Abstract
Fascioscapulohumeral muscular dystrophy (FSHD) is an autosomal dominant neuromuscular disorder linked to partial deletion of integral numbers of a 3.3 kb polymorphic repeat, D4Z4, within the subtelomeric region of chromosome 4q. Although the relationship between deletions of D4Z4 and FSHD is well established, how this triggers the disease remains unclear. We have mapped the DNA loop domain containing the D4Z4 repeat cluster in human primary myoblasts and in murine–human hybrids. A nuclear matrix attachment site was found located in the vicinity of the repeat. Prominent in normal human myoblasts and nonmuscular human cells, this site is much weaker in muscle cells derived from FSHD patients, suggesting that the D4Z4 repeat array and upstream genes reside in two loops in nonmuscular cells and normal human myoblasts but in only one loop in FSHD myoblasts. We propose a model whereby the nuclear scaffold/matrix attached region regulates chromatin accessibility and expression of genes implicated in the genesis of FSHD.
Keywords: D4Z4, nuclear matrix, heterochromatin, transcription
Fascioscapulohumeral muscular dystrophy (FSHD) is an autosomal dominant disease with a prevalence of 1:20,000 (1). FSHD is characterized by weakness and atrophy of muscles of the face, upper arms, and shoulder girdle. Linkage analysis has mapped the FSHD locus to the subtelomeric region of the long arm of chromosome 4 (2).
The disorder is related to a short repeat array that remains after deletion of an integral number of tandemly arrayed 3.3-kb repeat units on chromosome 4. The size of this polymorphic locus (D4Z4) varies in normal individuals from 35 to 300 kb, whereas in FSHD patients it is consistently shorter than 35 kb (3). Partial deletion of the D4Z4 array on chromosome 4 ultimately leads to FSHD and is currently used as a diagnostic tool in genetic counseling to predict the probability of the disease (1, 3–5). A correlation exists between the extent of the deletion and its clinical expression: Indeed, patients with one to three repeats develop an early FSHD, whereas individuals with nine to 10 repeats exhibit a weaker form of the disease (5).
Extensive efforts to identify gene transcripts associated with the 4q35-specific D4Z4 repeat, as potential FSHD candidate genes, have been largely unsuccessful (6). The 3.3-kb D4Z4 elements contain a cryptic DUX4 gene potentially coding for a double homeodomain protein (7), and an overall perturbation of mRNA expression profiles can be observed in FSHD patients (8–10), but the disease appears to result from an as yet unexplained mechanism with a genetic alteration not residing within a causative gene for the disease.
The 4q35 genomic region (Fig. 1) displays heterochromatic features and might exert repressive effects on neighboring genes with a mechanism similar to position effect variegation. A decreased D4Z4 repeat number consistently results in inappropriate up-regulation of adjacent FRG2, FRG1, and Ant1 in FSHD muscle (11–13). It has also been shown that a transcriptional repressor complex binds D4Z4, whose deletion would trigger overexpression by lack of repression (11). Indeed, overexpression of FRG1 in transgenic mice provokes a phenotype similar to that of FSHD (14). However, such a model of position effect has been recently challenged in two reports of an apparent lack of up-regulation of any 4q35 gene and because of the histone H4 acetylation state in FSHD lymphoid cells (9, 10). The region is also hypomethylated in FSHD patients myoblasts as compared with normal tissues, suggesting that the chromatin status may be different (15).
Fig. 1.
Organization of the 4q35 locus.
According to a recent study of histone H4 acetylation, D4Z4 exhibits properties of unexpressed euchromatin (16). Another study has not revealed any significant differences in chromatin organization at 4q35.2 between normal and FSHD myoblasts. It is difficult to draw conclusions from these data, however, because they relate to regions 300 and 850 kb away from the D4Z4 array on chromosome 4 (17). Recently, distal regions of 4q35 have been shown to be associated with peripheral heterochromatin (18). Earlier studies also had demonstrated an association of D4Z4 with heterochromatin (19). Extended studies of DNA domains show the existence of short blocks of heterochromatin that are easy to overlook by partial analysis but that nevertheless play an important role in gene regulation (20). Therefore, it appeared important to compare the large-scale chromatin organization of the 4q35 locus between cells from FSHD patients and normal human myoblasts.
In eukaryotic nuclei and metaphase chromosomes, DNA is organized into loop domains (21). These loops are anchored to the nuclear skeleton or matrix via specific sequences, called nuclear scaffold/matrix attached regions (S/MARs) (22, 23). S/MARs, generally A/T-rich, are located within fragments ranging from 200 to 1,000 bp. Some S/MARs reside in nontranscribed regions, sometimes within introns, whereas others are function-related and found in the vicinity of enhancers, insulators, replication origins, and transcribed genes (see ref. 24 for a review). We have shown earlier that association of S/MARs with the nucleoskeleton may change during development (25).
To evaluate the possible effect of chromatin organization of D4Z4 on genes located at a distance to the D4Z4 repeat array, we have studied the large-scale organization of the 4q35 locus. We have found that it contains an upstream S/MAR and forms an independent loop in nonmuscular cells and in normal myoblasts. In contrast, in myoblasts from FSHD patients, the D4Z4 array and the upstream genes form a single loop domain, thus allowing for cis-regulation of these genes.
Results
A Strong S/MAR Is Located Upstream of the D4Z4 Repeat in the 4q35 Locus.
Long-range chromatin organization plays an important role in the organization of the genome for replication and transcription (24). Chromatin loop domains represent one of the highest levels of DNA organization within the genome. The loops are delimited by the S/MARs.
