for probe present/absent calls, mapping to transcripts, and tissue data are given in Tables 10, 11, and 12, which are published as supporting information on the PNAS web site.
Motifs and Motif Modules.
Motifs are represented by using the position weight matrix model (18, 19), and putative motif occurrences are scored by using a log-likelihood function (51). Two distinct motifs are compared by using matcompare (52) and the information divergence measure. Motifs are taken from the vertebrate subset of TRANSFAC 8.3 (48) and identified de novo by using the original dme algorithm (49) and a modified version that considers only the best occurrences in each sequence. Given positive (Pos) and negative (Neg) sequence sets and the base composition f of Pos ∪ Neg, dme iteratively identifies the top motif M, ranked according to the ratio LPos,P(M, f)/LNeg,Q(M, f) of maximum likelihood scores, where LS,Z(M, f) is the likelihood of M and base composition f, given sequence set S with values for the missing data Z maximizing the equation:
The modified version of dme identifies motifs to maximize the difference between the number of sequences in Pos and Neg that contain sites that are more likely to be generated from M than from f, with ties broken in favor of sensitivity. Both algorithms consider sites on the forward and reverse complement strands. For each tissue, 150 motifs (widths 8, 10, and 12) were identified by using the two programs.
Motifs were evaluated, ranked, pruned, and grouped into modules using the cread programs motifclass, sortmotifs, uniqmotifs, and modclass. Motif quality was measured by using classification error rate based on the maximum scoring sequence in a given promoter and its reverse compliment and a threshold set to minimize this error using motifclass. Motifs found to be similar to higher-quality motifs were eliminated by using uniqmotifs. Motif modules (sets of motifs) were constructed by identifying motif sets with low classification error using independently set thresholds by modclass. We constructed modules of size 2 by exhaustive evaluation of motif pairs and modules of size 3 from which each pair of single motifs was a high-ranking size 2 module. More details on these programs can be found in Supporting Text, which is published as supporting information on the PNAS web site.
Predictor Construction.
Predictors are constructed with the MARS algorithm (9, 16, 50) by using motif and module features. We used MARS to construct predictors that include seven terms using stepwise forward addition of linear splines and their products. We then used backward elimination to reduce the size of predictors to two terms. Splines operate on basis functions of the form {1, max(sik−ξi, 0), max(ξi − sik, 0)}, where sik is the value of feature i on sequence k. Splines can be combined using addition or multiplication to model linear and nonlinear relationships. Supporting Text contains a full list of the features we tested. We provided MARS with a restricted set of features, including the max-score feature for motifs and the max-score-sum and max-score-product features for modules. For a given motif, the max-score feature assigns to each promoter the score of the highest matching substring in the promoter and its reverse complement. The max-score-sum and max-score-product features assign to each promoter the sum of max-scores and product of max-scores over all motifs in the module, respectively. Preliminary experiments indicated that the best models were most often built by using features of those types, and a general restriction in the complexity of the feature space is required to prevent overfitting.
MARS builds predictive models (in our case CRMs) by iteratively adding terms and factors to a spline-based polynomial. It makes greedy local moves, adding the best possible term at each forward step and removing the least effective term at each backward step. MARS may fail to identify the best combination of terms if these terms are not individually dominant. Although MARS is designed to perform regression, it will function as a classifier when given −1/1 response variables as input. Positive and negative responses correspond to positive and negative prediction. Supporting Text contains additional details on the MARS algorithm. Most predictors were constructed by using experimentally verified motifs and modules composed of experimentally verified motifs. Novel motifs were used only when they provided a significantly improved prediction.
Evaluating Predictive Ability.
We used 10-fold crossvalidation to estimate prediction error. The procedure is as follows: (i) the data are randomly partitioned into 10 subsets; (ii) each subset is removed exactly once from the data, and a predictor is trained on the remaining 90% and tested on the removed subset; and (iii) the classification errors are summed over each subset, producing the prediction error. We assume that an uninformed random predictor will make a correct prediction with probability 0.5. We stress that motif identification and optimization must be done only after removal of the testing subset. Approaches that identify and optimize motifs on the entire set, build predictors on a subset, and test on another subset grossly underestimate prediction errors. Our predictors contain no information about the test set, and their predictive error can be directly used to evaluate their quality. We used MARS to build predictors up to seven terms and iteratively removed the weakest term from each predictor until reaching a predictor of size 2, resulting in six predictors. The best model size with minimum prediction error was selected, and MARS was then applied to build a predictor of that size using the entire data set.
Supplementary Material
Acknowledgments
We thank J. Hogenesch and J. Walker for providing tissue-specific expression data; S. Kamalakaran, G. Chen, V. Agarwala, and X. Zhao for advice and suggestions; and BIOBASE (Wolfenbuettel, Germany) for providing access to TRANSFAC. This work is supported by National Institutes of Health Grants GM060513 and HG001696 and National Science Foundation Grants DBI-0306152 and EIA-0324292.
