Abstract
Activity of the serine-threonine protein kinase PINOID (PID) has been implicated in the asymmetrical localization of the membrane-associated PINFORMED (PIN) family of auxin transport facilitators. However, the means by which PID regulates PIN protein distribution is unknown. We have used recombinant PID protein to dissect the regulation of PID activity in vitro. We demonstrate that intramolecular PID autophosphorylation is required for the ability of PID to phosphorylate an exogenous substrate. PID-like mammalian AGC kinases act in a phosphorylation cascade initiated by the phospholipid-associated kinase, 3-phosphoinositide-dependent protein kinase 1 (PDK1), which binds to the C-terminal hydrophobic PDK1-interacting fragment (PIF) domain found in PDK1 substrates. We find that Arabidopsis PDK1 interacts with PID, and that transphosphorylation by PDK1 increases PID autophosphorylation. We show that a PID activation loop serine is required for PDK1-dependent PID phosphorylation. This activation is rapid and requires the PIF domain. Cell extracts from flowers and seedling shoots dramatically increase PID phosphorylation in a tissue-specific manner. A PID protein variant in which the PIF domain was mutated failed to be activated by the seedling shoot extracts. PID immunoprecipitated from Arabidopsis cells in which PDK1 expression was inhibited by RNAi showed a dramatic reduction in transphosphorylation of myelin basic protein substrate. These results indicate that AtPDK1 is a potent enhancer of PID activity and provide evidence that phospholipid signaling may play a role in the signaling processes controlling polar auxin transport.
Keywords: AGC kinase, auxin signaling
The plant hormone auxin is synthesized in rapidly dividing tissues and directionally transported to the plant apex and root to direct numerous developmental processes (1). Unidirectional or polar auxin transport correlates with the asymmetrical distribution of the PINFORMED (PIN) family of proteins that differentially localize to the apical, lateral, or basal membranes of auxin-transmitting cells (2, 3). Rapid and specific localization of PIN proteins is maintained by actin-dependent cycling of vesicles between the plasma membrane and endosomal compartments, suggesting that the recruitment of PIN proteins to targeted plasma membranes is required for polar auxin distribution (4, 5). Directional auxin flux associated with the asymmetrical distribution of transport components establishes local auxin gradients that direct patterning and initiate tropic responses (6–9). However, little is known about the mechanism by which the asymmetric distribution of auxin carrier proteins to a particular membrane domain is established.
PINOID (PID) has been shown to play a pivotal role in localizing members of the PIN protein family. Analysis of PIN polarity in wild-type plants, pid loss-of-function mutants, and PID overexpression lines indicates that relatively high PID levels result in apical localization of PIN proteins. Conversely, cells with low PID expression levels are associated with a pronounced basal localization pattern (10). Current models explain patterning of lateral organ development through the creation of regions of high relative auxin concentration produced by PIN-directed auxin flux (7, 9). Loss-of-function pinoid mutants are defective in initiating lateral organ development, and this phenotype can be rescued by local auxin application at the plant apex (9), suggesting that pinoid mutants are defective in establishing regions of maximal auxin concentration.
PINOID is a member of the AGC family of serine-threonine protein kinases. In yeast and animals, this class of second-messenger-activated kinases has been extensively studied and shown to regulate diverse cellular processes. One particularly well characterized example of AGC function is the involvement of mammalian PKC in the asymmetrical membrane localization of the GLUT4 glucose transporter in response to insulin signaling (11). During this process, PKC is recruited to the plasma membrane, where it is activated by the 3-phosphoinositide-dependent kinase 1 (PDK1) through the phosphorylation of a specific threonine residue within the catalytic activation loop (12). Examination of numerous other mammalian AGC kinases, including Akt, S6K, SGK, and PKA, indicate that activation by PDK1 is an evolutionarily conserved mechanism by which the AGC kinase family is regulated (13). Despite the progress made in understanding the roles of AGC kinases in yeast and metazoans, the signaling processes controlled by AGC kinases in plants remain poorly understood.
In Arabidopsis, the large AGC kinase family is subdivided into phylogenetically distinct categories (14). The largest and most diverse of these categories is subfamily VIII, which contains 23 members. The majority of Arabidopsis subfamily VIII kinases contain a C-terminal hydrophobic domain similar to that found in PKA, which mediates the interaction of the protein with PDK1 (Fig. 1A). In addition to PINOID, five AGC VIII kinases have been genetically or biochemically identified in Arabidopsis. KIPK was identified in a screen for proteins that bound to the kinesin-like motor (15). Two other Arabidopsis AGC kinases, AGC1-1 and AGC2-1, which are involved in root hair growth, have been shown to interact with PDK1 (16). The PHOT1 and PHOT2 genes encode blue-light photoreceptors that respond to light with a conformational change that stimulates autophosphorylation of the C-terminal kinase domain of both proteins. However, the PHOT proteins lack the PDK1-interacting fragment (PIF) regulatory domain (14, 17).