We have studied the organization of the D4Z4 array and surrounding sequences by analyzing the nuclear matrix attachment in the region. We prepared the fraction of nuclear matrix-associated DNA by DNase I treatment of the isolated nuclei followed by extraction of soluble proteins with 2 M NaCl. The resulting nuclear matrix contains the sites of DNA attachment to the nuclear matrix (for details, see ref. 26). The DNA associated with the nuclear matrix was purified, radiolabeled, and used as a probe to examine the organization of the nuclear matrix attachment sites within the D4Z4 loop domain.
A recombinant plasmid pGEM42 containing two D4Z4 repeats and 5' and 3' flanking sequences was digested with HincII and KpnI restriction endonucleases (Fig. 2A). The digested DNA was then separated on agarose gels (Fig. 2B). The gels were blotted and probed with either total human DNA (Fig. 2C) or nuclear matrix-associated DNA from HeLa cells (Fig. 2D).
Fig. 2.
Association of the 4q35 locus with the nuclear matrix in normal cells and in primary myoblasts derived from FSHD patients. (A) Map of recombinant DNA used for hybridization with nuclear matrix DNA (E, EcoRI; K, KpnI; H, HincII). (B) Electrophoretic pattern of restriction fragments. Numbers indicate the positions of the restriction fragments on the map in A. (C) Hybridization with total DNA. The blot is underexposed to reveal DNA repeats. (D–G) Hybridization with nuclear matrix-associated DNA from HeLa cells, nuclear matrix-associated DNA from GM10115A cells (E), nuclear matrix-associated DNA from normal human myoblasts (F), and nuclear matrix-associated DNA from myoblasts derived from an FSHD patient (G). Arrows indicate the positions of the human and murine c-myc S/MARs used as controls, the FRR MAR, and the pLAM.
HeLa matrix-associated DNA strongly hybridizes to two D4Z4 locus restriction fragments, corresponding to the flanking sequence and the D4Z4 array (Fig. 2D), whereas total human DNA strongly hybridizes with D4Z4 repeats and the repetitive sequence pLAM (Fig. 2C) and weakly hybridizes to the unique sequences (data not shown). Therefore, (i) the D4Z4 repeats are associated with the nuclear matrix, and (ii) they are flanked by an S/MAR on the centromeric side of the repeat array, thus physically separating the D4Z4 repeats from upstream genes. We designated the 400-bp S/MAR located in the proximal region of the D4Z4 repeat as FSHD-related region S/MAR (FRR MAR). A sequence flanking the D4Z4 array on the telomeric side, pLAM, is also weakly associated with the nuclear matrix.
The human genome contains sequences homologous to D4Z4 and pLAM on several chromosomes in addition to chromosome 4 (4, 19, 27), and FRR MAR is present in two copies in the genome: on chromosome 4 and in the homologous region on chromosome 10. It was therefore possible that the interaction detected between the nuclear matrix and the D4Z4 array in our assay could have taken place not at 4q35 but rather at one or more of these other chromosomal loci. To address this possibility, we used a human/rodent hybrid cell line, GM10115A, containing a single human chromosome 4. Indeed, the S/MARs are highly conserved between species (28, 29); therefore, one can study chromatin organization in such hybrids. The rodent genome lacks D4Z4 repeats (30); thus, the only genomic copy of D4Z4 and adjacent sequences originates from 4q35 of the human chromosome 4. Fig. 2E shows that, in the nuclear matrix preparations from GM10115A cells, D4Z4 is weakly associated with the nuclear matrix. The association of the distal S/MAR corresponding to the pLAM repeat was somewhat stronger in GM10115A cells than in HeLa cells. The attachment of the ubiquitous and non-species-specific S/MAR from the upstream region of the c-myc gene (29, 31) to the nuclear matrix was used to normalize the exposures. On the basis of these results, we conclude that the FRR MAR derived from chromosome 4 is indeed associated with the nuclear matrix and flanks the D4Z4 repeat array within the 4q35 locus.
FRR MAR Association with the Nuclear Matrix Is Strongly Diminished in Primary Myoblasts from FSHD Patients.
We have tested for potential changes in the association of the 4q35 locus to the nuclear matrix between cultured primary myoblasts from FSHD patients and from normal myoblasts. A significant decrease in the association of the FRR MAR with the nuclear matrix was observed in patients' myoblasts as compared with normal ones (Fig. 2 F and G). Interaction of pLAM with the nuclear matrix was diminished both in the normal primary human myoblasts and in myoblasts from FSHD patients, as compared with non-muscular HeLa and GM10115A cells, whereas association of the D4Z4 repeat was also much weaker than in HeLa cells. A similar pattern was observed in normal differentiated myoblasts and in differentiated myoblasts from FSHD patients (data not shown). The association of the FRR MAR was diminished ≈30 ± 6% in myoblasts from FSHD patients, suggesting that one FRR MAR of four on either chromosomes 4 or 10 is dissociated from the nuclear matrix.
Taken together, our data indicate that the loop domain organization at the 4q35 locus in myoblasts of normal subjects is different from that of FSHD patients.
Mapping the Chromatin Loop Domain at the 4q35 Locus by Using Genomic DNA Array.
Next we undertook the mapping of the chromatin loop organization in the 250-kb region between D4Z4 repeats and the upstream area of the FRG1 gene in normal myoblasts and in cells from FSHD patients by using genomic DNA arrays. We have used a technique for mapping the interactions of DNA with the nuclear matrix based on oligonucleotide DNA arrays (32). This method allows for rapid and accurate study of S/MARs over large sequenced areas of various genomes.
