Abstract
Mastitis, a mammary gland inflammation in response to bacterial infection, is a major problem in the dairy industry. We found that cows susceptible to mastitis have a three-base insertion in a glycine-coding stretch of the gene for forebrain embryonic zinc finger-like (FEZL), a transcription factor with a role in neuronal development. Mastitis induces FEZL expression in mammary glands, and induced FEZL promotes expression of the axon-attracting molecule semaphorin 5A (SEMA5A) through a GCAG sequence. FEZL also induces SEMA5A expression in susceptible cattle but at a lower level than in resistant cattle. Enhanced SEMA5A induces expression of at least nine genes related to the host’s immune response, including TNF-α and IL-8. We propose that susceptibility to mastitis results from an impaired immune response due to the lower transcription activity of susceptible FEZL. Our results provide an avenue to select for genetic improvement of resistance to mastitis and suggest that the FEZL–SEMA5A pathway might control both neuronal development and innate immunity.
Keywords: FEZL, quantitative trait locus, semaphorin 5A
Mastitis generates annual total losses of >69 billion yen in Japan (1) and 2 billion dollars in the United States (2) by reducing milk quantity and quality and increasing health costs. Just as current selective breeding programs based on population genetics have successfully increased milk yield, genetic selection for mastitis resistance could potentially be practiced, not only to reduce the cost of producing milk but also to improve the health and well-being of dairy cows. Recently Wall et al. (3) produced transgenic cows resistant to infection by inducing the cells of the mammary gland to secret an antibacterial protein. However, the public may not quickly accept milk from transgenic animals despite the improved health of dairy herds (4). On the other hand, several groups have begun quantitative trait locus (QTL) mapping using a granddaughter design to identify genes influencing mastitis resistance (e.g., ref. 2). Alternatively, we used six half-sibling families whose sires were derived from the same grandsire and performed selective genotyping. This method reduces the number of progeny genotyped, although phenotypic effects may be overestimated because of the biased selection of progeny (e.g., ref. 5). The function of a gene carrying a putative polymorphism positionally identified should significantly influence the phenotype.
Forebrain embryonic zinc finger-like (FEZL) (6) controls the development of monoaminergic neurons (7). Hirata et al. (8) reported that FEZL-deficient mice display hyperactive behavior, suggesting that FEZL has a role in neuronal development. Semaphorin 5A (SEMA5A), an axon guidance cue (9), belongs to the semaphorin family characterized by functionally conserved roles in neuronal development (10). Recent studies, however, revealed the existence of crosstalk between neuronal development and immunity. The neuronal repellent Slit regulates both neuronal and leukocyte migration (11). The semaphorin family members SEMA4A and SEMA4D activate T cells (12) and enhance B cell responses (13), respectively. Kumanogoh et al. (12, 13) called them “immune semaphorins,” but it is not known whether SEMA5A is an immune semaphorin.
We demonstrate here that susceptible cows have a three-base insertion in the FEZL gene. Mastitis enhances FEZL, FEZL promotes SEMA5A, and SEMA5A induces TNF-α and IL-8 expression. Because the resistant FEZL allele promotes higher SEMA5A expression than the susceptible FEZL allele, the susceptibility to mastitis might result from impaired immune response. Here we identify a gene influencing mastitis resistance at the molecular level and outline an unexpected role for the FEZL–SEMA5A signaling pathway in innate immunity.
Results
A Somatic Cell Score (SCS) Locus Is Linked to FEZL.
To determine QTL influencing mastitis resistance, we selected six half-sibling families with a total of 6,561daughters in the Tokachi county area of Hokkaido, Japan, to minimize possible environmental variations. We used the average SCS [log2 (somatic cell count/100) + 3] during the first lactation period as a phenotype parameter, because there is a strong genetic correlation between SCS and mastitis (14). The somatic cell count in milk did not normally distribute, whereas SCS did (skewness = 0.93; kurtosis = −0.42; Fig. 5, which is published as supporting information on the PNAS web site). We attempted to collect cows with SCS of >5, suspected to have clinical mastitis (15); however, most of them had been culled. The resulting families consisted of 181 daughters with high SCS of 4.82 and 297 daughters with low SCS of 0.65 (Table 1).
Table 1.
