Abstract
Cytosine methylation is an epigenetic mark that promotes gene silencing and plays important roles in development and genome defense against transposons. Methylation patterns are established and maintained by DNA methyltransferases that catalyze transfer of a methyl group from S-adenosyl-l-methionine to cytosine bases in DNA. Erasure of cytosine methylation occurs during development, but the enzymatic basis of active demethylation remains controversial. In Arabidopsis thaliana, DEMETER (DME) activates the maternal expression of two imprinted genes silenced by methylation, and REPRESSOR OF SILENCING 1 (ROS1) is required for release of transcriptional silencing of a hypermethylated transgene. DME and ROS1 encode two closely related DNA glycosylase domain proteins, but it is unknown whether they participate directly in a DNA demethylation process or counteract silencing through an indirect effect on chromatin structure. Here we show that DME and ROS1 catalyze the release of 5-methylcytosine (5-meC) from DNA by a glycosylase/lyase mechanism. Both enzymes also remove thymine, but not uracil, mismatched to guanine. DME and ROS1 show a preference for 5-meC over thymine in the symmetric dinucleotide CpG context, where most plant DNA methylation occurs. Nevertheless, they also have significant activity on both substrates at CpApG and asymmetric sequences, which are additional methylation targets in plant genomes. These findings suggest that a function of ROS1 and DME is to initiate erasure of 5-meC through a base excision repair process and provide strong biochemical evidence for the existence of an active DNA demethylation pathway in plants.
Keywords: Arabidopsis, demethylation, gene silencing, methylation
Methylation of cytosine at carbon 5 of the pyrimidine ring [5-methylcytosine (5-meC)] is an epigenetic modification that guides formation of transcriptionally silent chromatin and allows transmission of specific patterns of gene activity across cellular divisions (1, 2). In eukaryotes, DNA methylation is detected in protists, fungi, plants, and animals (3) and plays important roles in the establishment of developmental programs (4, 5) and in genome defense against parasitic mobile elements (6). Most of mammalian and plant DNA methylation is restricted to symmetrical CpG sequences, but plants also have significant levels of cytosine methylation in the symmetric context CpNpG (where N is any nucleotide) and even in asymmetric contexts (2, 7). Similarly to other biochemical modifications such as protein phosphorylation and acetylation, DNA methylation is also reversible. Demethylation may take place as a passive process because of lack of maintenance methylation during several cycles of DNA replication or as an active mechanism in the absence of replication (8). In mammalian preimplantation embryos the maternal genome is demethylated by a passive process along cleavage stages, whereas the paternal genome is demethylated by an active mechanism immediately after fertilization (9). In addition to global demethylation, site-specific local demethylation also occurs throughout development and tissue differentiation (10).
In contrast to the well studied genetics, biochemistry and biology of cytosine DNA methyltransferases, the enzymatic basis of active demethylation has remained elusive. Direct removal of the methyl group from 5-meC residues has been claimed (11), but this mechanism, which involves a thermodynamically unfavorable breakage of the carbon–carbon bond, has been questioned (12) and could not be independently reproduced (13, 14). Another proposed mechanism is disruption of the labile N-glycosidic bond between the 5-meC base and the deoxyribose moiety in DNA, followed by replacement with an unmodified cytosine. A 5-meC DNA glycosylase activity was first identified in chicken embryos (15) and found to copurify with a protein homologous to human thymine DNA glycosylase (TDG) (16, 17). It was latter reported that methyl-CpG binding protein (MBD) 4, another human DNA glycosylase with no sequence similarity to TDG, also has 5-meC DNA glycosylase activity (18). Both TDG and MBD4 are monofunctional DNA glycosylases that show a preference for U·G and T·G mismatches within a CpG context (19, 20). However, they have been shown to have a very weak activity on 5-meC·G pairs compared with their activities toward U·G and T·G mismatches (18, 20, 21), and hence their roles in DNA demethylation remain unclear. It has been also suggested that demethylation might be achieved indirectly through deamination of 5-meC by activation-induced cytidine deaminase and repair of the ensuing T·G mismatch (22).
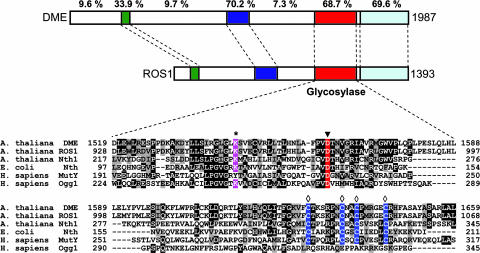
Recent work in plants has added genetic evidence for a role of DNA glycosylases in an active demethylation pathway. DEMETER (DME) was identified in a search for mutations causing parent-of-origin effects on seed viability (23). DME is expressed primarily in the central cell of the female gametophyte, where it is required for the expression of the maternal alleles of the imprinted genes MEA and FWA (23, 24). At least in the case of MEA imprinting, mutations in the methyltransferase gene MET1 suppress the requirement for DME (25). ROS1 was identified in a screen for mutants with deregulated expression of the repetitive RD29A-LUC transgene (26). Whereas in wild plants the transgene and the homologous endogenous gene are expressed, ros1 mutants display transcriptional silencing and hypermethylation of both loci (26). ROS1 and DME are predicted to encode large proteins containing a DNA glycosylase domain with significant sequence similarity to base excision DNA repair proteins in the HhH-GPD superfamily (27) (Fig. 1). The presence of a conserved invariant lysine in the HhH motif suggests that DME and ROS1 belong to the subset of DNA glycosylases/lyases, able both to hydrolyze the N-glycosidic bond linking bases to DNA and to cleave the phosphodiester backbone at the site where a base has been removed. A recombinant truncated form of ROS1 was found to have strand-breaking activity of methylated plasmid DNA (26), and ectopic DME expression in plants results in nicks in the MEA promoter (23). Furthermore, mutation of an invariant aspartic acid in the HhH-GPD motif of DME suppresses its capacity to activate MEA gene transcription in the central cell (28).