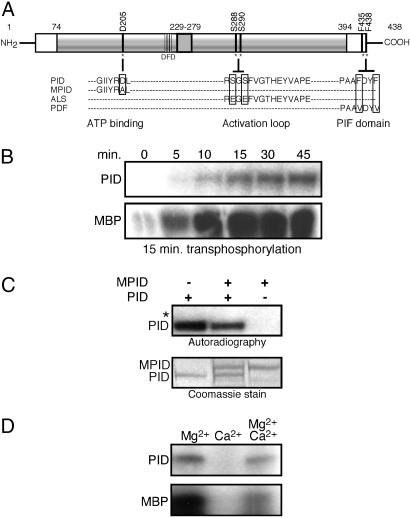
Fig. 1.
Intramolecular autophosphorylation activates PID transphosphorylation activity. (A) Schematic representation of PINOID, showing the wild-type sequence of the ATP binding domain, activation loop, and PIF domain, and the corresponding changes introduced in the MPID, ALS, and PDF derivatives. (B) Autoradiograph showing that autophosphorylation of PID is required for its transphosphorylation of MBP. (Upper) PID was incubated in kinase buffer for the indicated times. (Lower) For each time point, unlabeled, autophosphorylated PID was mixed with MBP and 5 μCi of [γ-32P]ATP (1Ci = 37 GBq) and incubated for an additional 15 min. The results shown are representative of three independent experiments. (C) PID autophosphorylates intramolecularly. His-tagged PID and GST-tagged MPID were coincubated with 10 μCi of [γ-32P]ATP and kinase buffer. The asterisk marks the location of nonphosphorylated GST-MPID visible in the Coomassie-stained gel. All autoradiographs shown are 24-h exposures. Identical results were obtained in two independent experiments. (D) Calcium inhibition of PID activity. PID activity was assayed in the presence of 15 mM MgCl2, 15 mM CaCl2, or both. For each reaction, PID was allowed to autophosphorylate (Upper) for 45 min before the addition of 3 μg of MBP (Lower). Experiments were performed in triplicate.
To understand how PID might function in the localization of the auxin transport machinery, we have investigated how PID activity is regulated. We have examined the regulation of PID in response to tissue-specific cell extracts, changes in cation concentration, and binding by the Arabidopsis homologue of the mammalian AGC activating kinase, PDK1. We find that Ca2+ directly inhibits PID activity in vitro and that AtPDK1 interacts with and activates PID both in vitro and in plant cell cultures. These results provide a link between auxin activity and membrane-associated signaling processes.
Results
Intramolecular Autophosphorylation Activates PID Transphosphorylation Activity.
PID has previously been shown to have autocatalytic activity in vitro (18, 19). To determine whether the observed autophosphorylation is required for activation or stabilization of PID activity, we used time course experiments to examine the effect of PID autophosphorylation on transphosphorylation of the nonspecific substrate myelin basic protein (MBP). After undergoing autophosphorylation for increasing time intervals, PID was mixed with MBP and incubated for 15 min (Fig. 1B). In the absence of prior autophosphorylation (0 min), PID exhibited only weak transphosphorylation activity. Over a 45-min time course, PID showed an essentially linear increase in autophosphorylation activity. This increase was accompanied by a corresponding increase in transphosphorylation of MBP.
We asked whether PID autophosphorylation involved intra- or intermolecular reactions. GST-tagged MPID (Fig. 1A), a kinase defective derivative containing a mutated ATP binding site (20) that has been shown to be inactive both in vitro and in vivo (10, 18), was mixed with wild-type His-tagged recombinant PID. After 45 min of coincubation, no radiolabeled phosphate was incorporated by the MPID protein (Fig. 1C). These data provide evidence that intramolecular autophosphorylation positively regulates the ability of PID to transphosphorylate substrates in vitro.
Calcium Directly Inhibits PID Activity in Vitro.
It has been suggested that calcium is a negative regulator of PID activity in vivo, although it is unclear whether this observation reflects a direct inhibition of PID activity by calcium ions or an indirect effect through the binding of PID to calcium-binding proteins (19). To distinguish between these possibilities, we tested the effect of Mg2+ and Ca2+ ions on PID activity in vitro. PID is similar to other known kinases in its absolute requirement for Mg2+ in nucleotide binding (Fig. 1D). Increasing levels of Mg2+ stimulated both autophosphorylation of PID and transphosphorylation of MBP (Fig. 7A, which is published as supporting information on the PNAS web site).
By contrast, PID was unable to use CaCl2 as divalent cation donor (Fig. 1D). Coincubation of PID with equimolar concentration of Mg2+ and Ca2+ resulted in a moderate reduction in PID autophosphorylation and effectively inhibited the ability of PID to transphosphorylate MBP as compared with PID activity in the presence of Mg2+ alone (Fig. 1D). In the presence of 15 mM Mg2+, calcium levels higher than 2.5 mM clearly inhibited both the auto- and transphosphorylation activities of PID (Fig. 7B). These data suggest that Ca2+ may inhibit PID activity by competitive binding and displacement of the Mg2+ cation.
Transactivation of PID by Tissue-Specific Cell Extracts.
PID is transiently expressed in embryos, young floral organs, and in root and shoot vascular tissues (18, 21, 22). To test whether the PID protein is differentially activated in these tissues, we measured the ability of plant extracts derived from flowers, siliques, leaves, seedling roots, or seedling shoots to stimulate PID activity. Total-protein extracts from each tissue type were coincubated with recombinant GST:PID in the presence of [γ-32P]ATP. Cell extracts from flowers and seedling shoots dramatically increased levels of PID phosphorylation (Fig. 2A). To distinguish between transphosphorylation by an endogenous kinase or increased autophosphorylation induced by a phosphorylation independent interaction, the experiment was repeated by using recombinant MPID protein as a substrate. MPID is phosphorylated in a pattern similar to that observed for PID, indicating that the differential phosphorylation is likely to be because of the tissue-specific activity of an upstream regulatory kinase present in tissue extracts.