SCS of the half-sibling families with the number of collected samples in parentheses
| Sire | SCS ≤ 1 | 1 < SCS < 4 | 4 ≤ SCS | Total | Average SCS ± SD |
|---|---|---|---|---|---|
| 1 | 163 (16) | 1,340 | 751 (7) | 2,254 | 2.67 ± 1.37 |
| 2 | 215 (65) | 1,124 | 601 (30) | 1,940 | 2.56 ± 1.49 |
| 3 | 62 (51) | 418 | 208 (27) | 688 | 2.55 ± 1.39 |
| 4 | 57 (57) | 285 | 269 (72) | 611 | 2.97 ± 1.60 |
| 5 | 88 (41) | 441 | 183 (23) | 712 | 2.41 ± 1.39 |
| 6 | 67 (67) | 203 | 86 (22) | 356 | 2.28 ± 1.48 |
| Total | 652 (297) | 3,811 | 2,098 (181) | 6,561 | 2.60 ± 1.45 |
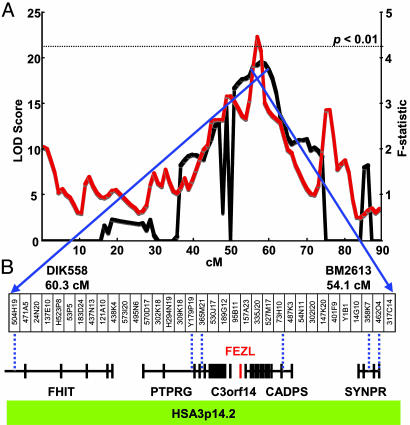
With our 478 samples, we embarked on a whole-genome scan of 29 autosomal chromosomes by typing 272 microsatellite markers. The analysis using the dismult program of the diseq package, which is suitable for a multipoint analysis of selective genotyping data, revealed significant linkage of SCS loci to Bos taurus autosomal chromosomes BTA21 and BTA22 with a likelihood of odds (LOD) score of >5 (Fig. 6, which is published as supporting information on the PNAS web site). Further analysis with an additional 64 markers in BTA22 supported the linkage (Fig. 1A), although the LOD scores by diseq are likely overestimates because all pedigrees were related. We also confirmed significant linkage as an F-statistic of 4.45 (P < 0.01) in a total of 6,561 daughters of the families using qtlexpress, which was developed for the analysis of QTL from outbred populations and was able to include the phenotype information of untyped samples (Fig. 1A). Next we assumed that the 6.2-centimorgan interval with a LOD score of >18 by diseq is critical for SCS, and we constructed a bacterial artificial chromosome contig containing the region (Fig. 1B). Partial sequencing of the contig indicated that the 6.2-centimorgan region was equivalent to Homo sapiens autosomal chromosome 3p14.2, where six known genes were assigned (ref. 16 and Fig. 1B).
Fig. 1.
An SCS locus is linked to FEZL. (A) Likelihood of odds score profile by diseq (black) and F-statistic profile by qtlexpress (red) on BTA22 obtained by the second screening. A dotted line indicates the chromosome-wide threshold of P < 0.01 by qtlexpress. (B) A bacterial artificial chromosome contig. The names of clones beginning with H were from CHORI-240, names of clones beginning with Y were from Wagyu, and others were from RPCI-42. Blue dotted lines indicate human homologue locations.
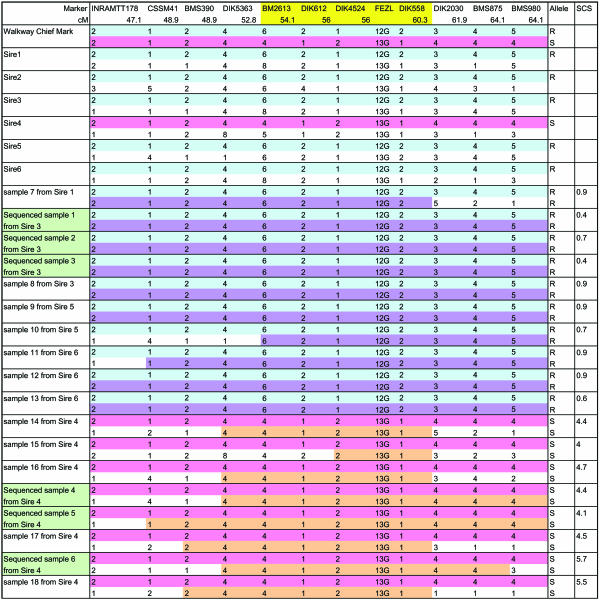
We determined phase of the 6.2-centimorgan region in 478 daughters using four microsatellites and found that Sire 4 had susceptible (S) alleles whereas the remaining sires had resistant (R) alleles, suggesting that Walkway Chief Mark, a grandsire of our families, would have been heterozygous (Fig. 2). Sequencing exons of the six genes in three cows having homozygous R alleles with low SCS and three cows having homozygous S alleles with high SCS, we identified neither nonsense nor missense mutations except for a three-base insertion in the FEZL S allele. FEZL contains six C2H2 type zinc-finger domains and a glycine stretch where the insertion resulted in an extension of 12 glycine (12G) to 13 glycine (13G) (Fig. 7, which is published as supporting information on the PNAS web site). Although we cannot exclude the possibility that one or more of the other five genes may affect SCS, it is valuable to examine whether the polymorphism in FEZL affects SCS.
Fig. 2.