The array covers 250 kb of the 4q35.2 locus between the D4Z4 and the upstream area of the FRG1 gene (Fig. 1). The oligonucleotides have been numbered according to their distance (in kilobases) from the first proximal D4Z4 repeat in the array. The oligonucleotides were quantified and slot-blotted onto a nylon membrane as described in ref. 32. As a control, the chosen oligonucleotides in the array were tested for the presence of repetitive DNA by hybridization with total DNA. Total human DNA hybridizes almost equally to the 4q35 locus array (Fig. 3A). We concluded that the chosen oligonucleotides do not contain multiple-copy DNA repeats.
Fig. 3.
Mapping the nuclear matrix attachment site in the 4q35 locus by DNA array technique. (A and B) Hybridization of the total human DNA (A) and total DNA from the mouse–human hybrid cell line GM11015A (B) with a genomic DNA array covering 250 kb in the 4q35 locus. (C) The results of hybridization of nuclear matrix-associated DNA from cultured primary myoblasts from FSHD patients (white columns) and normal subjects (black columns). The hybridization data were normalized against a positive control. Hybridization to a S/MAR from the human c-myc gene locus in A and C and against murine c-myc S/MAR in B was assigned a value of 1. The average of three independent experiments (two hybridizations per experiment) is presented.
Chromosome 4 contains large regions of homology with chromosome 10 within the 4q35 locus. To double-check the array, we hybridized it with the total DNA extracted from the GM10115A hybrid cell line containing human chromosome 4. The pattern of hybridization was largely similar to that of total human DNA, thus validating the specificity of our array (Fig. 3B). Somewhat stronger hybridization of the murine–human hybrid cell line with the oligonucleotides at 163 and 151 bp can be explained by the homologies of the oligonucleotides and the murine DNA.
In contrast, the nuclear matrix-associated DNA from the normal myoblasts shows a specific hybridization pattern: The association of the 4q35 locus with the nuclear matrix becomes extremely specific, restricted to two strong sites located at 4 and 165 bp relative to the D4Z4 array (Fig. 3C). The attachment at 4 bp corresponds to the FRR MAR, in perfect agreement with our previous experiments. Consistently, a decrease was observed in the attachment of the nuclear matrices from FSHD patients at 4 bp. Interestingly, the distal border of the DNA loop was also different and located at 171 kb in FSHD patients, compared with 165 kb in normal human myoblasts.
Hybridization of the soluble DNA fraction with the array revealed a pattern similar to that obtained with total DNA (data not shown). This similarity is not surprising given that the non-matrix-associated DNA constitutes 95–98% of the genome.
We conclude that there exists a specific organization of the 4q35 locus in human myoblasts with the loop domain ends located at 171–165 and 4 kb relative to the D4Z4 array. Therefore, the D4Z4 repeats and neighboring genes (FRG2 and FRG1) are located in two distinct loop domains and are physically separated by the FRR MAR. Dissociation of the FRR MAR on the defective chromosome 4 in myoblasts from the FSHD patients from the nuclear matrix may change the overall configuration of the loop domain, bringing together the D4Z4 array and the neighboring genes. This evidence conclusively identifies a changed chromatin organization between the cells of FSHD patients and the nonaffected cells.
D4Z4 and Upstream Genes Within the 4q35 Locus Are Located in Two Distinct Loops in Non-Myoblast Cells and on “Normal” Chromosome 4 in FSHD Patients.
The DNA loops fixed at the nuclear matrix or scaffold can be visualized in histone-depleted nuclei as a halo of DNA surrounding the nuclear matrix (33). Using FISH on high-salt extracted nuclei (nuclear halos) or chromosomes, one can detect individual loops and determine whether short individual sequences are located on the nuclear matrix or in the DNA loops (32, 34–38).
The experiments on biochemical mapping of the S/MAR in the proximity of the D4Z4 repeats suggest that D4Z4 and the upstream genes are located in two distinct DNA loop domains in normal primary myoblasts, whereas they comprise a single domain in the damaged chromosome in FSHD patients.
We have used FISH to further confirm these observations. Nuclear halos were prepared by salt extraction of nuclei immobilized on glass slides. The blue halo corresponds to DNA loops originating from the nuclear matrix (Fig. 4). The nuclear matrix can also be visualized in these preparations by staining with lamins, a signal that coincides with bright DAPI staining (data not shown).
Fig. 4.
FISH on the nuclear halos from the primary myoblasts from normal subjects and FSHD patients. Shown are the results of hybridization of the FRR MAR (red) and the D4Z4 repeat (green) to the nuclear halos from human primary myoblasts from unaffected individuals (A) and those from FSHD patients (B). The positions of the FISH signals corresponding to FRR MAR are indicated by arrows. The first image in each row shows the nuclei and nuclear halos/matrices counterstained with DAPI (blue); the second image shows FISH signals from FRR MAR; the third image shows FISH signals corresponding to D4Z4; the fourth image shows the merged results of the two previous images; and the fifth image shows paths of chromatin loops (green) relative to the nuclear matrices and halos (blue).
A PCR-amplified DNA fragment corresponding to the FRR MAR was labeled and used as a probe in FISH with the nuclear halos or matrices from normal myoblasts. Hybridization was virtually restricted to the nuclear matrix (Fig. 4A; see also Fig. 7 A and B, which is published as supporting information on the PNAS web site). In 57 cells inspected, 89% of the FRR MAR probe was detected on the nuclear matrix, thus validating the results of S/MAR analysis (Table 1, which is published as supporting information on the PNAS web site).