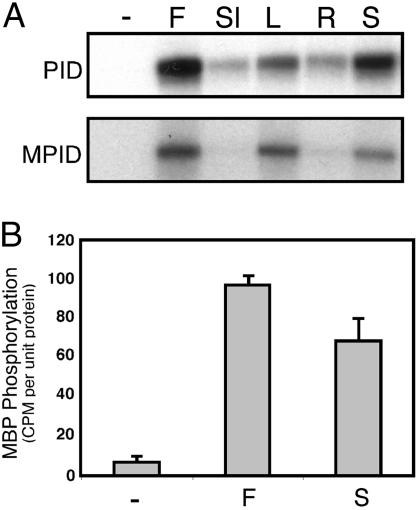
Fig. 2.
Transactivation of PID by tissue-specific cell extracts. (A) Total protein purified from mixed stage flowers (F), siliques (Sl), rosette leaves (L), 10-day-old seedling shoots (S), or roots (R) was incubated with GST:PID or GST:MPID. PID labeling was detected by autoradiography after PAGE. (B) GST-PID was phosphorylated by tissue-specific protein extracts as described in A, washed, and incubated with MBP to assess PID activity. Incorporation of γ-32P by MBP was quantified by using a liquid scintillation counter. Data are representative of three independent experiments; error bars represent standard error. Autoradiographs were exposed for 12 h.
The ability of untreated wild-type PID protein to phosphorylate MBP was compared with that of protein treated with either flower or seedling shoot extracts. Fig. 2B shows that PID activity was increased 14- and 10-fold, respectively, by the endogenous kinase present in flower and seedling shoot extracts.
Autophosphorylation Activity of PID Is Induced by PDK1 Phosphorylation.
Because mammalian AGC kinases are known substrates for the upstream regulatory kinase PDK1 (13), we tested whether PID and Arabidopsis PDK1 interact in vitro. Recombinant His-tagged PDK1 was incubated with GST:PID. Western blots of bound proteins probed with anti-His antibody show that both PID and MPID bind PDK1 (Fig. 3A). The interaction between PID and PDK1 results in a dramatic increase in PID phosphorylation (Fig. 3B). PID showed a 6.5-fold increase in measured phosphate incorporation as compared with MPID. This difference must represent increased PID autocatalysis in response to activation by PDK1, suggesting that transphosphorylation by PDK1 activates PID through up-regulation of autophosphorylation activity (Fig. 3B). Time course experiments show that the increase in PDK1-dependent PID autophosphorylation is rapid, with peak levels of activation occurring after 30 min of coincubation (Fig. 8, which is published as supporting information on the PNAS web site).
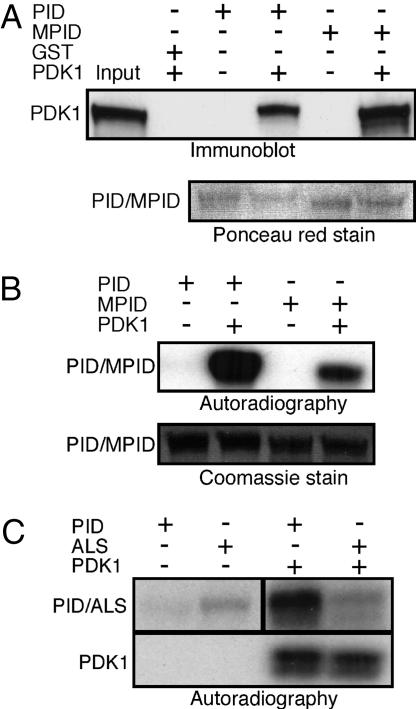
Fig. 3.
PID autophosphorylation is induced by PDK1 phosphorylation. (A) His:PDK1-containing bacterial lysate was incubated with GST, GST:PID, or GST:MPID bound to glutathione-agarose beads. Half of the sample was used to confirm PDK1 binding by Western blot analysis by using anti-polyhistidine antibody. (B) The remaining sample was used in a kinase assay in the presence of [γ-32P]ATP and subjected to PAGE. Incorporation of radiolabeled phosphate was determined by autoradiography. Binding and phosphorylation analyses were confirmed by three independent experiments. (C) An activation loop serine is required for PDK1 activation of PID. Wild-type and ALS mutant protein (see Fig. 1A) phosphorylation was analyzed in the absence or presence of PDK1 protein. Results shown were representative of two independent experiments. Autoradiographs were exposed for 1 h in B and C.
Regulation of kinases often involves phosphorylation of residues within the activation loop (23). There are two serine residues, S288 and S290, within the region of the PID activation loop corresponding to the putative PDK1 phosphorylation domain. Because elimination of one potential phosphorylation site within the PDK1 activation domain of PKC results in compensatory phosphorylation at an adjacent threonine (24), we used a PID construct, ALS, in which both extant serines were replaced by glutamic acid to mimic the negative charge conferred by phosphorylation. Substitution of both serines with glutamic acid resulted in an increase in the basal autophosphorylation level of the mutant protein over that observed in wild-type PID, suggesting that one or more of these amino acids function as positive regulatory elements for PID function (Fig. 3C). This conclusion is supported by the observation that elimination of serine residues within the regulatory domain abolishes the ability of PDK1 to phosphorylate PID (Fig. 3C). Together, these data indicate that PDK1 activates PID by phosphorylating one or more of the serines within the PID activation loop.