Haplotype map of selected samples. Blue and purple boxes indicate homozygous R alleles, and pink and beige boxes indicate homozygous S alleles. Green boxes indicate sequenced samples, and yellow boxes indicate the 6.2-centimorgan region.
In our half-sibling families born during 1990–1996, 8.7% of 12G/12G FEZL cattle had SCS of >5, whereas 17.0% of 13G/13G FEZL cattle did (Table 2), suggesting that 13G/13G FEZL cattle have twice the rate of susceptibility to mastitis than 12G/12G FEZL cattle. Because one sire had 13G/13G FEZL alleles, whereas five had 12G/13G FEZL alleles in our families (Fig. 2), we were able to determine the effect of FEZL genotypes in the six sires on SCS of their offspring. As shown in Table 2, one 12G allele was sufficient to decrease SCS. We also randomly collected and typed 492 samples derived from 217 sires in the Tokachi county area. Although these samples for animals born during 1997–2001 did not include 12G/12G FEZL cattle, the average SCS of 13G/13G FEZL cattle was higher than that of cattle with 12G/13G FEZL (Table 2), supporting the observations in our half-sibling families. Together, these results indicate that 12G FEZL influences resistance to clinical mastitis.
Table 2.
The effects of FEZL on SCS
| Animals | 5 ≤ SCS (%) | Average SCS ± SD | P value (t test) |
|---|---|---|---|
| 12G/12G daughters | 8/92 (8.7) | 1.62 ± 1.84 | <0.0005∗ |
| 12G/13G daughters | 18/162 (11.1) | 2.12 ± 1.98 | |
| 13G/13G daughters | 38/224 (17.0) | 2.56 ± 2.17 | <0.05∗∗ |
| Daughters from 12G/13G sire | 468/5,950 (7.9) | 2.56 ± 1.42 | |
| Daughters from 13G/13G sire | 73/611 (12.0) | 2.97 ± 1.60 | <0.000001 |
| 12G/13G random samples | 0/44 (0) | 2.00 ± 0.69 | |
| 13G/13G random samples | 4/448 (0.9) | 2.31 ± 0.89 | <0.01 |
∗, P value vs. 13G/13G daughters.
∗∗, P value vs. 12G/13G daughters.
Mastitis Induces FEZL.
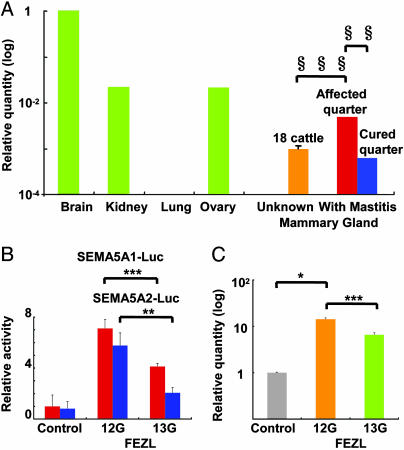
Consistent with a role for FEZL as a transcription factor that regulates forebrain development, bovine FEZL was expressed strongly in the brain and weakly in the mammary gland (Fig. 3A). Nevertheless, FEZL expression is greater in the quarter affected with mastitis, confirmed by the California Mastitis Test, than in the cured quarter of the same cattle (Fig. 3A). Because it is possible that infection enhances FEZL expression in the mammary gland, we hypothesized that FEZL has an important role in antimicrobial response and that the polymorphism we described affects function.
Fig. 3.
Mastitis induces SEMA5A through FEZL. (A) FEZL expression. Results are means of quantity relative to brain. Green bars indicate the mean (n = 3) from the same cattle, and red and blue bars indicate the mean (n = 3) from other cattle with mastitis. The orange bar indicates the mean ± SEM (n = 54) from 18 randomly collected cattle with unknown status. Symbols denote statistically significant results as determined by Student’s t test (∗∗, P < 0.0005; ∗∗∗, P < 0.0001). (B) Luciferase assay of FEZL with SEMA5A1 (red) and SEMA5A2 (blue). Results are means ± SEM of relative activity to COS7 cells transfected without FEZL. Asterisks denote statistically significant results as determined by Student’s t test (∗∗, P < 0.01; ∗∗∗, P < 0.005). (C) SEMA5A expression in OCUB-M cells transfected with FEZL. Results are means ± SEM (n = 9; three experiments) of quantity relative to OCUB-M cells transfected without FEZL after adjusting for the FEZL expression level. Asterisks denote statistically significant results as determined by Student’s t test (∗, P < 0.05; ∗∗∗, P < 0.005).
FEZL Binds GCAG.
To elucidate the effect of the polymorphism on function, V5-tagged FEZL were transfected into monkey kidney COS-7 cells or human breast cancer-derived OCUB-M cells. Both 12G and 13G FEZL localized to the nucleus (Fig. 8, which is published as supporting information on the PNAS web site), indicating that the polymorphism does not affect protein localization.