In contrast, hybridization of the same probe with nuclear halos and matrices isolated from FSHD-derived myoblasts resulted in a different distribution of the signal (Figs. 4B and 7 C and D). In 51 nuclei inspected, only 67% of signals were located within the nuclear matrix, whereas 33% of signals were found on loop region of the halos. Similar results have been obtained by using in situ detection of the FRR MAR on the DNaseI-treated nuclear matrices: four FRR MAR signals were detected in nuclear matrices from normal cells vs. three in FSHD cells (data not shown).
The present FISH analysis shows that, in 89% of the cases, the delocalized FRR MAR is associated with a short D4Z4 array, suggesting that it is the deleted chromosome 4 that is delocalized from the nuclear matrix. The probe corresponding to D4Z4 gave signals preferentially within loop DNA in both types of myoblasts, again as one would expect on the basis of the results of biochemical analysis. In 51 cells inspected, 88% of the signals given by the probe were detected in loop DNA. The presence of 12% of the signals on the nuclear matrix area most probably reflects distortion of the three-dimensional loop halos during immobilization on the microscopic slide. Indeed, in human–murine hybrid cell line GM10115A containing a single D4Z4 array, the signals derived from the D4Z4 probe in contrast to the FRR MAR probe are present in 98% of the nuclear halos.
Together, these observations confirm the delocalization of defective chromosome 4 from the nuclear matrix, which may result in drastic changes in both chromatin structure and transcriptional regulation with a probable role in FSHD.
Visualization of a Chromatin Loop Domain in the 4q35 Locus.
We then attempted to visualize individual loops containing the D4Z4 upstream genes by using as a probe a bacterial artificial chromosome clone with an insert of 160 kb covering the 4q35 locus between positions 191108570 and 191272691 (≈90–250 kb relative to the D4Z4 array) (see Fig. 1 for details).
In these conditions, most of the observed loops were V-shaped (Fig. 5; see also Fig. 8, which is published as supporting information on the PNAS web site). This shape corresponds to two incomplete loops on each side of a S/MAR in the middle, consistent with the fact that the bacterial artificial chromosome clone used did not cover the entire loop, as mapped by the array technique.
Fig. 5.
Visualization of DNA loops from the 4q35 locus. Shown are the results of hybridization of the bacterial artificial chromosome probe that contains an insert covering the region between 90 and 250 kb relative to the D4Z4 array (green) of the nuclear halos from human primary myoblasts of unaffected individuals (A–A‴) or FSHD patients (B–B‴). (A and B) DNA stained with DAPI. (A' and B') The results of hybridization. (A″ and B″) Merged images. (A‴ and B‴) The paths of chromatin loops (green) relative to the nuclear matrices and halos (blue).
Incomplete loops were observed in 73% of inspected halos. In 27% of the nuclei, the signals were distributed in a disordered fashion over both the core and halo (data not shown), which is likely to result from distortion of the halo during preparation and/or from an unfavorable loop position on the plane surface of the slide. Only one S/MAR was observed in this region. No visible difference in loop organization of the distal end of the loop was observed between primary myoblasts from healthy subjects and FSHD patients, thus confirming the results of our biochemical analysis. Indeed, the resolution of the FISH technique is not sufficient to detect a subtle difference (6 kb) between the attachment sites located at positions 171 and 165, respectively.
We have roughly estimated the size of incomplete loop branches in case of both types of studied myoblasts, measured by tracing their contour (Fig. 5). Each loop of such shape reveals almost equivalent arms, which would correspond to the MAR position at ≈170 kb relative to D4Z4 array, whereas we have observed the S/MARs positioned at positions 165–171 by using the genomic DNA array technique.
Discussion
Drastic Changes in Chromatin Loop Domain Organization of the 4q35 Locus in Myoblasts from Patients: Implications for the FSHD.
The results presented here clearly demonstrate that normal human primary myoblasts and myoblasts derived from FSHD patients present with specific organizations of the 4q35 locus into DNA loop domains and that these organizations differ.
Several models for the molecular basis of FSHD have been proposed. One model is based on the existence of a repressive element (D4Z4-binding element) within the D4Z4 repeat that binds a repressor complex (D4Z4-repressing complex), resulting in transcriptional repression spreading onto neighboring sequences. According to this model, deletion of an integral number of D4Z4 repeats in FSHD patients would reduce the amount of bound repressor complexes and consequently decrease (or abolish) transcriptional repression of 4q35 genes (11). Thus, the deletion of repeated elements in the subtelomeric 4q region would act in cis on neighboring genes, derepressing transcription and starting a cascade of events that ultimately leads to FSHD.
According to other studies, no significant difference in chromatin organization can be detected in the 4q35.2 locus between normal human primary myoblasts and myoblasts derived from FSHD patients. It appears difficult to draw any conclusions from these data, because they were focused on regions 300 and 850 kb away, respectively, from the chromosome 4 D4Z4 array (16, 17).
According to another study, the D4Z4 array was found to be associated with the periphery of the nucleus (18). Unfortunately, these investigations carried out in cells derived from FSHD patients were done in cells of lymphoid, not muscular, origin, making it less relevant for exploring the mechanistic substratum of defects expressed in myoblasts only.