The PIF Domain Is Required for PDK1-Mediated PID Phosphorylation.
Plant AGC kinases contain a carboxyl-terminal PIF similar to that found in PKA. Two conserved phenylalanine residues in this domain have been shown to be required for PDK1 binding to mammalian AGC kinases (12). To test whether these amino acids are also required for the interaction of PID with PDK1, we used a PID variant, PDF, in which both of the phenylalanines were replaced with valine (Fig. 1A) in GST pull-down experiments. A comparison of normalized protein levels shows that the amount of PDK1 recovered by PDF interaction was not appreciably different from that recovered by using wild-type PID (Fig. 4A), suggesting that these residues are not critical for protein interaction. However, phosphorylation assays showed that substitution of the phenylalanine residues blocks PDK1 phosphorylation of the PDF substrate (Fig. 4B). These data indicate that the conserved PIF domain is required for a functional interaction between PID and PDK1.
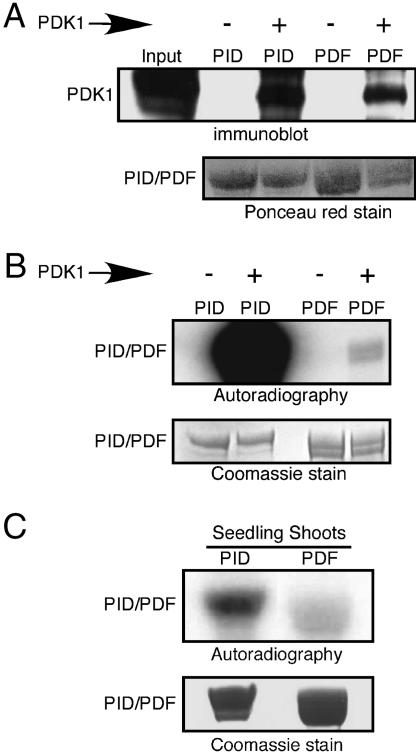
Fig. 4.
The PIF domain is required for PDK1-mediated PID phosphorylation in vitro and in vivo. (A) Binding of PDK1 to PID and PDF protein was visualized by Western blotting as described in the legend for Fig. 3. (B) PID phosphorylation level was compared with that of PDF in the absence or presence of bound PDK1. (C) Equivalent amounts of GST:PID or GST:PDF proteins were incubated with seedling shoot extract in kinase buffer in the presence of [γ-32P]ATP for 30 min. Protein phosphorylation was visualized by autoradiography. All experiments were performed in triplicate. Autoradiographs were exposed for 1 h in B and 12 h in C.
We examined the effect of the PDF mutation on the ability of seedling shoot extract to activate PID autophosphorylation. Fig. 4C shows that PDF autophosphorylation is reduced by 2.5-fold as compared with autophosphorylation of the wild-type protein after treatment with seedling extracts. The observed decrease in PDF phosphorylation suggests that PDK1 acts upstream of PID and may play an important role in PID activation.
PID and PDK1 Interact in Vivo.
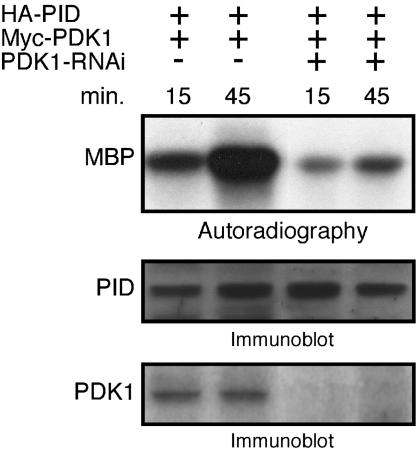
To determine whether PDK1 is required to activate PID in vivo, we tested the ability of PDK1 to activate PID in Arabidopsis protoplasts. Because cells showing stable expression of a PDK1 RNAi construct were difficult to propagate, protoplast transient assays were used. Cells were cotransfected with hemagglutinin (HA)-tagged PID and myc-tagged PDK1 in the presence or absence of a PDK1 RNAi construct. Immunoprecipitated PID was allowed to autophosphorylate for 15 or 45 min and then incubated with MBP, which is not a substrate for PDK1 (data not shown), to assess the transphosphorylation activity of PID. PID extracted from cells expressing PDK1 showed robust transphosphorylation activity as assayed by MBP labeling. In contrast, MBP was only weakly phosphorylated in cells coexpressing PID, PDK1, and the PDK1-RNAi constructs (Fig. 5). These results demonstrate that AtPDK1 is a potent upstream activator of PID in vivo.
Fig. 5.
In vivo activation of PID by PDK1. PID was immunoprecipitated from cells cotransfected with HA-PID and Myc-PDK1 in the presence or absence of PDK1-RNAi. After PID autophosphorylation for the indicated times, MBP was added to the kinase reaction for an additional 30 min to asses PID transphosphorylation activity (Top). Equal amounts of proteins from the above transfected cells were used to confirm PID and PDK1 expression by immunoblotting by using HA (Middle) and Myc (Bottom) antibodies, respectively. Data are representative of two independent experiments.