In many transcription factors, the zinc fingers mediate binding to promoter elements of their target genes (17). To determine the optimal binding sequences for the FEZL protein, we performed chromatin immunoprecipitation assays of cells transfected with His-tagged FEZL. Comparison of the selected sequences by anti-His antibody yielded an optimal binding site of GCAG, and there was no difference between the 12G and 13G FEZL consensus in COS-7 and OCUB-M cells (Fig. 9, which is published as supporting information on the PNAS web site). Gel mobility shift and competition experiments showed that binding was abolished by coincubation with unlabeled consensus (lanes 4 and 9 in Fig. 10, which is published as supporting information on the PNAS web site) but not by AP2 (lanes 5 and 10 in Fig. 10), demonstrating that DNA binding activities of 12G and 13G FEZL are similar.
FEZL Activates SEMA5A.
Androgen receptors contain a glycine stretch, and its complete deletion reduces transactivation in vitro (18), suggesting that the length of the glycine stretch in FEZL might affect its transcription activity. To compare 12G and 13G FEZL transcription activities, we selected the first (SEMA5A1) and second (SEMA5A2) introns of SEMA5A as binding sequences obtained from the chromatin immunoprecipitation assays (Fig. 11, which is published as supporting information on the PNAS web site). 12G FEZL induces both the SEMA5A1 and SEMA5A2 reporters to higher levels than 13G FEZL (Fig. 3B). 12G FEZL stimulates greater SEMA5A expression than 13G FEZL in OCUB-M cells (Fig. 3C). Conversely, small interfering RNA (siRNA) targeting FEZL reduces SEMA5A expression (Fig. 12, which is published as supporting information on the PNAS web site). Moreover, SEMA5A expression is greater in 12G/13G FEZL cattle than in 13G/13G FEZL cattle, indicating that SEMA5A is the target of FEZL in vivo (Fig. 13, which is published as supporting information on the PNAS web site). Furthermore, mastitis induces SEMA5A expression, demonstrating that infection enhances SEMA5A through FEZL in vivo (Fig. 13). Thus, the glycine stretch in FEZL appears to influence transcription activity of SEMA5A after infection.
FEZL Induces TNF-α and IL-8 Through SEMA5A.
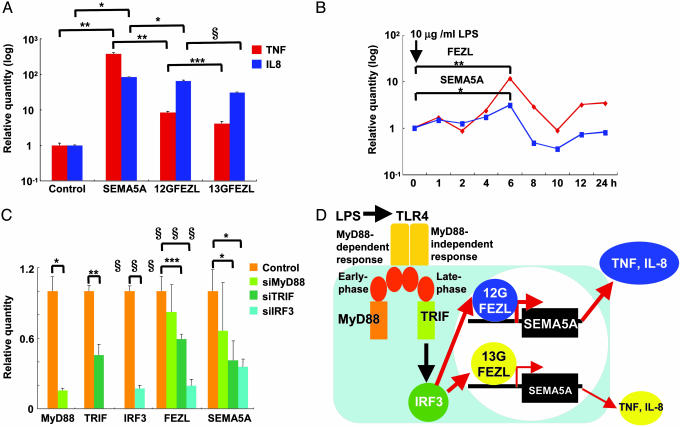
To investigate potential genes that might be induced by SEMA5A, we screened OCUB-M cells transfected with SEMA5A by microarray analysis. The candidate genes induced by SEMA5A included nine genes related to chemotaxis and inflammatory/immune response, but not plexin (PLXN) B3, a gene coding a known receptor for SEMA5A (ref. 19 and Fig. 14, which is published as supporting information on the PNAS web site). The induction of two of the nine genes, TNF-αand IL-8, was confirmed by quantitative RT-PCR (Fig. 4A). In addition, 12G FEZL induced greater TNF-α and IL-8 expression than 13G FEZL (Fig. 4A). In contrast, RNA interference of FEZL or SEMA5A reduced TNF-α and IL-8 expression (Fig. 12). Impaired TNF-α and IL-8 expression controlled by FEZL and SEMA5A could explain susceptibility of 13G FEZL cattle to mastitis.
Fig. 4.
FEZL is in the TLR4 signaling pathways. (A) TNF-α (red) and IL-8 (blue) expression in OCUB-M cells transfected with SEMA5A or FEZL. Results are means ± SEM (n = 9; three experiments) of quantity relative to OCUB-M cells transfected without constructs after adjusting for the FEZL expression level. Symbols denote statistically significant results as determined by Student’s t test (∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.005; ∗, P < 0.001). (B) FEZL (red) and SEMA5A (blue) expression in OCUB-M cells treated by 10 μg/ml LPS. Results are means (n = 3), and asterisks denote statistically significant results as determined by Student’s t test (∗, P < 0.05; ∗∗, P < 0.01). (C) MyD88, TRIF, IRF3, FEZL, and SEMA5A expression in OCUB-M cells transfected with siRNAs. Results are means ± SEM (n = 9; three experiments) of quantity relative to OCUB-M cells transfected without constructs. Symbols denote statistically significant results as determined by Student’s t test (∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.005; ∗∗∗, P < 0.0001). (D) Schematic diagram of the TLR4 signaling pathways. Red arrows indicate the pathway revealed by this work.