The present data therefore provide evidence of clear-cut differences between the deleted and nondeleted chromosomes 4 in myoblasts from FSHD patients as well as differences between a deleted chromosome 4 from FSHD patients and the corresponding subtelomeric region of chromosome 4q in cultured primary myoblasts derived from normal individuals. The D4Z4 array and its neighboring genes, namely FRG2 and FRG1, were found to be included in two distinct loop domains in control primary myoblasts as well as in the normal chromosome 4 in FSHD patients. In contrast, in the defective chromosome 4q of FSHD patients, these genes were located within the same chromatin loop domain as the partially deleted D4Z4 repeat array.
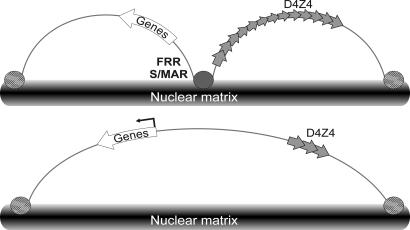
We postulate that these differences may account for changes observed in transcriptional pattern at the 4q35 locus. Indeed, our previous data suggest that D4Z4 has the properties of a transcriptional enhancer (ref. 39 and A.P. and Y.S.V., unpublished data) and might enhance transcription from the FRG2 promoter (13). The presence of the D4Z4 enhancer within the same loop domain as the FSHD candidate genes FRG2 and FRG1, along with the reduction in size of the repeat array containing the repressive D4Z4-binding element, could up-regulate transcription of neighboring genes in affected individuals, thereby resulting in FSHD in these patients (Fig. 6).
Fig. 6.
Loop domain organization and transcriptional control in the 4q35 locus.
It is also noteworthy that a subtle difference exists at the distal end of the loop domain. In primary myoblasts of normal individuals, the 5' S/MAR is located at position 171 in the array, whereas it lies at position 165 in myoblasts from FSHD patients. This phenomenon, referred to as “sliding,” may be linked to the selection of an S/MAR better adapted to a specific function or transcriptional pattern, as suggested by Heng et al. (40). Further studies will be necessary to determine whether this phenomenon plays a role in the genesis of FSHD.
4qA/4qB Genotypes and Long-Range Chromatin Organization.
A polymorphic segment of 10 kb directly distal to D4Z4 was shown to exist in two allelic forms, 4qA and 4qB. Although both alleles are equally common in the general population, it has been reported that FSHD is associated only with the 4qA allele (41, 42). It would be interesting to see whether this polymorphism affects the association with the nuclear matrix of the region located between the D4Z4 array and the telomere. Unfortunately, no coherent sequence data exist on this region, making it impossible to use the array technique; however, classical biochemical technique may prove useful.
S/MARs and Reduction of the D4Z4 Array by Recombination.
The near identity of the subtelomeric parts of chromosomes 4q and 10q makes it likely that the mechanism of partial deletion of the D4Z4 array on chromosome 4 occurs through a translocation or recombination with chromosome 10 (43–45). The presence of a strong S/MAR near the D4Z4 array may favor such a recombination.
In many recombination-prone regions, including the MLL gene and the IFN-II loci, the recombination hot spots are located in the vicinity of S/MARS (for reviews, see refs. 46 and 47). Recombination enhanced by adjacent matrix attachment regions may result in the loss of D4Z4 repeats. This hypothesis clearly calls for further investigation.
Relationship Between Heterochromatin and Matrix Attachment.
The question of a relationship between matrix attachment and heterochromatin has been widely discussed (for a review, see ref. 48). It is generally believed that S/MARs may stop the expansion of heterochromatin and protect neighbor genes that are being transcribed from heterochromatinization. Indeed, inserting a S/MAR into a construct carrying a (reporter) gene generally results in enhanced transcription in transgenic animals or plants, in relation with changes observed in the chromatin structure proximal and distal to the integrated gene (49). The assembly of heterochromatin at S/MARs has also been implicated in the function of the imprinting center at human chromosome 15q11–15q13 (50). Contrasting with these views, our results suggest that S/MARs do not constitute efficient barriers for heterochromatinization. Indeed, heterochromatin was found to spread over the FRR MAR in normal human myoblasts (19). In FSHD patients, in whom the heterochromatin structure seems to be altered under the effect of a reduced D4Z4 array (11), the FRR MAR appears not to be associated with the nuclear matrix.
The changes in long-range chromatin organization between normal myoblasts and those from FSHD patients reported in the present study may reveal perspectives in the study of the disease.
Materials and Methods
Cells.
HeLa cell line was purchased from American Type Culture Collection. GM10115A hybrid murine cell line containing human chromosome 4 was the kind gift of R. Tupler (Universitá delgi Studi di Modena e Reggio Emilia, Modena, Italy).
Primary myoblasts from two healthy individuals and two FSHD patients with 5.5 D4Z4 repeats and seven repeats in the 4q35 array were cultured on collagen-coated support in DMEM supplemented with 20% bovine fetal serum.
Nuclei and Nuclear Matrices.
Nuclei from primary human myoblasts and HeLa and GM10115A cells were isolated as described in ref. 26. Nuclear matrices were prepared by treatment of the isolated nuclei with NaCl as follows: digestion buffer (100 mM NaCl/25 mM KCl/10 mM Tris·HCl, pH 7.5/0.25 mM spermidine) was added to 105 nuclei to a final volume of 400 μl. DNase I was added to a final concentration of 100 μg/ml, and the samples were digested for 2 h at 4°C, followed by the addition of CuCl2 to a final concentration of 1 mM for 10 min at 4°C. The nuclei were then extracted by addition of one volume of an EB buffer (4 M NaCl/20 mM EDTA/40 mM Tris·HCl, pH 7.5). The resulting nuclear matrices were washed (2 M NaCl/10 mM EDTA/20 mM Tris·HCl, pH 7.5), and nuclear matrix-associated DNA was extracted after proteinase K treatment and either radioactively labeled by using the Ready-to-Go kit (AP Biotech) or labeled with DIG by using a DIG-High Prime kit (Roche Diagnostics) and used as a probe for hybridization.