Fig. 1.
DME and ROS1 contain a DNA glycosylase domain and are closely related. (Upper) Diagram of DME and ROS1 showing the conserved domains as colored sections. The percentage of identical residues between both proteins is shown above each region. (Lower) Amino acid sequence alignment of DME (amino acids 1519–1659) and ROS1 (amino acids 928-1068) with A. thaliana Nth1, E. coli Nth, and Homo sapiens MutY and Ogg1. GenBank accession nos. are ABC61677, AAP37178, CAC16135, P20625, Q9UIF7, and O15527, respectively. An asterisk marks the lysine residue that is diagnostic of a glycosylase/lyase activity, the filled triangle indicates the conserved aspartic acid residue in the active site, and the open diamonds label the cysteine residues that in E. coli Nth1 ligate a [4Fe–4S] cluster.
The effects of ROS1 and DME in counteracting DNA methylation and gene silencing may be explained by distinct mechanisms. One possibility is that they exert an indirect effect because of interactions with transcription factors and/or chromatin-modifying proteins, perhaps promoted by DNA incisions that facilitate local chromatin remodeling. A second possibility is that ROS1 and DME directly excise 5-meC from DNA, initiating replacement by an unmethylated cytosine and leading to hypomethylation and gene activation. Our biochemical characterization of the enzymatic activity of ROS1 and DME supports the latter mechanism.
Results
DME and ROS1 Excise 5-meC and Mismatched Thymine from DNA.
We cloned full-length cDNAs corresponding to Arabidopsis DME and ROS1 genes. The protein sequence encoded by the full-length DME cDNA is identical to that reported by Choi et al. (23) but includes 258 additional amino acids at the amino terminus (Fig. 1). Comparison of the DME cDNA sequence reported here (GenBank accession no. DQ335243) and the sequence previously described (23) reveals that the latter comprises two additional segments with consensus donor and acceptor splice sites that probably represent introns. Hence, both cDNA sequences may correspond to alternative splicing forms of the DME transcript.
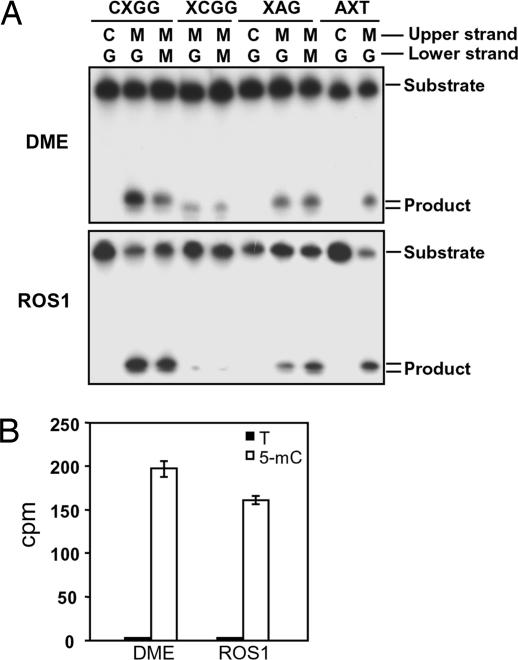
We expressed recombinant full-length DME and ROS1, and the purified proteins were used in oligonucleotide incision assays on DNA substrates containing 5-meC in diverse sequence contexts. Both DME and ROS1 incised a radioactively labeled oligonucleotide duplex containing a single 5-meC at an internal position in a CpCpGpGp sequence (Fig. 2A). Methylated cytosines located at the external position were also processed, but with a reduced efficiency. Incision was also observed in the symmetrical context CpApG and in an asymmetric context. In all symmetric contexts the incision activity on hemimethylated and bimethylated DNA was not significantly different (Fig. 2A and data not shown). Hydrolytic deamination of cytosine produces uracil mismatched to guanine, whereas deamination of 5-meC generates mismatched thymine (29). To determine whether DME and ROS1 excise 5-meC as a free base or whether it is transformed in thymine before removal, we analyzed the excision products by two-dimensional TLC (see Materials and Methods). Incubation of DME or ROS1 with a DNA substrate containing 3H-labeled 5-meC resulted in release of ethanol-soluble radioactive material that comigrated with a nonradioactive 5-meC standard in both dimensions, whereas no radioactivity was detected comigrating with thymine (Fig. 2B). From these experiments we conclude that both DME and ROS1 are 5-meC DNA glycosylases.
Fig. 2.