Infection Induces FEZL Through Toll/IL-1-Receptor-Domain-Containing Adapter Protein-Inducing IFN-β (TRIF).
Escherichia coli causes 13.9% of mastitis (1). To build a model of mammary glands infected by Gram-negative bacteria, we conducted quantitative RT-PCR using OCUB-M cells after LPS treatment. Consistent with the results in bovine mammary gland (Figs. 3A and 13), treatment with 10 μg/ml LPS from E. coli induced both FEZL and SEMA5A expression in OCUB-M cells (Fig. 4B). The maximum induction was achieved at 6 h, suggesting that FEZL–SEMA5A signaling is involved in the late phase of the infection. LPS facilitates the myeloid differentiation primary-response protein 88 (MyD88)-dependent response in the early phase and the MyD88-independent response in the late phase through Toll-like receptor (TLR) 4 (ref. 20 and Fig. 4D). RNA interference experiments targeting MyD88, TRIF, and IFN-regulatory factor (IRF) 3 revealed that both small interfering TRIF and siIRF3 reduce FEZL and SEMA5A expression 6 h after LPS treatment whereas siMyD88 does not (Fig. 4C). These observations indicated that LPS activates FEZL expression in a MyD88-independent manner through the late phase of infection mediated by TLR4, TRIF, and IRF3 (Fig. 4D).
Discussion
We have presented multiple lines of evidence demonstrating that FEZL is one of the genes responsible for mastitis resistance in cattle. First, the FEZL region was mapped as a QTL on BTA22 by using six half-sibling families having the same grandsire. It is important to note that Heyen et al. (21) identified seven putative QTL for SCS, including two on chromosome 22. Second, the polymorphism of the length of the glycine stretch of FEZL was significantly associated with SCS. Third, the length of the glycine stretch of FEZL impacts its transcription activity, leading to control of cytokine expression. Our linkage study and cell-based assay clearly indicated that a single glycine insertion into the glycine stretch of FEZL has a large effect on its downstream pathway. In our 1990–1996 samples, the incidence of 13G/13G FEZL homozygotes was 46.9% (224/478). In the 1997–2001 samples, it was 91.1% (448/492). The incidence of clinical mastitis in the Tokachi County area has increased from 19.7–22.5% during 1997–1998 to 24.8–28.2% during 2002–2003 (22), suggesting that changes in FEZL genotypes might have an impact on the incidence of clinical mastitis.
Many studies have documented the importance of neutrophil migration for innate immunity. Neutrophils are highly effective phagocytes for clearing infecting bacterial pathogens from host tissue (23). The number of neutrophils migrating to the site of infection has been associated with the production of IL-8 (24). TNF-α induces IL-8 expression in the late phase of infection (25), and the IL-8 receptor is also involved in transepithelial neutrophil migration (26). Moreover, lung-specific transgenic mice with KC, one of the IL-8 homologues in the mouse, are resistant to Klebsiella pneumoniae infection (27). On the other hand, IL-8 receptor knockout mice are susceptible to acute experimental pyelonephritis (28). Thus, the ability of FEZL to promote TNF-α and IL-8 expression through SEMA5A triggered by LPS represents a plausible mechanism to explain the enhanced antimicrobial activity of FEZL-expressing cells. In addition, we identified FEZL as a QTL influencing mastitis resistance. Our results suggest that FEZL is positioned at the crossroads of neuronal development and innate immunity.
Materials and Methods
Mapping.
Genomic DNA was isolated from blood by using NA-1000/48S (Kurabo, Tokyo). Fluorescently labeled (CA)n microsatellite markers were selected from the Shirakawa–U.S. Department of Agriculture genetic map (29). Genotyping was performed by using the ABI 3700 sequencer, genescan v.3.1.2, and genotyper v.2.1 (Applied Biosystems). Linkage analysis was done with diseq (30) and qtlexpress (31). To construct a bacterial artificial chromosome contig, we screened the bovine CHORI-240, RPCI-42, and Wagyu bacterial artificial chromosome libraries (32, 33) by PCR. Sequence analysis was conducted by sequencher v.4.1 (Gene Codes, Ann Arbor, MI).
Transfection.