DNA Array.
The DNA array consisted of 102 35- to 45-mer oligonucleotides spaced ≈2 kb apart spanning the region from D4Z4 repeat array to the far upstream region of the FRG1 gene. The list of oligonucleotides is available upon request. The distal part of the array surrounding the FRG1 gene contained the unique chromosome 4-specific sequences. The proximal part contained sequences specific for chromosomes 4 and 10 due to 99% homology between these regions. The oligonucleotides were slot-blotted onto Hybond N+ filters (Amersham Pharmacia) and hybridized at 40.5°C overnight. The blot was incubated with the anti-DIG antibodies (Roche) and revealed by using an enhanced chemiluminescence kit (ECL+; Amersham Pharmacia). The films were scanned and quantified with image gauge 4.0 (Fuji). The hybridization data were normalized against S/MAR from the human or murine c-myc gene loci. The experiments were carried out in triplicate. Data from two independent experiments are presented.
Nuclear Halo/Matrices Preparation for FISH.
Nuclei were prepared as described in ref. 26. To obtain the nuclear halos, nuclei were pelleted at 200 × g for 10 min onto glass slides. Slides were then incubated in buffer H1 (10 mM Pipes, pH 6.8/0.1 M NaCl/0.3 M sucrose/3 mM MgCl2/0.5% Triton X-100/0.1 mM CuSO4/1 mM PMSF) for 10 min on ice, followed by treatment in buffer H2 (1 mM Pipes, pH 6.8/2 M NaCl/10 mM EDTA/0.1% digitonin/0.05 mM spermine/0.125 mM spermidine) for 4 min. Slides were then passed through 10×, 5×, 2×, and 1× PBS followed by 10%, 30%, 50%, 70%, and 95% ethanol solutions, air-dried, and finally fixed at 70°C for 2 h.
Preparation of nuclear matrices on the microscope slides was carried out essentially as described in ref. 51.
FISH Analysis.
FRR MAR was amplified by PCR with the pGEM42 plasmid and labeled with DIG-11-dUTP (Roche Diagnostics). The D4Z4 probe was derived from pGEM42 (51) and labeled with biotin-14 as well as BAC RP11521G19 located 100 kb proximal to D4Z4 array on 4q35.
Hybridization to slides was performed as described in ref. 52 by using anti-biotin mouse antibodies conjugated with Alexa Fluor 488 (Molecular Probes) or anti-DIG sheep antibodies conjugated with TAMRA (Roche). The nuclei, nuclear halos, and matrices were counterstained by DAPI in VECTASHIELD mounting medium (Vector Laboratories). Slides were examined on an Olympus Provis fluorescence microscope with a 60× oil immersion objective and appropriate filters. Images were captured with a charge-coupled device camera (Photometrics, Tucson, AZ), using rsimage software (Scanalytics, Billerica, MA).
Supplementary Material
Acknowledgments
We thank Dr. A. Belayev (Universite de Mons-Hainaut, Mans, Belgium) for the kind gift of the pGEM42 plasmid, Dr. R. Tupler for the kind gift of the GM10115A strain, and Drs. A. Hair and V. Ogryzko for critical readings of the manuscript. This work was supported by grants from the Association Francaise contre les Myopathies (to Y.S.V.). A.P. was supported by postdoctoral fellowships from The Federation of Biochemical Societies and the Fondation pour la Recherche Médicale. I.P. was supported by a postdoctoral fellowship from the Association Francaise contre les Myopathies. D.L. was suppored by the Association Francaise contre les Myopathies.
Abbreviations
- FSHD
facioscapulohumeral muscular dystrophy
- S/MAR
nuclear scaffold/matrix attached region
- FRR MAR
FSHD-related region S/MAR
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Lunt P. W., Harper P. S. J. Med. Genet. 1991;28:655–664. doi: 10.1136/jmg.28.10.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wijmenga C., Padberg G. W., Moerer P., Wiegant J., Liem L., Brouwer O. F., Milner E. C., Weber J. L., van Ommen G. B., Sandkuyl L. A., et al. Genomics. 1991;9:570–575. doi: 10.1016/0888-7543(91)90348-i. [DOI] [PubMed] [Google Scholar]
- 3.van Deutekom J. C., Wijmenga C., van Tienhoven E. A., Gruter A. M., Hewitt J. E., Padberg G. W., van Ommen G. J., Hofker M. H., Frants R. R. Hum. Mol. Genet. 1993;2:2037–2042. doi: 10.1093/hmg/2.12.2037. [DOI] [PubMed] [Google Scholar]
- 4.Wijmenga C., Hewitt J. E., Sandkuijl L. A., Clark L. N., Wright T. J., Dauwerse H. G., Gruter A. M., Hofker M. H., Moerer P., Williamson R., et al. Nat. Genet. 1992;2:26–30. doi: 10.1038/ng0992-26. [DOI] [PubMed] [Google Scholar]