DME and ROS1 are 5-meC DNA glycosylases. (A) Double-stranded oligonucleotide substrates containing 5-meC (M) in CpG (CXGG) and non-CpG (XCGG, XAG, and AXT) sequence contexts or the corresponding unmethylated controls were incubated with purified DME (Upper) or ROS1 (Lower) as described in Materials and Methods. Reaction products were separated in a 12% denaturing polyacrylamide gel and visualized by autoradiography. (B) Release of 5-meC from methylated DNA. Plasmid DNA methylated with S-adenosyl-l-[methyl-3H]methionine (1,200 cpm) was incubated with purified DME or ROS1, and the released ethanol-soluble material was analyzed by two-dimensional TLC (see Materials and Methods). Distribution of radioactivity between thymine (filled bars) and 5-meC (open bars) was assayed by scintillation counting. The mean of duplicate experiments and their standard errors are shown.
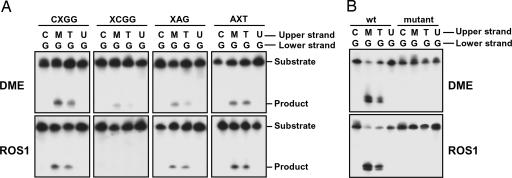
In addition to 5-meC paired to guanine, DME and ROS1 also removed thymine from a T·G mismatch located in either CpG or non-CpG sequence contexts (Fig. 3A). However, no activity was detected on uracil either mismatched to guanine (Fig. 3A) or paired to adenine (data not shown). To verify that the observed DNA incision activity is intrinsic to DME and ROS1 we generated mutant proteins in which a conserved aspartic acid residue in the glycosylase domain (Fig. 1) was changed to alanine. Preparations of both mutant proteins, purified by the same procedure as the WT proteins described above, lacked any detectable incision activity (Fig. 3B).
Fig. 3.
DME and ROS1 excise 5-meC and thymine but not uracil. (A) Double-stranded oligonucleotide substrates containing 5-mec (M), T·G, or U·G mismatches in CpG (CXGG) and non-CpG (XCGG, XAG, and AXT) sequence contexts were incubated with purified DME (Upper) or ROS1 (Lower) as described in Materials and Methods. Reaction products were separated in a 12% denaturing polyacrylamide gel and visualized by autoradiography. (B) Double-stranded oligonucleotide substrates containing 5-meC (M), T·G, or U·G mismatches in a CpG sequence context were incubated for 24 h with purified WT DME or mutant DME D1562A (Upper) or WT ROS1 or mutant ROS1 D971A (Lower). Reaction products were separated in a 12% denaturing polyacrylamide gel and visualized by autoradiography.
DME and ROS1 Have a Preference for 5-meC in a CpG Sequence Context.
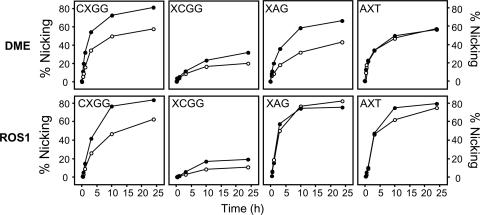
We wanted to determine the relative activities of DME and ROS1 toward substrates containing a 5-meC·G pair or a T·G mispair positioned in different sequence contexts. Time course experiments revealed that 5-meC and T·G mismatches were efficiently processed by both proteins when located in a CpG sequence context but were removed at a notably slower rate when present at the external position of the sequence CpCpGpG (Fig. 4). The incision activity of DME and ROS1 was also significant in the symmetrical sequence CpApG and in an asymmetric context (Fig. 4). For both proteins, base preference depended on the sequence context. Thus, DME processed 5-meC at a higher rate than thymine in all symmetrical sequence contexts, whereas in the asymmetrical context both bases were removed with similar efficiency (Fig. 4). ROS1 showed a very similar behavior, with the difference that excised 5-meC and thymine from a CpApG context at comparable rates. We conclude that both DME and ROS1 show a preference for 5-meC over thymine in a CpG context and have significant activity on both base substrates at CpApG and asymmetric sequences, which are additional but less frequent sites of cytosine methylation in plants.
Fig. 4.
Kinetics of DME and ROS1 action on 5-meC and thymine in CpG and non-CpG sequence contexts. The time-dependent generation of incised oligonucleotides was measured by incubating purified DME or ROS1 with double-stranded oligonucleotide substrates containing a 5-meC·G pair (•) or a T·G mismatch (○) in CpG (CXGG) and non-CpG (XCGG, XAG, and AXT) sequence contexts. Reactions were stopped at the indicated times, products were separated in a 12% denaturing polyacrylamide gel, and the relative amount of incised oligonucleotide was quantitated by phosphor imaging.
DME and ROS1 Process 5-meC Through a DNA Glycosylase/Lyase Mechanism.