Bovine FEZL coding sequences were derived by RT-PCR for exons 1–4 of FEZL by using primers CCTCGGCGGCCACCTCTTCC (forward) and GGAATTCCAGCTCTGCACTGTCCGAGTC (reverse) and by genomic PCR for exon 1, including the glycine stretch of resistant or susceptible FEZL using primers GGGGATCCGCGCCATGGCAAGCTC (forward) and TGACGCCGCCGCGCTCAGCA (reverse). These coding sequences were connected by BglI (TOYOBO, Osaka) and T4 ligase (New England Biolabs, Beverly, MA). The human SEMA5A coding sequence was derived by RT-PCR using primers AAAAAGCAGGCTCCATGAAGGGAACCTGTGTTATAGCATG (forward) and AGAAAGCTGGGTAGTACTCATCATAATTATTGAGATCTGTAAAGTA (reverse) and human fetal kidney Marathon-ready cDNA (BD Biosciences). Full-length bovine FEZL or human SEMA5A cDNAs were cloned into pcDNA-DEST40 (Invitrogen) to produce V5-His-tagged protein. COS-7 and OCUB-M (34) cells were provided by the RIKEN Bio-Resource Center (Tsukuba, Japan). Transfection was done with Lipofectamine 2000 (Invitrogen) for 24 h for COS-7 and 48 h for OCUB-M cells. Immunofluorescence was performed by using fluorescein isothiocyanate-conjugated V5 antibody (Invitrogen), Vectorshield (Vector Laboratories), Axioplan 2, and Metasystems Isis (Carl Zeiss, Oberkochen, Germany).
Chromatin Immunoprecipitation Assays.
Cells transfected with FEZL were processed with the chromatin immunoprecipitation assay kit (Upstate Biotechnology, Lake Placid, NY). Sequences selected by anti-His (C-terminal) antibody (Invitrogen) were amplified in ligation-mediated PCR (35), and the consensus was determined by comparing 20 sequences for 12G FEZL and 16 sequences for 13G FEZL in COS-7 cells and 27 sequences for 12G FEZL and 21 sequences for 13G FEZL in OCUB-M cells.
Gel Mobility Shift Assay.
Nuclear protein of COS-7 cells transfected with FEZL was extracted by using the CelLytic NuClear extraction kit (Sigma) and examined by the gel shift assay system (Promega). Protein concentration was measured by Bio-Rad protein assay using BSA as the standard.
Luciferase Assay.
Luciferase assay was performed by using a FEZL- pcDNA-DEST40 vector, SEMA5A-pGL3 (R2.2) basic vector, and pRL-TK vector as an internal control (ratio of 10:10:1) based on the dual-luciferase reporter assay system (Promega). Assays were repeated five times each, and the average measurements were calculated after subtracting the background signals based on 10 readings for Renilla luciferase.
Microarray Analysis.
RNA of OCUB-M cells transfected with SEMA5A was extracted by using an RNeasy Mini kit (Qiagen, Valencia, CA) and hybridized to Human Genome U133 Plus 2.0 GeneChips according to standard Affymetrix (Santa Clara, CA) protocols. Assays were duplicated.
Real-Time Quantitative RT-PCR.
RNA of bovine brain, kidney, lung, mammary gland, ovary, and transfected OCUB-M cells was extracted by using TRIzol (Invitrogen). Quantitative RT-PCR was conducted by using an ABI 7900HT sequence detection system using the comparative Ct method and glyceraldehyde-3-phosphate dehydrogenases (GAPD) as internal controls (Applied Biosystems). RT-PCR with siRNAs was conducted with RNA extracted 6 h after 10 μg/ml LPS treatment. LPS was purchased from Sigma. Primers for RT-PCR are CTACAAGCCCTTCGTCTGTGAAT (forward), GCTGTGGGTCAGCTTGTGATT (reverse), and ACCAAAAAGGGAACTAC (probe) for bovine FEZL; TGTGGGACCAACGCTTTCA (forward), TCATGGATCTCCGTCAGGTTACT (reverse), and CCTGTCTGCACGAACCGCACGTT (probe) for bovine SEMA5A; and GCCCTCAACGACCACTTTGT (forward), CCTGTTGCTGTAGCCAAATTCA (reverse), and AAGCTCATTTCCTGGTACGA (probe) for bovine GAPD. Sequences of primers for human FEZL, SEMA5A, TNF, IL-8, MyD88, TRIF, IRF3, and GAPD could be purchased as TaqMan Gene Expression Assays (P/N: Hs00375188_m1, Hs00187651_m1, 4327055M, 4327042M, Hs00182082_m1, Hs00706140_s1, Hs00155574_m1, and 4326317E, Applied Biosystems). Sequences for siRNAs for human MyD88 could be purchased as Stealth RNAi (P/N: 46-1564 and 46-1565, Invitrogen). Sequences for siRNAs for human FEZL, SEMA5A, TRIF, and IRF3 could be purchased as siGENOM SMARTpool reagent (P/N: M-016985-00, M-019490-00, M-012833-00, and M-006875-01; Dharmacon, Lafayette, CO).