- 5.Tawil R., van der Maarel S. Muscle Nerve. 2006 Feb 6; doi: 10.1002/mus.20522. 10.1002/mus.20522. [DOI] [PubMed] [Google Scholar]
- 6.Hewitt J. E., Lyle R., Clark L. N., Valleley E. M., Wright T. J., Wijmenga C., van Deutekom J. C., Francis F., Sharpe P. T., Hofker M., et al. Hum. Mol. Genet. 1994;3:1287–1295. doi: 10.1093/hmg/3.8.1287. [DOI] [PubMed] [Google Scholar]
- 7.van Geel M., Heather L. J., Lyle R., Hewitt J. E., Frants R. R., de Jong P. J. Genomics. 1999;61:55–65. doi: 10.1006/geno.1999.5942. [DOI] [PubMed] [Google Scholar]
- 8.Tupler R., Perini G., Pellegrino M. A., Green M. R. Proc. Natl. Acad. Sci. USA. 1999;96:12650–12654. doi: 10.1073/pnas.96.22.12650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winokur S. T., Chen Y. W., Masny P. S., Martin J. H., Ehmsen J. T., Tapscott S. J., Van Der Maarel S. M., Hayashi Y., Flanigan K. M. Hum. Mol. Genet. 2003;12:2895–2907. doi: 10.1093/hmg/ddg327. [DOI] [PubMed] [Google Scholar]
- 10.Wohlgemuth M., Lemmers R. J., Van Der Kooi E. L., Van Der Wielen M. J., Van Overveld P. G., Dauwerse H., Bakker E., Frants R. R., Padberg G. W., Van Der Maarel S. M. Neurology. 2003;61:909–913. doi: 10.1212/wnl.61.7.909. [DOI] [PubMed] [Google Scholar]
- 11.Gabellini D., Green M. R., Tupler R. Cell. 2002;110:339–348. doi: 10.1016/s0092-8674(02)00826-7. [DOI] [PubMed] [Google Scholar]
- 12.Laoudj-Chenivesse D., Carnac G., Bisbal C., Hugon G., Bouillot S., Desnuelle C., Vassetzky Y., Fernandez A. J. Mol. Med. 2005;83:216–224. doi: 10.1007/s00109-004-0583-7. [DOI] [PubMed] [Google Scholar]
- 13.Rijkers T., Deidda G., van Koningsbruggen S., van Geel M., Lemmers R. J., van Deutekom J. C., Figlewicz D., Hewitt J. E., Padberg G. W., Frants R. R., van der Maarel S. M. J. Med. Genet. 2004;41:826–836. doi: 10.1136/jmg.2004.019364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gabellini D., D'Antona G., Moggio M., Prelle A., Zecca C., Adami R., Angeletti B., Ciscato P., Pellegrino M. A., Bottinelli R., Green M. R., Tupler R. Nature. 2005;439:973–977. doi: 10.1038/nature04422. [DOI] [PubMed] [Google Scholar]
- 15.van Overveld P. G., Lemmers R. J., Sandkuijl L. A., Enthoven L., Winokur S. T., Bakels F., Padberg G. W., van Ommen G. J., Frants R. R., van der Maarel S. M. Nat. Genet. 2003;35:315–317. doi: 10.1038/ng1262. [DOI] [PubMed] [Google Scholar]
- 16.Jiang G., Yang F., van Overveld P. G., Vedanarayanan V., van der Maarel S., Ehrlich M. Hum. Mol. Genet. 2003;12:2909–2921. doi: 10.1093/hmg/ddg323. [DOI] [PubMed] [Google Scholar]
- 17.Yang F., Shao C., Vedanarayanan V., Ehrlich M. Chromosoma. 2004;112:350–359. doi: 10.1007/s00412-004-0280-x. [DOI] [PubMed] [Google Scholar]
- 18.Tam R., Smith K. P., Lawrence J. B. J. Cell Biol. 2004;167:269–279. doi: 10.1083/jcb.200403128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winokur S. T., Bengtsson U., Feddersen J., Mathews K. D., Weiffenbach B., Bailey H., Markovich R. P., Murray J. C., Wasmuth J. J., Altherr M. R., et al. Chromosome Res. 1994;2:225–234. doi: 10.1007/BF01553323. [DOI] [PubMed] [Google Scholar]
- 20.Prioleau M. N., Nony P., Simpson M., Felsenfeld G. EMBO J. 1999;18:4035–4048. doi: 10.1093/emboj/18.14.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buongiorno-Nardelli M., Micheli G., Carri M. T., Marilley M. Nature. 1982;298:100–102. doi: 10.1038/298100a0. [DOI] [PubMed] [Google Scholar]
- 22.Cockerill P. N., Garrard W. T. Cell. 1986;44:273–282. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- 23.Mirkovitch J., Mirault M. E., Laemmli U. K. Cell. 1984;39:223–232. doi: 10.1016/0092-8674(84)90208-3. [DOI] [PubMed] [Google Scholar]
- 24.Vassetzky Y., Lemaitre J. M., Mechali M. Crit. Rev. Eukaryotic Gene Expression. 2000;10:31–38. [PubMed] [Google Scholar]
- 25.Vassetzky Y., Hair A., Mechali M. Genes Dev. 2000;14:1541–1552. [PMC free article] [PubMed] [Google Scholar]
- 26.Gasser S. M., Vassetzky Y. S. In: Chromatin: A Practical Approach. Gould H., editor. Oxford: Oxford Univ. Press; 1998. pp. 111–124. [Google Scholar]
- 27.Lyle R., Wright T. J., Clark L. N., Hewitt J. E. Genomics. 1995;28:389–397. doi: 10.1006/geno.1995.1166. [DOI] [PubMed] [Google Scholar]