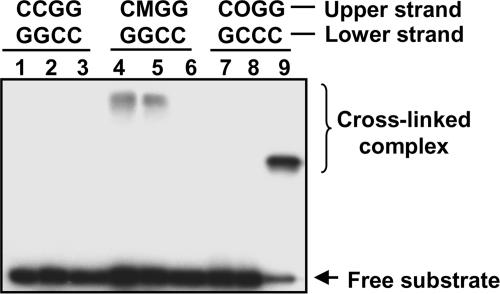
Repair reactions catalyzed by DNA glycosylases/lyases proceed through a transient imine intermediate (Schiff base) that can be reduced by borohydride, leading to the formation of an irreversibly crosslinked complex between enzyme and substrate. This borohydride-dependent “trapping assay” is a convincing test for catalytic processing of a substrate by a DNA glycosylase/lyase (30). To determine whether the excision of 5-meC catalyzed by DME and ROS proceeds through a DNA glycosylase/lyase mechanism, reactions with a methylated DNA substrate were performed in the presence of NaBH4 and products were analyzed by SDS/PAGE (Fig. 5). A DNA duplex with a single 7,8-dihydro-8-oxoguanine (8-OG) paired to cytosine located at the same position as the 5-meC·G pair was used as a control in reactions with Arabidopsis thaliana 8-OG-DNA glycosylase (AtOGG1) (31). The upper shifted bands indicate formation of the crosslinked complex between the labeled oligonucleotide containing 5-meC and DME (Fig. 5, lane 4) or ROS1 (Fig. 5, lane 5). As expected, the complex for AtOGG1 (molecular mass = 44 kDa) migrated faster than those for DME (227 kDa) or ROS1 (200 kDa). No shifted bands were observed when DME or ROS1 was incubated in the presence of nonmethylated DNA or the duplex containing 8-OG, indicating that the Schiff base formation was specific to the DNA substrate containing 5-meC. These results indicate that DME and ROS1 process 5-meC through a DNA glycosylase/lyase mechanism.
Fig. 5.
DME and ROS1 process 5-meC by a DNA glycosylase/lyase mechanism proceeding through a transient imine intermediate. Double-stranded oligonucleotide substrates containing a C·G, 5-meC·G, or 8-oxoG·C pair in a CpG context were incubated with purified DME (lanes 1, 4, and 7), ROS1 (lanes 2, 5, and 8), or AtOGG1 (lanes 3, 6, and 9) in the presence of NaBH4. After incubation, the reaction mixture was subjected to SDS/PAGE, and products were visualized by autoradiography. Free substrates and crosslinked enzyme–substrate complexes are indicated by arrows. M, 5-meC; O, 8-oxoG.
Analysis of DNA Termini Generated upon Excision of 5-meC.
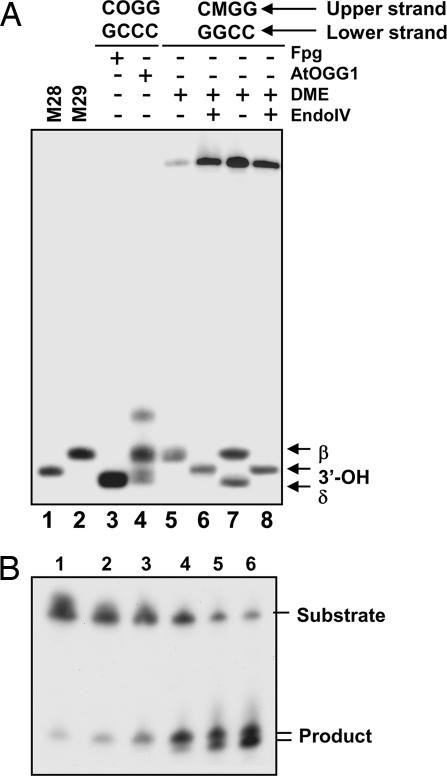
To identify the products of the 5-meC excision reaction, we compared the activity of DME on a single 5-meC·G pair in a CpG context to the activities of the 8-OG DNA glycosylases Fpg (32) and AtOGG1 on an 8-OG·C pair placed at the same position. As expected, the incubation of the 8-OG substrate with Fpg exclusively resulted in a fragment (Fig. 6A, lane 3) migrating slightly faster than the 28-mer marker. This fragment is the product of successive β- and δ-elimination reactions at the abasic site resulting from the excision of 8-OG, leaving a single nucleoside gap limited by 3′- and 5′-phosphate ends in DNA. Incubation with AtOGG1 generated a main product with a mobility similar to that of the 29-mer marker (Fig. 6A, lane 4), the result of a β-elimination that leaves an α,β-unsaturated aldehyde at the 3′ terminus of the nucleoside gap. The additional minor bands are presumably due to isomerization of the 3′-hydroxypentenal terminus (33). Incubation of the 5-meC substrate with DME during 60 min generated a fragment comigrating with the AtOGG1 β-elimination product (Fig. 6A, lane 5). However, we found that longer incubation times generated an additional fragment (Fig. 6B) comigrating with the Fpg β,δ-elimination product (Fig. 6A, lane 7). Upon addition of Escherichia coli endonuclease IV, which has a 3′-phosphodiesterase activity, both products were converted to 3′-OH fragments comigrating with the 29-mer marker. Similar results were obtained with ROS1 (data not shown). Therefore, the products generated by DME and ROS1 upon excision of 5-meC are likely to be a mixture of β- and β,δ-elimination products.
Fig. 6.
Products formed by DME upon excision of 5-meC. (A) Double-stranded oligonucleotide substrates containing a 5-meC·G (lanes 5–8) or an 8-oxoG·C pair (lanes 3 and 4) in a CpG context were incubated with purified Fpg (lane 3), AtOGG1 (lane 4), or DME (lanes 5–8), and reaction mixtures were separated in a denaturing polyacrylamide sequencing gel (40 × 20 cm). The products formed by DME were further treated with endonuclease IV (lanes 6 and 8) to analyze the nature of 3′ termini. Substrates and enzymes used are indicated at the top of the gel. Oligonucleotide markers of 28 and 29 nucleotides were loaded in lanes 1 and 2, respectively. The β- and δ-elimination products and those carrying 3′-OH termini are indicated by arrows. (B) A double-stranded oligonucleotide substrate containing a 5-meC·G pair in a CpG sequence context was incubated with purified DME, and reactions were stopped at different times. Lanes 1–6 correspond to reaction times of 0.25, 0.5, 1, 2, 10, and 24 h, respectively. Products were separated in a 12% denaturing polyacrylamide gel and visualized by autoradiography.