Discussion
Autophosphorylation of regulatory kinases is an important means by which these proteins are conformationally stabilized to maximize substrate recognition (23). Time course experiments showed that PID has a low basal level of autophosphorylation that increases linearly over a 30- to 40-min period. This increase in PID autophosphorylation correlates with the ability of PID to transphosphorylate the exogenous substrate MBP, suggesting that autophosphorylation is a required step in PID activation. Both intra- and intermolecular interactions have been implicated in the autophosphorylation-dependent activation of protein kinases. We used differentially tagged recombinant PID proteins to examine the interaction between wild-type PID and a kinase defective derivative, MPID (18). The failure of MPID to be phosphorylated by wild-type PID indicates that intramolecular phosphorylation is sufficient to explain the observed basal autophosphorylation levels. It has been suggested that intramolecular autocatalytic reactions may play a relevant role in facilitating the rapid activation of low abundance proteins (25), consistent with the observed requirement for the abrupt repositioning of auxin transport proteins at specific developmental stages (26).
Auxin-mediated regulation of cell elongation and root orientation in response to gravistimulation have been associated with transient changes in cytoplasmic calcium levels (27, 28). Recently, Benjamins and colleagues showed that PID interacts with two calcium-binding proteins, and that both a calmodulin inhibitor and calcium-channel blockers similarly enhanced the effect of PID overexpression on root collapse, suggesting that calcium may inhibit PID activity (19). However, it is not clear whether this interaction reflects a direct inhibitory effect or an indirect role of calcium inhibition through secondary protein interaction. We find that Ca2+ levels >2.5 mM are sufficient to interfere with kinase activity in vitro. Our data suggest that Ca2+ may directly compete with Mg2+ for binding to PID. Interestingly, the conserved DFG sequence that is replaced by DFD in the plant ACG-VIII kinase subfamily has been implicated in the coordinate binding of Mg2+-ATP (20). This change may facilitate the exchange of Mg2+ for Ca2+, leading to loss of ATP binding.
Here we present in vitro data showing that PDK1 is able to bind and phosphorylate PID, resulting in a rapid and significant increase in PID autocatalytic activity. Consistent with this result, PID immunoprecipitated from Arabidopsis protoplasts with reduced levels of PDK1 expression showed decreased transphosphoryation of MBP. In the plant, PID expression is detected in the embryo, shoot vasculature, and transiently in developing floral organs (18, 21, 22), which correspond to those tissues in which PID is preferentially activated by protein extracts. By contrast, PDK1 has been shown to be uniformly transcribed throughout the plant body (16). The specific activation of PID by PDK1 in seedling shoots and flowers suggests that PDK1 is preferentially translated in these tissues, or that it is selectively activated in response to localized signals. Our data do not rule out the possibility that other regulatory proteins function in tandem with PDK1 to potentiate PID activity in vivo.
The signature activation loop sequence recognized and phosphorylated by PDK1 is evolutionarily conserved across the plant, animal, and fungal kingdoms (15, 24). Our data provide strong evidence that at least one of the serines within the PID activation loop is a substrate for PDK1 phosphorylation. S290 is very likely to be the functional PDK1 phosphorylation site (P) based on the presence of a conserved hydrophobic residue F291 in the P + 1 site as predicted by the PDK1 phosphorylation consensus motif (29). The AGC kinases share a second required element for PDK1 activation, the PIF domain, which mediates substrate-specific interaction (12). We find that substitution of the two conserved phenylalanine residues for valine within the PID FXXF motif results in a substantial decrease in activation of PDF when compared with wild-type PID protein. The physiological significance of PDK1-dependent PID activation is demonstrated by the finding that, in comparison with wild-type PID, the ability of seedling shoot extract to activate the PDF mutant is dramatically reduced. Residual phosphorylation of PDF by cell extracts suggests that proteins other than PDK1 may also activate PID in vivo. This result would be consistent with an earlier report in which a PID:GUS fusion lacking the PIF domain was able to rescue a partial loss-of-function pid mutant (21). Nevertheless, the magnitude of the observed change in phosphorylation suggests that PDK1 is a primary component of the endogenous transphosphorylation activity detected in seedling extracts.
It has been proposed that binding the PIF domain is a required step in the conformational changes leading to stimulation of PDK1 activity (12). Earlier studies in which the interaction between two AGC kinases, AGC1-1 and AGC2-1, were examined in both yeast and Arabidopsis protoplasts indicated that the PIF motif is required for binding of these kinases to AtPDK1 (16). The discrepancy between the previous result and the data presented here might reflect the fact that numerous AGC kinases, including PID, AGC1-1, and AGC2-1, contain cryptic FXXF motifs that may allow PDK1 binding under the in vitro conditions used here, but these sites may not be recognized under cellular conditions. Interestingly, we find that two related Arabidopsis AGC class VIII kinases that do not contain the hydrophobic PDK1 docking sequence also bind to, but are not activated by, PDK1 in vitro (H.Z and S.K.C., unpublished data), suggesting that domains other than the PIF motif may be responsible for the affinity between PDK1 and plant AGC kinases.