- 28.Cockerill P. N., Garrard W. T. FEBS Lett. 1986;204:5–7. doi: 10.1016/0014-5793(86)81377-1. [DOI] [PubMed] [Google Scholar]
- 29.Girard-Reydet C., Gregoire D., Vassetzky Y., Mechali M. Gene. 2004;332:129–138. doi: 10.1016/j.gene.2004.02.031. [DOI] [PubMed] [Google Scholar]
- 30.Clark L. N., Koehler U., Ward D. C., Wienberg J., Hewitt J. E. Chromosoma. 1996;105:180–189. doi: 10.1007/BF02509499. [DOI] [PubMed] [Google Scholar]
- 31.Gromova I. I., Thomsen B., Razin S. V. Proc. Natl. Acad. Sci. USA. 1995;92:102–106. doi: 10.1073/pnas.92.1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ioudinkova E., Petrov A., Razin S. V., Vassetzky Y. S. Genomics. 2005;85:143–151. doi: 10.1016/j.ygeno.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 33.Vogelstein B., Pardoll D. M., Coffey D. S. Cell. 1980;22:79–85. doi: 10.1016/0092-8674(80)90156-7. [DOI] [PubMed] [Google Scholar]
- 34.Bickmore W. Exp. Cell Res. 1996;229:198–200. doi: 10.1006/excr.1996.0359. [DOI] [PubMed] [Google Scholar]
- 35.Byrd K., Corces G. J. Cell Biol. 2003;162:565–574. doi: 10.1083/jcb.200305013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iarovaia O. V., Bystritskiy A., Ravcheev D., Hancock R., Razin S. V. Nucleic Acids Res. 2004;32:2079–2086. doi: 10.1093/nar/gkh532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kikyo N., Wade P. A., Guschin D., Ge H., Wolffe A. P. Science. 2000;289:2360–2363. doi: 10.1126/science.289.5488.2360. [DOI] [PubMed] [Google Scholar]
- 38.Ratsch A., Joos S., Kioschis P., Lichter P. Exp. Cell Res. 2002;273:12–20. doi: 10.1006/excr.2001.5429. [DOI] [PubMed] [Google Scholar]
- 39.Petrov A. P., Laoudj D., Vassetzky Y. S. Genetics. 2003;39:147–151. [Google Scholar]
- 40.Heng H. H., Goetze S., Ye C. J., Liu G., Stevens J. B., Bremer S. W., Wykes S. M., Bode J., Krawetz S. A. J. Cell Sci. 2004;117:999–1008. doi: 10.1242/jcs.00976. [DOI] [PubMed] [Google Scholar]
- 41.Lemmers R. J., de Kievit P., Sandkuijl L., Padberg G. W., van Ommen G. J., Frants R. R., van der Maarel S. M. Nat. Genet. 2002;32:235–236. doi: 10.1038/ng999. [DOI] [PubMed] [Google Scholar]
- 42.Lemmers R. J., Wohlgemuth M., Frants R. R., Padberg G. W., Morava E., van der Maarel S. M. Am. J. Hum. Genet. 2004;75:1124–1130. doi: 10.1086/426035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Overveld P. G., Lemmers R. J., Deidda G., Sandkuijl L., Padberg G. W., Frants R. R., van der Maarel S. M. Hum. Mol. Genet. 2000;9:2879–2884. doi: 10.1093/hmg/9.19.2879. [DOI] [PubMed] [Google Scholar]
- 44.van Geel M., Dickson M. C., Beck A. F., Bolland D. J., Frants R. R., van der Maarel S. M., de Jong P. J., Hewitt J. E. Genomics. 2002;79:210–217. doi: 10.1006/geno.2002.6690. [DOI] [PubMed] [Google Scholar]
- 45.Lemmers R. J., van der Maarel S. M., van Deutekom J. C., van der Wielen M. J., Deidda G., Dauwerse H. G., Hewitt J., Hofker M., Bakker E., Padberg G. W., Frants R. R. Hum. Mol. Genet. 1998;7:1207–1214. doi: 10.1093/hmg/7.8.1207. [DOI] [PubMed] [Google Scholar]
- 46.Bode J., Benham C., Ernst E., Knopp A., Marschalek R., Strick R., Strissel P. J. Cell. Biochem. 2000;35(Suppl.):3–22. doi: 10.1002/1097-4644(2000)79:35+<3::aid-jcb1121>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 47.Bystritskiy A. A., Razin S. V. Crit. Rev. Eukaryotic Gene Expression. 2004;14:65–77. [PubMed] [Google Scholar]
- 48.Podgornaya O. I., Voronin A. P., Enukashvily N. I., Matveev I. V., Lobov I. B. Int. Rev. Cytol. 2003;224:227–296. doi: 10.1016/s0074-7696(05)24006-8. [DOI] [PubMed] [Google Scholar]
- 49.Allen G. C., Spiker S., Thompson W. F. Plant Mol. Biol. 2000;43:361–376. doi: 10.1023/a:1006424621037. [DOI] [PubMed] [Google Scholar]
- 50.Greally J. M., Gray T. A., Gabriel J. M., Song L., Zemel S., Nicholls R. D. Proc. Natl. Acad. Sci. USA. 1999;96:14430–14435. doi: 10.1073/pnas.96.25.14430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gabriels J., Beckers M. C., Ding H., De Vriése A., Plaisance S., van der Maarel S. M., Padberg G. W., Frants R. R., Hewitt J. E., Collen D., Belayew A. Gene. 1999;236:25–32. doi: 10.1016/s0378-1119(99)00267-x. [DOI] [PubMed] [Google Scholar]
- 52.Cai S., Kohwi-Shigematsu T. Methods. 1999;19:394–402. doi: 10.1006/meth.1999.0875. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.