Discussion
There is accumulating evidence of multiple connections between DNA glycosylases and the processes that regulate gene expression. A primary mechanism involves interactions with proteins that regulate transcription. It has been reported that methyl purine DNA glycosylase associates with the transcriptional repressor MBD1 to cooperatively inhibit gene expression (34), and 3-methyladenine DNA glycosylase interacts with estrogen receptor α to modulate estrogen-mediated transcription (35). There is also evidence that TDG modulates transcription through interactions with hormone receptors (36, 37), transcription factors (38), and coactivators such as the CPB/p300 acetylase complex (39). MBD4 has the ability to repress transcription through an interaction with Sin3A and HDAC1 (40).
But, in addition to protein–protein interactions, DNA glycosylases may also modulate gene expression by direct modification of the methylation status of DNA. A role for DNA glycosylases in genome demethylation during cell differentiation in vertebrates has been previously suggested (15), and an active demethylation pathway initiated by TDG and/or MBD4 DNA glycosylases has been proposed in animal cells (16, 18, 41, 42). However, it has been argued that the main in vivo role for both proteins is to counteract the mutagenic potential of 5-meC and C deamination in CpG sequences (20, 43), given their high efficiency on U·G and T·G mismatches (17, 19, 20), compared with their weak activity on 5-meC·G base pairs (18, 21). DME and ROS1 are structurally unrelated to TDG, which belongs to a large family of uracil DNA glycosylases different from the HhH-GPD family (44), but share with MBD4 an HhH-GPD DNA glycosylase domain located at the C-terminal half of the protein. However, unlike MBD4, neither DME nor ROS1 has a methyl-CpG binding domain (20). In contrast to the strong substrate specificity of TDG and MBD4 for T·G and U·G mismatches, DME and ROS1 show a preference for 5-meC over a T·G mismatch in a CpG sequence context, the most frequent DNA methylation target in plant and animal genomes, and they do not display detectable activity on U·G mispairs. Thus, the biochemical properties of DME and ROS1, together with the available genetic evidence, suggest that an important role for both enzymes in vivo is excision of 5-meC. We cannot rule out the possibility that DME and ROS1 also play a role in neutralizing the mutagenic deamination of 5-meC to thymine through their activity on T·G mismatches. The effect of dme and ros1 mutations on mutagenesis in vivo has not been assessed, but it could be compounded by their epigenetic effects on plant development.
We have found that DME and ROS1 cleave the phosphodiester backbone at the 5-meC removal site by successive β,δ-elimination, leaving an abasic site that has to be further processed to generate a 3′-OH terminus suitable for polymerization and ligation. It has been questioned whether a base excision repair mechanism would explain the active global demethylation observed in early mammalian embryogenesis, because that would require a base excision/substitution at a global scale (45). Unlike vertebrates, in plants there is no clear evidence for a global demethylation during the life cycle (7). We propose that the base excision process initiated by DME and ROS1 may be rather involved in local demethylation of selected loci during development. In this scenario, DME would be required to specifically initiate erasure of 5-meC at MEA and FWA, and perhaps other unidentified loci, in female gametes before fertilization. ROS1 is needed to prevent transcriptional gene silencing and hypermethylation of a repetitive transgene, but the observation of developmental abnormalities in ros1 mutants after inbreeding (26) suggests that it also regulates expression of endogenous loci. Thus, the available evidence suggests that the two enzymes may have specialized roles in counteracting methylation and gene silencing at specific loci.
This line of reasoning leads to the important question of how the 5-meC DNA glycosylase activity of DME and ROS1 is directed to specific target genes. Both DME and ROS1 are unusually large proteins, much bigger than typical DNA glycosylases (23, 26). In addition to the DNA glycosylase domain, DME and ROS1 show a high sequence similarity at the carboxyl-terminal region (Fig. 1). However, they are mostly unrelated in the rest of the sequence, where the only recognizable feature is a region rich in basic residues at the amino-terminal region that displays a weak similarity to H1 histones. It is possible that the divergent domains of DME and ROS1 play a role in targeting the two proteins to different genome loci, either directly or through interactions with associated cofactors, such as chromatin components. The biochemical demonstration that DME and ROS1 can initiate an active demethylation process through 5-meC excision will help to unravel the intricacies of this mechanism of epigenetic regulation.
Materials and Methods
Cloning of DME and ROS1 Full-Length cDNAs.
Truncated clones lacking the 5′ portions of DME and ROS1 cDNAs were isolated from an Arabidopsis cDNA library (46) (Arabidopsis Biological Resource Center, Ohio State University, Columbus). The 5′ ends of both cDNAs were obtained by RT-PCR on total RNA isolated from Arabidopsis plants (ecotype Columbia) by using primers designed according to the sequence of a 3′-truncated DME cDNA clone and to ROS1 genome sequence information. After sequencing, a PCR error at position 1330 of DME cDNA was corrected by site-directed mutagenesis (QuikChange Site-Directed Mutagenesis Kit; Stratagene). Verified fragments were connected by digestion and ligation, and a full-length cDNA was assembled for each gene.