Supplementary Material
Acknowledgments
We thank the Tokachi Agriculture Cooperative Society, Ayako Shiroto, Kenji Hashiba, Tetsuro Beppu, and Shigeru Kudo for collecting samples; the Hokkaido Association of Dairy Industry, Misao Kanemaki, Yamato Atagi, and Tatsuo Shirai for providing the SCS data; Tomohito Itoh, Hiroko Tsukazawa, Fumie Mafune, Yumi Hoshi, Tsuyoshi Ohtake, and Yukio Oyamada for helping with experiments; and Akiko Takasuga and Toshio Watanabe for helpful discussion during this work.
Abbreviations
- QTL
quantitative trait locus
- SCS
somatic cell score
- SEMA5A
semaphorin 5A
- siRNA
small interfering RNA
- TLR
Toll-like receptor
- MyD88
myeloid differentiation primary-response protein 88
- TRIF
Toll/IL-1-receptor-domain-containing adapter protein-inducing IFN-β
- IRF
IFN-regulatory factor.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AY644769 (resistant) and AY644770 (susceptible) for B. taurus forebrain embryonic zinc finger-like (FEZL) protein mRNA; accession nos. DQ335870, DQ335872, DQ335871, and DQ335873 for B. taurus fragile histidine triad protein mRNA, B. taurus protein tyrosine phosphatase, receptor type G protein mRNA, B. taurus hypothetical protein LOC57415 (C3orf14) mRNA, and B. taurus synaptoporin protein mRNA, respectively; and accession nos. DQ335869 (resistant) and DQ335868 (susceptible) for B. taurus Ca2+-dependent secretion activator protein mRNA].
References
- 1.Arita T. Mastitis in Cattle: Clinical Diagnosis and Treatment (Tikusan, Tokyo) 1991:1–26. [Google Scholar]
- 2.Ashwell M. S., Rexroad C. E., Jr., Miller R. H., VanRaden P. M. Anim. Genet. 1996;27:235–242. doi: 10.1111/j.1365-2052.1996.tb00484.x. [DOI] [PubMed] [Google Scholar]
- 3.Wall R. J., Powell A. M., Paape M. J., Kerr D. E., Bannerman D. D., Pursel V. G., Wells K. D., Talbot N., Hawk H. W. Nat. Biotechnol. 2005;23:445–451. doi: 10.1038/nbt1078. [DOI] [PubMed] [Google Scholar]
- 4.Rainard P. Nat. Biotechnol. 2005;23:430–432. doi: 10.1038/nbt0405-430. [DOI] [PubMed] [Google Scholar]
- 5.Lander E. S., Botstein D. Genetics. 1989;121:185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsuo-Takasaki M., Lim J. H., Beanan M. J., Sato S. M., Sargent T. D. Mech. Dev. 2000;93:201–204. doi: 10.1016/s0925-4773(00)00264-1. [DOI] [PubMed] [Google Scholar]
- 7.Levkowitz G., Zeller J., Sirotkin H. I., French D., Schilbach S., Hashimoto H., Hibi M., Talbot W. S., Rosenthal A. Nat. Neurosci. 2003;6:28–33. doi: 10.1038/nn979. [DOI] [PubMed] [Google Scholar]
- 8.Hirata T., Suda Y., Nakao K., Narimatsu M., Hirano T., Hibi M. Dev. Dyn. 2004;230:546–556. doi: 10.1002/dvdy.20068. [DOI] [PubMed] [Google Scholar]
- 9.Kantor D. B., Chivatakarn O., Peer K. L., Oster S. F., Inatani M., Hansen M. J., Flanagan J. G., Yamaguchi Y., Sretava D. W., Giger R. J., et al. Neuron. 2004;44:961–975. doi: 10.1016/j.neuron.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 10.Goodman C. S., Kolodkin A. L., Luo Y., Püchel A. W., Raper J. A. Cell. 1999;97:551–552. [Google Scholar]
- 11.Wu J. Y., Feng L., Park H. T., Havlioglu N., Wen L., Tang H., Bacon K. B., Jiang Z., Zhang X., Rao Y. Nature. 2001;410:948–952. doi: 10.1038/35073616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumanogoh A., Marukawa S., Suzuki K., Takegahara N., Watanabe C., Ch’ng E., Ishida I., Fujimura H., Sakoda S., Yoshida K., et al. Nature. 2002;419:629–633. doi: 10.1038/nature01037. [DOI] [PubMed] [Google Scholar]
- 13.Kumanogoh A., Kikutani H. J. Cell Sci. 2003;116:3463–3470. doi: 10.1242/jcs.00674. [DOI] [PubMed] [Google Scholar]
- 14.Shook G. E., Schutz M. M. J. Dairy Sci. 1994;77:648–658. doi: 10.3168/jds.S0022-0302(94)76995-2. [DOI] [PubMed] [Google Scholar]