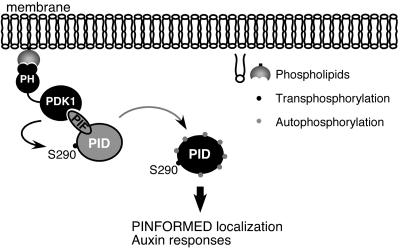
The functional similarity between the localization of PIN proteins during auxin transport and the animal glucose transporter GLUT4 in response to insulin has led to the suggestion that these processes may be mechanistically analogous (30). During insulin signaling, two AGC kinases, PKB and PKC, are recruited to the membrane and activated by PDK1 to generate a secondary signaling cascade, resulting in asymmetrical localization of GLUT 4 within the plasma membrane (11). AtPDK1 has a pleckstrin homology (PH) domain that mediates its binding to phospholipids including phosphatidic acid and phosphatidylinositol-4, 5-diphosphate (PIP2) (16). Our data raise the possibility that PID could be recruited to phospholipid-enriched membrane locations and activated by PDK1, in a manner similar to that of PKB/PKC in glucose transport, to initiate a signaling cascade resulting in membrane localization of a polar auxin transport complex (Fig. 6). The data presented here provide a link between membrane-localized phospholipid ligands and downstream auxin-mediated signaling events. Remaining questions include the role of auxin in triggering the signaling cascade and how the resulting signals are transduced to effect the cellular changes by which targeted-vesicle transport is achieved.
Fig. 6.
Proposed role of PID in phospholipid-mediated membrane targeting. PDK1 is localized to specific membrane regions by binding through the pleckstrin homology (PH) domain to locally produced phosphoinositide ligands. PID is recruited to the membrane through the interaction of PDK1 with the PID PIF domain. PDK1 phosphorylation of Ser 290 within the activation loop stimulates PID autophosphorylation. Active PID phosphorylates substrate proteins, resulting in the targeted localization of the polar auxin transport complex machinery.
Materials and Methods
Expression and Purification of Recombinant Proteins.
The PDF variant was generated by PCR amplification and subcloned into the EcoRI–NotI sites of pGEX4T-1 vector (Amersham). Polyhistidine-tagged PID and PDK1 (At5g04510) were PCR amplified and subcloned into pET15b vector (Novagen) by using NdeI–XhoI and XhoI–BamHI sites, respectively. Protein expression was induced in transformed BL21 (DE3) cells with 1 mM isopropyl β-d-thiogalactoside when the OD600 reached 0.7. The culture was grown for 18 h at 24°C, and the bacterial pellet was harvested by centrifugation.
Bacterial cells expressing polyhistidine-tagged PID (HIS:PID), were solubilized in buffer A (50 mM NaH2PO4/300 mM NaCl, pH 8.0) containing 1 mg/ml lysozyme, 10 mM imidazole (Sigma), and EDTA-free protease inhibitors (Roche) and incubated on ice for 30 min. Triton X-100 was added to 1%, and the soluble fraction was isolated by centrifuging for 30 min at 8,000 × g in an SS-34 rotor (Sorvall). Next, 2-mercaptoethanol was added to the lysate at a concentration of 14 mM. One milliliter of the soluble fraction was added to 250 μl of Ni-NTA resin (Qiagen) preequilibrated with 10 mM imidazole in buffer A, and the mixture was agitated at 4°C for 2 h. The resin-protein complex was washed four times with buffer B (buffer A with 20 mM imidazole) and two times with buffer B supplemented with 15% ethanol. GST-tagged proteins were purified as described in ref. 18.
PID Autophosphorylation and Kinase Assays.
Twenty microliters of resin-bound HIS:PID or GST:PID proteins were washed once with kinase buffer (KB) (20 mM Tris, pH 7.5/15 mM MgCl2/1 mM DTT), resuspended in 30 μl of KB containing 50 μM ATP and 10 μCi of [γ-32P]ATP [Amersham; 3,000 Ci/mmol (1 Ci = 37 GBq)], and incubated for 45 min at 30°C. Reactions were terminated by adding 10 μl of 4× Laemmli loading buffer, and the samples were separated on 12% polyacrylamide gels (Life Therapeutics, Frenchs Forest, Australia). Gels were stained with Coomassie brilliant blue (Bio-Rad) and visualized by autoradiography. To determine PID transactivity, 3 μg of MBP and 5 μCi of [γ-32P]ATP were added to the autophosphorylation reaction after 45 min, and the mixture was incubated at 30°C for 15 min. Samples containing MBP were separated on 4–20% gradient gel. Divalent cations were added to the above reactions as described in the text.
PID Activation by Plant Protein Extract.
Fresh Arabidopsis tissue was homogenized on ice by grinding in KB supplemented with protease inhibitors. Supernatant containing 20 μg of total plant protein was applied to recombinant GST:PID protein immobilized on beads, and phosphorylation assays were performed as described earlier. After 30-min incubation, the beads were washed two times with KB buffer, resuspended in 30 μl of KB containing 3 μg of MBP, 50 μM ATP, and 5 μCi of [γ-32P] ATP, then incubated at 30°C for 15 min before the reaction was terminated. Samples were separated by PAGE and visualized by autoradiography. The band corresponding to MBP was excised from the gel, and incorporation of radioactivity was quantitated by liquid scintillation counting.