Protein Expression and Purification.
The full-length DME cDNA was inserted into the pET30b expression vector (Novagen) to add a polyhistidine (His-6) tag at the N terminus of DME protein. Expression in E. coli BL21(DE3) dcm− Codon Plus cells (Stratagene) was induced by the addition of isopropyl-1-thio-β-d-galactopyranoside, and the His-6-tagged DME protein was purified by affinity chromatography on a Ni2+-NTA column (Amersham Pharmacia Biotech) by using standard protocols. The full-length ROS1 cDNA was cloned into the pMAL-c2X expression vector (New England Biolabs) to obtain a malE-ROS1 in-frame fusion. Expression was induced in E. coli strain BL21(DE3) dcm− Codon Plus cells as indicated above, and the MBP-ROS1 fusion protein was purified by amylose affinity chromatography by using standard protocols.
Site-Directed Mutagenesis.
Site-directed mutagenesis was performed by using the QuikChange XL site-directed mutagenesis kit (Stratagene). The D1562A mutation was introduced into pET-DME by using the oligonucleotides DMED1562A forward (5′-CACAATCTTGCTTTCCCTGTTGCCACGAATGTTGGAAGG-3′) and DMED1562A reverse (5′-CCTTCCAACATTCGTGGCAACAGGGAAAGCAAGATTGTG-3′). The D971A mutation was introduced into pMAL-ROS1 by using the primers ROSD971A forward (5′-CACCATCTTGCCTTTCCAGTTGCTACAAATGTTGGGCGC-3′) and ROSD971A reverse (5′-GCGCCCAACATTTGTAGCAACTGGAAAGGCAAGATGGTG-3′). The mutant sequences were confirmed by DNA sequencing, and the constructs were used to transform E. coli strain BL21(DE3) dcm− CodonPlus. Mutant proteins were overexpressed and purified as described above.
DNA Substrates.
Oligonucleotides used (see Table 1, which is published as supporting information on the PNAS web site) were synthesized by operon and purified by PAGE. Upper-strand oligonucleotides were labeled at the 5′ end by using T4 polynucleotide kinase (Roche) and [γ-32P]ATP (Amersham Pharmacia Biotech). Double-stranded substrates were prepared by mixing a 0.05 μM solution of the labeled upper strand with a 0.1 μM solution of the complementary lower strand, heating to 95°C for 5 min, and slowly cooling to room temperature.
Enzymatic Activity Assay.
Double-stranded oligodeoxynucleotides (50 fmol) were incubated at 30°C for 1 h (unless indicated otherwise) in a reaction mixture containing 50 mM Tris·HCl (pH 8.0), 1 mM EDTA, 1 mM DTT, 0.1 mg/ml BSA, and 25 pmol of protein in a total volume of 100 μl. Reactions were stopped by adding EDTA to 20 mM, SDS to 0.6%, and proteinase K to 200 μg/ml, and the mixtures were incubated for 30 min at 37°C. DNA was extracted with phenol:chloroform:isoamyl alcohol (25:24:1) and ethanol-precipitated at −20°C in the presence of 0.3 mM NaCl and 16 μg/ml glycogen. When required, the DNA was resuspended in 10 μl of EF buffer (10 mM Tris·HCl, pH 7.5/1 mM EDTA/50 mM NaCl), incubated with 10 units of endonuclease IV (New England Biolabs) at 37°C for 30 min, and finally extracted and precipitated again as above. Samples were resuspended in 10 μl of formamide dye mix (80% formamide, 1 mg/ml bromophenol blue, and 10 mM EDTA), and heated at 95°C for 5 min. Reaction products were separated in a 12% denaturing polyacrylamide gel containing 7 M urea, visualized by autoradiography, and quantitated by using a phosphor imager (Bio-Rad).
TLC Analysis.
Plasmid pBluescript DNA (2 μg; Stratagene) was methylated with SssI methylase (New England Biolabs) in the presence of 5 μCi (1 Ci = 37 GBq) of 15 Ci/mmol S-adenosyl-l-[methyl-3H]methionine (Amersham Pharmacia Biosciences), precipitated in ethanol, and dissolved in 10 mM Tris·HCl (pH 8.0) and 1 mM EDTA. Purified DME or ROS1 (100 pmol) were incubated at 30°C for 1 h with methylated plasmid (1,200 cpm) in a reaction mixture containing 50 mM Tris·HCl (pH 8.0), 1 mM EDTA, 1 mM DTT, and 0.1 mg/ml BSA in a total volume of 100 μl. After precipitating in the presence of glycogen, the ethanol-soluble material was concentrated by centrifugation under vacuum and dissolved in 10 μl of a saturated equimolar solution of 5-meC and thymine in methanol. The mixture was spotted on a silica gel TLC plate (Silicagel 60 F254, 20 × 20 cm; Merck). The first dimension was run in chloroform/methanol/water (4:2:1, vol/vol/vol). The plate was dried in air and developed in the second dimension with ethylacetate:propanol:water (75:16:9, vol/vol/vol). Spots corresponding to 5-meC and thymine were visualized under UV light, scraped off, and counted in 5 ml of scintillation fluid.