- 15.Ward G. E., Schultz L. H. J. Dairy Sci. 1972;55:1428–1431. doi: 10.3168/jds.S0022-0302(72)85689-3. [DOI] [PubMed] [Google Scholar]
- 16.Kent W. J., Sugnet C. W., Furey T. S., Roskin K. M., Pringle T. H., Zahler A. M., Haussler D. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bieker J. J. J. Biol. Chem. 2001;276:34355–34358. doi: 10.1074/jbc.R100043200. [DOI] [PubMed] [Google Scholar]
- 18.Lee D. K., Chang C. J. Clin. Endocrinol. Metab. 2003;88:4043–4054. doi: 10.1210/jc.2003-030261. [DOI] [PubMed] [Google Scholar]
- 19.Artigiani S., Conrotto P., Fazzari P., Gilestro G. F., Barberis D., Giordano S., Comoglio P. M., Tamagnone L. EMBO Rep. 2004;5:710–714. doi: 10.1038/sj.embor.7400189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akira S., Takeda K. Nat. Rev. Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 21.Heyen D. W., Weller J. I., Ron M., Band M., Beever J. E., Feldmesser E., Da Y., Wiggans G. R., VanRaden P. M., Lewin H. A. Physiol. Genomics. 1999;1:165–175. doi: 10.1152/physiolgenomics.1999.1.3.165. [DOI] [PubMed] [Google Scholar]
- 22.Hokkaido Agriculture Cooperative Society. Statistical Report of the Livestock Cooperative Society. Hokkaido, Japan: Hokkaido Agriculture Cooperative Soc.; 2003. [Google Scholar]
- 23.Saad A. M., Concha C., Astrom G. J. Vet. Med. B. 1989;36:337–345. doi: 10.1111/j.1439-0450.1989.tb00612.x. [DOI] [PubMed] [Google Scholar]
- 24.Agace W. W., Hedges S. R., Ceska M., Svanborg C. J. Clin. Invest. 1993;92:780–785. doi: 10.1172/JCI116650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lafleur R. L., Abrahamsen M. S., Maheswaran S. K. Infect. Immun. 1998;66:4087–4092. doi: 10.1128/iai.66.9.4087-4092.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Godaly G., Hang L., Frendéus B., Svanborg C. J. Immunol. 2000;165:5287–5294. doi: 10.4049/jimmunol.165.9.5287. [DOI] [PubMed] [Google Scholar]
- 27.Tsai W. C., Strieter R. M., Wilkowski J. M., Bucknell K. A, Burdick M. D., Lora S. A., Standiford T. J. J. Immunol. 1998;161:2435–2440. [PubMed] [Google Scholar]
- 28.Frendéus B., Godaly G., Hang L., Karpman D., Lundstedt A. C., Svanborg C. J. Exp. Med. 2000;192:881–890. doi: 10.1084/jem.192.6.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ihara N., Takasuga A., Mizoshita A., Takeda H., Sugimoto M., Mizoguchi Y., Reed K. M., Snelling W. M., Kappes S. M., Beattie C. W., et al. Genome Res. 2004;14:1987–1998. doi: 10.1101/gr.2741704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Terwilliger J. D. Am. J. Hum. Genet. 1995;56:777–787. [PMC free article] [PubMed] [Google Scholar]
- 31.Seaton G., Haley C. S., Knott S. A., Kearsey M., Visscher P. M. Bioinformatics. 2002;18:339–340. doi: 10.1093/bioinformatics/18.2.339. [DOI] [PubMed] [Google Scholar]
- 32.Warren W., Smith T. P., III, Rexroad C. E., Fahrenkrug S. C., Allison T., Shu C. L., Catanese J., de Jong P. J. Mamm. Genome. 2000;11:662–663. doi: 10.1007/s003350010126. [DOI] [PubMed] [Google Scholar]
- 33.Fujisaki S., Mizoguchi Y., Takahashi S., Chen Y. Z., Suzuki K., Asakawa S., Soeda E., Shimizu N., Sugimoto Y., Yasue H. Anim. Genet. 2002;33:379–381. doi: 10.1046/j.1365-2052.2002.00896_3.x. [DOI] [PubMed] [Google Scholar]
- 34.Sawada T., Chung Y. S., Nakata B., Kubo T., Kondo Y., Sogabe T., Onoda N., Ogawa Y., Yamada N., Sowa M. Hum. Cell. 1994;7:138–144. [PubMed] [Google Scholar]
- 35.Odom D. T., Zizlsperger N., Gordon D. B., Bell G. W., Rinaldi N. J., Murray H. L., Vilkert T. L., Schreiber J., Rolfe P. A., Gifford D. K., et al. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.