Abbreviations
- CRM
cis-regulatory module
- TFBS
transcription factor-binding site
- MARS
multivariate adaptive regression splines
- CSHLmpd
Cold Spring Harbor Laboratory Mammalian Promoter Database.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
S.I. is a guest editor invited by the Editorial Board.
The negative set in the ovary is enriched for an experimentally verified E2F-1-binding motif, and negative sets in skeletal muscle, salivary gland, and trigeminal ganglion are enriched for an experimentally verified E2F-1-, -3-, and -4-binding motif.
References
- 1.Istrail S., Davidson E. H. Proc. Natl. Acad. Sci. USA. 2005;102:4954–4959. doi: 10.1073/pnas.0409624102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orphanides G., Reinberg D. Cell. 2002;108:439–451. doi: 10.1016/s0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- 3.Levine M., Tjian R. Nature. 2003;424:147–151. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- 4.Davidson E. H., McClay D. R., Hood L. Proc. Natl. Acad. Sci. USA. 2003;100:1475–1480. doi: 10.1073/pnas.0437746100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kellis M., Patterson N., Endrizzi M., Birren B., Lander E. S. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 6.Beer M. A., Tavazoie S. Cell. 2004;117:185–198. doi: 10.1016/s0092-8674(04)00304-6. [DOI] [PubMed] [Google Scholar]
- 7.Xie X., Lu J., Kulbokas E. J., Golub T. R., Mootha V, Lindblad-Toh K, Lander E. S., Kellis M. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harbison C. T., Gordon D. B., Lee T. I., Rinaldi N. J., Macisaac K. D., Danford T. W., Hannett N. M., Tagne J.-B, Reynolds D. B., Yoo J., et al. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Das D., Banerjee N., Zhang M. Q. Proc. Natl. Acad. Sci. USA. 2004;101:16234–16239. doi: 10.1073/pnas.0407365101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hochheimer A., Tjian R. Genes Dev. 2003;17:1309–1320. doi: 10.1101/gad.1099903. [DOI] [PubMed] [Google Scholar]
- 11.Davidson E. H. Development (Cambridge, U.K.) 1993;118:665–690. doi: 10.1242/dev.118.3.665. [DOI] [PubMed] [Google Scholar]
- 12.Davidson E. H. Genomic Regulatory Systems. New York: Academic; 2001. [Google Scholar]
- 13.Bussemaker H. J., Li H., Siggia E. D. Nat. Genet. 2001;27:167–171. doi: 10.1038/84792. [DOI] [PubMed] [Google Scholar]
- 14.Conlon E. M., Liu X. S., Lieb J. D., Liu J. S. Proc. Natl. Acad. Sci. USA. 2003;100:3339–3344. doi: 10.1073/pnas.0630591100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Odom D. T., Zizlsperger N., Gordon D. B., Bell G. W., Rinaldi N. J., Murray H. L., Volkert T. L., Schreiber J., Rolfe P. A., Gifford D. K., et al. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith A. D., Sumazin P., Das D., Zhang M. Q. Bioinformatics. 2005;21:i403–i412. doi: 10.1093/bioinformatics/bti1043. [DOI] [PubMed] [Google Scholar]
- 17.Papatsenko D., Levine M. Proc. Natl. Acad. Sci. USA. 2005;102:4966–4971. doi: 10.1073/pnas.0409414102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schneider T. D., Stormo G. D., Gold L., Ehrenfeucht A. Nucleic Acids Res. 1982;10:2997–3011. doi: 10.1093/nar/10.9.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J. S., Lawrence C. E., Neuwald A. J. Am. Stat. Assoc. 1995;90:1156–1170. [Google Scholar]
- 20.Eisen M. B., Spellman P. T., Brown P. O., Botstein D. Proc. Natl. Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hollenhorst P. C., Jones D. A., Graves B. J. Nucleic Acids Res. 2004;32:5693–5702. doi: 10.1093/nar/gkh906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho I. C., Bhat N. K., Gottschalk L. R., Lindsten T., Thompson C. B., Papas T. S., Leiden J. M. Science. 1990;250:814–818. doi: 10.1126/science.2237431. [DOI] [PubMed] [Google Scholar]
- 23.Bhat N. K., Thompson C. B., Lindsten T., June C. H., Fujiwara S., Koizumi S., Fisher R. J., Papas T. S. Proc. Natl. Acad. Sci. USA. 1990;87:3723–3727. doi: 10.1073/pnas.87.10.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang C., Shapiro L. H., Rivera M., Kumar A., Brindle P. K. Mol. Cell Biol. 1998;18:2218–2229. doi: 10.1128/mcb.18.4.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sawada J.-i., Simizu N., Suzuki F., Sawa C., Goto M., Hasegawa M., Imai T., Watanabe H., Handa H. J. Biol. Chem. 1999;274:35475–35482. doi: 10.1074/jbc.274.50.35475. [DOI] [PubMed] [Google Scholar]