Crosslink Reaction with NaBH4.
Crosslink reactions between enzymes and substrates were performed by incubating 25 pmol of protein with double-stranded oligodeoxynucleotides (50 fmol) at 30°C for 1 h in a reaction mixture containing 50 mM Tris·HCl (pH 8.0), 1 mM EDTA, 1 mM DTT, 0.1 mg/ml BSA, and 50 mM NaBH4 in a total volume of 10 μl. After incubation, the samples were mixed with SDS/PAGE loading buffer, heated at 100°C for 5 min, and electrophoresed on a 12% SDS polyacrylamide gel. The reaction products were visualized by autoradiography.
Supplementary Material
Acknowledgments
We thank Emilio Fernández for generous support with radiation safety facilities. M.I.P.-M. is the recipient of a Ph.D. Fellowship from Junta de Andalucía. This research was supported by Ministerio de Educación y Ciencia Grant BFU2004-05303 (to T.R.-A.).
Abbreviations
- 5-meC
5-methylcytosine
- 8-OG
7,8-dihydro-8-oxoguanine
- TDG
thymine DNA glycosylase
- MBD
methyl-CpG binding protein
- AtOGG1
Arabidopsis thaliana 8-OG-DNA glycosylase.
Note.
While this work was under review, an article based on independent research reporting similar results for DME was published (47).
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper for DME cDNA has been deposited in the GenBank database (accession no. DQ335243).
References
- 1.Bender J. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2004;55:41–68. doi: 10.1146/annurev.arplant.55.031903.141641. [DOI] [PubMed] [Google Scholar]
- 2.Bird A. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 3.Colot V., Rossignol J. L. BioEssays. 1999;21:402–411. doi: 10.1002/(SICI)1521-1878(199905)21:5<402::AID-BIES7>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 4.Holliday R., Pugh J. E. Science. 1975;187:226–232. [PubMed] [Google Scholar]
- 5.Riggs A. D. Cytogenet. Cell Genet. 1975;14:9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- 6.Yoder J. A., Walsh C. P., Bestor T. H. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 7.Finnegan E. J., Genger R. K., Peacock W. J., Dennis E. S. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998;49:223–247. doi: 10.1146/annurev.arplant.49.1.223. [DOI] [PubMed] [Google Scholar]
- 8.Kress C., Thomassin H., Grange T. FEBS Lett. 2001;494:135–140. doi: 10.1016/s0014-5793(01)02328-6. [DOI] [PubMed] [Google Scholar]
- 9.Reik W., Dean W., Walter J. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 10.Frank D., Keshet I., Shani M., Levine A., Razin A., Cedar H. Nature. 1991;351:239–241. doi: 10.1038/351239a0. [DOI] [PubMed] [Google Scholar]
- 11.Bhattacharya S. K., Ramchandani S., Cervoni N., Szyf M. Nature. 1999;397:579–583. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- 12.Smith S. S. J. Mol. Biol. 2000;302:1–7. doi: 10.1006/jmbi.2000.4046. [DOI] [PubMed] [Google Scholar]
- 13.Ng H. H., Zhang Y., Hendrich B., Johnson C. A., Turner B. M., Erdjument-Bromage H., Tempst P., Reinberg D., Bird A. Nat. Genet. 1999;23:58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- 14.Wade P. A., Gegonne A., Jones P. L., Ballestar E., Aubry F., Wolffe A. P. Nat. Genet. 1999;23:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- 15.Jost J. P., Siegmann M., Sun L., Leung R. J. Biol. Chem. 1995;270:9734–9739. doi: 10.1074/jbc.270.17.9734. [DOI] [PubMed] [Google Scholar]
- 16.Zhu B., Zheng Y., Hess D., Angliker H., Schwarz S., Siegmann M., Thiry S., Jost J. P. Proc. Natl. Acad. Sci. USA. 2000;97:5135–5139. doi: 10.1073/pnas.100107597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neddermann P., Gallinari P., Lettieri T., Schmid D., Truong O., Hsuan J. J., Wiebauer K., Jiricny J. J. Biol. Chem. 1996;271:12767–12774. doi: 10.1074/jbc.271.22.12767. [DOI] [PubMed] [Google Scholar]
- 18.Zhu B., Zheng Y., Angliker H., Schwarz S., Thiry S., Siegmann M., Jost J. P. Nucleic Acids Res. 2000;28:4157–4165. doi: 10.1093/nar/28.21.4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sibghat U., Gallinari P., Xu Y. Z., Goodman M. F., Bloom L. B., Jiricny J., Day R. S., III Biochemistry. 1996;35:12926–12932. doi: 10.1021/bi961022u. [DOI] [PubMed] [Google Scholar]
- 20.Hendrich B., Hardeland U., Ng H. H., Jiricny J., Bird A. Nature. 1999;401:301–304. doi: 10.1038/45843. [DOI] [PubMed] [Google Scholar]