GST Pull-Down and Immunoblotting.
HIS:PDK1 bacterial lysate was prepared as described by using a binding buffer (BB) (50 mM Tris, pH 6.8/200 mM NaCl/0.1% Tween). GST or GST-tagged PINOID proteins bound to agarose beads were incubated with equal volume of HIS:PDK1 lysate for 1–2 h at 4°C with gentle shaking. The beads were washed four times in BB and two times in BB with 300 mM NaCl and 2% glycerol, and the bound proteins were resuspended in Laemmli loading buffer. After separation by SDS/PAGE, the proteins were transferred to nitrocellulose membrane and the bound PDK1 was visualized by Western blotting with an anti-Penta-His primary antibody (Qiagen) and an horseradish peroxidase (HRP)-conjugated anti-mouse IgG secondary antibody. The HRP reaction was developed with an enhanced chemiluminescence Western blotting analysis system (Amersham).
Phosphorylation of PID Proteins by PDK1.
Twenty microliters of HIS:PDK1 bound to GST::PID beads was washed once with KB, resuspended in 30 μl of KB containing 50 μM ATP and 10 μCi of [γ-32P]ATP, and incubated for 30 min at 30°C. The reactions were terminated by adding 10 μl of 4× Laemmli loading buffer. The proteins were resolved by SDS/PAGE and visualized by autoradiography.
Protoplasts Transient Expression Assays.
cMyc-PDK1 and PDK1-RNAi were generated as described in ref. 16. The full-length PID cDNA was cloned into pDONR207 Gateway compatible vector via the BP reaction according to manufacturer’s protocols (Invitrogen). PID was transferred into the destination vector pGWB15 to generate a 3× N-terminal HA-tagged PID fusion construct 35S:HA-PID. Arabidopsis cell suspension cultures were maintained as described (31). Protoplast isolation and polyethylene glycol-mediated transfection were performed according to Meskiene et al. (32). For each time point shown, 5 μg each of HA-PID and cMyc-PDK1, with or without 5 μg of PDK1-RNAi were transfected into 7 × 105 cells, and the cells were cultured for 24 h before harvesting.
Arabidopsis Immunocomplex Kinase Assays.
Cells were lysed by vortexing frozen samples for 30 s in 100 μl of protein extraction buffer as described by Meskiene et al. (32). After centrifugation, 80 μl of protein from the cleared supernatant was preincubated in the presence of 15 μl of protein G Sepharose beads. The supernatant was immunoprecipitated with 20 μl of protein G beads and 5 μl of HA antibody (clone 3F10; Roche Molecular Biochemicals) at 4°C. The beads were washed three times with wash buffer (50 mM Tris, pH 7.4/250 mM NaCl/5 mM EGTA/5 mM EDTA/0.1% Nonidet P-40/0.1% Tween 20) and once with kinase buffer. The bound proteins were resuspended in 20 μl of kinase buffer and 1 mM ATP. After PID autophosphorylation for 15 or 45 min, the kinase reactions were mixed with 10 μg of MBP and 2 μCi of [γ-32P]ATP and incubated for 30 min at room temperature. Phosphorylation of MBP was analyzed by autoradiography.
For protein gel blot analysis, equal amounts of total protein extracts from each transfection were separated by SDS/PAGE, transferred to membrane, and probed with either monoclonal HA antibody or cMyc antibody (clone 9E10; Santa Cruz Biotechnology). The horseradish peroxidase reaction was developed with an enhanced chemiluminescence Western blotting analysis system.
Supplementary Material
Acknowledgments
We thank Kelly Smith for expert technical assistance and Gary Gardner and Elaine Tobin for helpful discussions. The pGWB15 vector was a generous gift from T. Nakagawa (Shimane University, Matsue, Japan). This research was supported by National Institutes of Health Grant R01GM64584 (to S.K.C.) and Biotechnology and Biological Sciences Research Council Grant 111/C18199 (to R.G.A. and L.B.).