- 26.Tsai J., Zhang J., Minami T., Voland C., Zhao S., Yi X., Lassalle P., Oettgen P., Aird W. J. Vasc. Res. 2002;39:148–159. doi: 10.1159/000057763. [DOI] [PubMed] [Google Scholar]
- 27.Shi Y., Seto E., Chang L. S., Shenk T. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 28.Seto E., Shi Y., Shenk T. Nature. 1991;354:241–245. doi: 10.1038/354241a0. [DOI] [PubMed] [Google Scholar]
- 29.Yang W. M., Inouye C., Zeng Y., Bearss D., Seto E. Proc. Natl. Acad. Sci. USA. 1996;93:12845–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji H.-B., Gupta A., Okamoto S., Blum M. D., Tan L., Goldring M. B., Lacy E., Roy A. L., Terhorst C. J. Biol. Chem. 2002;277:47898–47906. doi: 10.1074/jbc.M201025200. [DOI] [PubMed] [Google Scholar]
- 31.Krivan W., Wasserman W. W. Genome Res. 2001;11:1559–1966. doi: 10.1101/gr.180601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elkon R., Linhart C., Sharan R., Shamir R., Shiloh Y. Genome Res. 2003;13:773–780. doi: 10.1101/gr.947203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Attwooll C., Lazzerini Denchi E., Helin K. EMBO J. 2004;23:4709–4716. doi: 10.1038/sj.emboj.7600481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dyson N. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 35.DeGregori J. Biochim. Biophys. Acta. 2002;1602:131–150. doi: 10.1016/s0304-419x(02)00051-3. [DOI] [PubMed] [Google Scholar]
- 36.Radhakrishnan S. K., Feliciano C. S., Najmabadi F, Haegebarth A., Kandel E. S., Tyner A. L., Gartel A. L. Oncogene. 2004;23:4173–4176. doi: 10.1038/sj.onc.1207571. [DOI] [PubMed] [Google Scholar]
- 37.Edgar B. A., Orr-Weaver T. L. Cell. 2001;105:297–306. doi: 10.1016/s0092-8674(01)00334-8. [DOI] [PubMed] [Google Scholar]
- 38.Lilly M. A., Duronio R. J. Oncogene. 2005;24:2765–2775. doi: 10.1038/sj.onc.1208610. [DOI] [PubMed] [Google Scholar]
- 39.Banerjee N., Zhang M. Q. Curr. Opin. Microbiol. 2002;5:313–317. doi: 10.1016/s1369-5274(02)00322-3. [DOI] [PubMed] [Google Scholar]
- 40.Blais A., Dynlacht B. D. Genes Dev. 2005;19:1499–1511. doi: 10.1101/gad.1325605. [DOI] [PubMed] [Google Scholar]
- 41.Wyrick J. J., Young R. A. Curr. Opin. Genet. Dev. 2002;12:130–136. doi: 10.1016/s0959-437x(02)00277-0. [DOI] [PubMed] [Google Scholar]
- 42.Blackwood E. M., Kadonaga J. T. Science. 1998;281:60–63. doi: 10.1126/science.281.5373.60. [DOI] [PubMed] [Google Scholar]
- 43.Ohtsuki S., Levine M., Cai H. N. Genes Dev. 1998;12:547–556. doi: 10.1101/gad.12.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharpe J., Nonchev S., Gould A., Whiting J., Krumlauf R. EMBO J. 1998;17:1788–1798. doi: 10.1093/emboj/17.6.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xuan Z., Zhao F., Wang J. H., Chen G. X., Zhang M. Q. Genome Biol. 2005;6:R72. doi: 10.1186/gb-2005-6-8-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Su A. I., Wiltshire T., Batalov S., Lapp H., Ching K. A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., et al. Proc. Natl. Acad. Sci. USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McKenna N. J., O’Malley B. W. Cell. 2002;108:465–474. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- 48.Matys V., Fricke E., Geffers R., Gossling E., Haubrock M., Hehl R., Hornischer K., Karas D., Kel A. E., Kel-Margoulis O. V., et al. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith A. D., Sumazin P., Zhang M. Q. Proc. Natl. Acad. Sci. USA. 2005;102:1560–1565. doi: 10.1073/pnas.0406123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Friedman J. H. Ann. Stat. 1991;19:1–142. [Google Scholar]
- 51.Hertz G., Stormo G. Bioinformatics. 1999;15:563–577. doi: 10.1093/bioinformatics/15.7.563. [DOI] [PubMed] [Google Scholar]
- 52.Schones D., Sumazin P., Zhang M. Q. Bioinformatics. 2005;21:307–313. doi: 10.1093/bioinformatics/bth480. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.