- 21.Hardeland U., Bentele M., Jiricny J., Schar P. Nucleic Acids Res. 2003;31:2261–2271. doi: 10.1093/nar/gkg344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morgan H. D., Dean W., Coker H. A., Reik W., Petersen-Mahrt S. K. J. Biol. Chem. 2004;279:52353–52360. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- 23.Choi Y., Gehring M., Johnson L., Hannon M., Harada J. J., Goldberg R. B., Jacobsen S. E., Fischer R. L. Cell. 2002;110:33–42. doi: 10.1016/s0092-8674(02)00807-3. [DOI] [PubMed] [Google Scholar]
- 24.Kinoshita T., Miura A., Choi Y., Kinoshita Y., Cao X., Jacobsen S. E., Fischer R. L., Kakutani T. Science. 2004;303:521–523. doi: 10.1126/science.1089835. [DOI] [PubMed] [Google Scholar]
- 25.Xiao W., Gehring M., Choi Y., Margossian L., Pu H., Harada J. J., Goldberg R. B., Pennell R. I., Fischer R. L. Dev. Cell. 2003;5:891–901. doi: 10.1016/s1534-5807(03)00361-7. [DOI] [PubMed] [Google Scholar]
- 26.Gong Z., Morales-Ruiz T., Ariza R. R., Roldan-Arjona T., David L., Zhu J. K. Cell. 2002;111:803–814. doi: 10.1016/s0092-8674(02)01133-9. [DOI] [PubMed] [Google Scholar]
- 27.Nash H. M., Bruner S. D., Scharer O. D., Kawate T., Addona T. A., Spooner E., Lane W. S., Verdine G. L. Curr. Biol. 1996;6:968–980. doi: 10.1016/s0960-9822(02)00641-3. [DOI] [PubMed] [Google Scholar]
- 28.Choi Y., Harada J. J., Goldberg R. B., Fischer R. L. Proc. Natl. Acad. Sci. USA. 2004;101:7481–7486. doi: 10.1073/pnas.0402328101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindahl T. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 30.McCullough A. K., Dodson M. L., Lloyd R. S. Annu. Rev. Biochem. 1999;68:255–285. doi: 10.1146/annurev.biochem.68.1.255. [DOI] [PubMed] [Google Scholar]
- 31.Garcia-Ortiz M. V., Ariza R. R., Roldan-Arjona T. Plant Mol. Biol. 2001;47:795–804. doi: 10.1023/a:1013644026132. [DOI] [PubMed] [Google Scholar]
- 32.Boiteux S., O’Connor T. R., Laval J. EMBO J. 1987;6:3177–3183. doi: 10.1002/j.1460-2075.1987.tb02629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bailly V., Verly W. G. Biochem. J. 1987;242:565–572. doi: 10.1042/bj2420565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watanabe S., Ichimura T., Fujita N., Tsuruzoe S., Ohki I., Shirakawa M., Kawasuji M., Nakao M. Proc. Natl. Acad. Sci. USA. 2003;100:12859–12864. doi: 10.1073/pnas.2131819100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Likhite V. S., Cass E. I., Anderson S. D., Yates J. R., Nardulli A. M. J. Biol. Chem. 2004;279:16875–16882. doi: 10.1074/jbc.M313155200. [DOI] [PubMed] [Google Scholar]
- 36.Um S., Harbers M., Benecke A., Pierrat B., Losson R., Chambon P. J. Biol. Chem. 1998;273:20728–20736. doi: 10.1074/jbc.273.33.20728. [DOI] [PubMed] [Google Scholar]
- 37.Chen D., Lucey M. J., Phoenix F., Lopez-Garcia J., Hart S. M., Losson R., Buluwela L., Coombes R. C., Chambon P., Schar P., Ali S. J. Biol. Chem. 2003;278:38586–38592. doi: 10.1074/jbc.M304286200. [DOI] [PubMed] [Google Scholar]
- 38.Missero C., Pirro M. T., Simeone S., Pischetola M., Di Lauro R. J. Biol. Chem. 2001;276:33569–33575. doi: 10.1074/jbc.M104963200. [DOI] [PubMed] [Google Scholar]
- 39.Tini M., Benecke A., Um S. J., Torchia J., Evans R. M., Chambon P. Mol. Cell. 2002;9:265–277. doi: 10.1016/s1097-2765(02)00453-7. [DOI] [PubMed] [Google Scholar]
- 40.Kondo E., Gu Z., Horii A., Fukushige S. Mol. Cell. Biol. 2005;25:4388–4396. doi: 10.1128/MCB.25.11.4388-4396.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jost J. P., Oakeley E. J., Zhu B., Benjamin D., Thiry S., Siegmann M., Jost Y. C. Nucleic Acids Res. 2001;29:4452–4461. doi: 10.1093/nar/29.21.4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu B., Benjamin D., Zheng Y., Angliker H., Thiry S., Siegmann M., Jost J. P. Proc. Natl. Acad. Sci. USA. 2001;98:5031–5036. doi: 10.1073/pnas.091097298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barnes D. E., Lindahl T. Annu. Rev. Genet. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- 44.Aravind L., Koonin E. V. Genome Biol. 2000;1:research0007.1–research0007.8. doi: 10.1186/gb-2000-1-4-research0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Szyf M. Biochemistry. 2005;70:533–549. doi: 10.1007/s10541-005-0147-7. [DOI] [PubMed] [Google Scholar]
- 46.Kieber J. J., Rothenberg M., Roman G., Feldmann K. A., Ecker J. R. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- 47.Gehring M., Huh J. H., Hsieh T. F., Penterman J., Choi Y., Harada J. J., Goldberg R. B., Fischer R. L. Cell. 2006;124:495–506. doi: 10.1016/j.cell.2005.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.