Abbreviations
- HA
hemagglutinin
- HIS
polyhistidine
- KB
kinase buffer
- MBP
myelin basic protein
- PDK1
phosphoinositide-dependent protein kinase 1
- PID
PINOID
- PIF
PDK1-interacting fragment
- PIN
PINFORMED.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Friml J., Palme K. Plant Mol. Biol. 2002;49:273–284. [PubMed] [Google Scholar]
- 2.Gälweiler L., Guan C., Müller A., Wisman E., Mendgen K., Yephremov A., Palme K. Science. 1998;282:2226–2230. doi: 10.1126/science.282.5397.2226. [DOI] [PubMed] [Google Scholar]
- 3.Steinmann T., Geldner N., Grebe M., Mangold S., Jackson C. L., Paris S., Gälweiler L., Palme K., Jürgens G. Science. 1999;286:316–318. doi: 10.1126/science.286.5438.316. [DOI] [PubMed] [Google Scholar]
- 4.Geldner N., Friml J., Stierhof Y. D., Jurgens G., Palme K. Nature. 2001;413:425–428. doi: 10.1038/35096571. [DOI] [PubMed] [Google Scholar]
- 5.Geldner N., Anders N., Wolters H., Keicher J., Kornberger W., Muller P., Delbarre A., Ueda T., Nakano A., Jürgens G. Cell. 2003;112:219–230. doi: 10.1016/s0092-8674(03)00003-5. [DOI] [PubMed] [Google Scholar]
- 6.Friml J., Wisniewska J., Benková E., Mendgen K., Palme K. Nature. 2002;415:806–809. doi: 10.1038/415806a. [DOI] [PubMed] [Google Scholar]
- 7.Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., Friml J. Cell. 2003;115:591–602. doi: 10.1016/s0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- 8.Noh B., Bandyopadhyay A., Peer W. A., Spalding E. P., Murphy A. S. Nature. 2003;423:999–1002. doi: 10.1038/nature01716. [DOI] [PubMed] [Google Scholar]
- 9.Reinhardt D., Pesce E.-R., Stieger P., Mandel T., Baltensperger K., Bennett M., Traas J., Friml J., Kuhlemeier C. Nature. 2003;426:255–260. doi: 10.1038/nature02081. [DOI] [PubMed] [Google Scholar]
- 10.Friml J., Yang X., Michniewicz M., Weijers D., Quint A., Tietz O., Benjamins R., Ouwerkerk P. B. F., Ljung K., Sandberg G., et al. Science. 2004;306:862–865. doi: 10.1126/science.1100618. [DOI] [PubMed] [Google Scholar]
- 11.Watson R. T., Kanzaki M., Pessin J. E. Endocr. Rev. 2004;25:177–204. doi: 10.1210/er.2003-0011. [DOI] [PubMed] [Google Scholar]
- 12.Biondi R. M., Nebreda A. R. Biochem. J. 2003;15:1–13. doi: 10.1042/BJ20021641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mora A., Komander D., van Aalten D. M., Alessi D. R. Semin. Cell Dev. Biol. 2004;15:161–170. doi: 10.1016/j.semcdb.2003.12.022. [DOI] [PubMed] [Google Scholar]
- 14.Bögre L., Okresz L., Henriques R., Anthony R. G. Trends Plant Sci. 2003;8:424–431. doi: 10.1016/S1360-1385(03)00188-2. [DOI] [PubMed] [Google Scholar]
- 15.Day I. S., Miller C., Golovkin M., Reddy A. S. J. Biol. Chem. 2000;275:13737–13745. doi: 10.1074/jbc.275.18.13737. [DOI] [PubMed] [Google Scholar]
- 16.Anthony R. G., Henriques R., Helfer A., Meszaros T., Rios G., Testerink C., Munnik T., Deak M., Koncz C., Bögre L. EMBO J. 2004;23:572–581. doi: 10.1038/sj.emboj.7600068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Briggs W. R., Christie J. M. Trends Plant Sci. 2002;7:204–210. doi: 10.1016/s1360-1385(02)02245-8. [DOI] [PubMed] [Google Scholar]
- 18.Christensen S. K., Degenais N., Chory J., Weigel D. Cell. 2000;100:469–478. doi: 10.1016/s0092-8674(00)80682-0. [DOI] [PubMed] [Google Scholar]
- 19.Benjamins R., Galván Ampudia C. S., Hooykaas P. J. J., Offringa R. Plant Physiol. 2003;132:1623–1630. doi: 10.1104/pp.103.019943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanks S. K., Quinn A. M., Hunter T. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 21.Benjamins R., Quint A., Weijers D., Hooykaas P., Offringa R. Development. 2001;128:4057–4067. doi: 10.1242/dev.128.20.4057. [DOI] [PubMed] [Google Scholar]
- 22.Furutani M., Vernoux T., Trass J., Kato T., Tasaka M., Aida M. Development. 2004;131:5021–5030. doi: 10.1242/dev.01388. [DOI] [PubMed] [Google Scholar]
- 23.Huse M., Kuriyan J. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 24.Newton A. C. Chem. Rev. 2001;101:2353–2364. doi: 10.1021/cr0002801. [DOI] [PubMed] [Google Scholar]
- 25.Hallenbeck P. C., Walsh D. A. J. Biol. Chem. 1983;258:13493–13501. [PubMed] [Google Scholar]
- 26.Friml J., Vieten A., Sauer M., Weijers D., Schwartz H., Hamann T., Offringa R., Jürgens G. Nature. 2003;426:147–153. doi: 10.1038/nature02085. [DOI] [PubMed] [Google Scholar]
- 27.Chen R., Rosen E., Masson P. Plant Physiol. 1999;120:343–350. doi: 10.1104/pp.120.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang T., Poovaiah B. W. J. Biol. Chem. 2000;275:3137–3143. doi: 10.1074/jbc.275.5.3137. [DOI] [PubMed] [Google Scholar]
- 29.Cheng X., Ma Y., Moore M., Hemmings B. A., Taylor S. S. Proc. Natl. Acad. Sci. 1998;95:9849–9854. doi: 10.1073/pnas.95.17.9849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muday G. K., Murphy A. S. Plant Cell. 2002;14:293–299. doi: 10.1105/tpc.140230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mathur J., Koncz C. Methods Mol. Biol. 1998;82:35–42. doi: 10.1385/0-89603-391-0:35. [DOI] [PubMed] [Google Scholar]
- 32.Meskiene I., Baudouin E., Schweighofer A., Liwosz A., Jonak C., Rodriguez P. L., Jelinek H., Hirt H. J. Biol. Chem. 2003;278:18945–18952. doi: 10.1074/jbc.M300878200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.