Abstract
Protein phosphorylation plays a key role in vasopressin signaling in the renal-collecting duct. Large-scale identification and quantification of phosphorylation events triggered by vasopressin is desirable to gain a comprehensive systems-level understanding of this process. We carried out phosphoproteomic analysis of rat inner medullary collecting duct cells by using a combination of phosphopeptide enrichment by immobilized metal affinity chromatography and phosphorylation site identification by liquid chromatography-mass spectrometryn neutral loss scanning. A total of 714 phosphorylation sites on 223 unique phosphoproteins were identified from inner medullary collecting duct samples treated short-term with either calyculin A or vasopressin. A number of proteins involved in cytoskeletal reorganization, vesicle trafficking, and transcriptional regulation were identified. Previously unidentified phosphorylation sites were found for membrane proteins essential to collecting duct physiology, including eight sites among aquaporin-2 (AQP2), aquaporin-4, and urea transporter isoforms A1 and A3. Through label-free quantification of phosphopeptides, we identified a number of proteins that significantly changed phosphorylation state in response to short-term vasopressin treatment: AQP2, Bclaf1, LRRC47, Rgl3, and SAFB2. In the presence of vasopressin, AQP2 monophosphorylated at S256 and diphosphorylated AQP2 (pS256/261) increased in abundance, whereas AQP2 monophosphorylated at S261 decreased, raising the possibility that both sites are involved in vasopressin-dependent AQP2 trafficking. This study reveals the practicality of liquid chromotography-mass spectrometryn neutral loss scanning for large-scale identification and quantification of protein phosphorylation in the analysis of cell signaling in a native mammalian system.
Keywords: LC-MS/MS, collecting duct, kidney, Collecting Duct Phosphoprotein Database (CDPD), neutral loss
Elucidation of cellular signaling networks requires methodologies for large-scale quantitative phosphoproteomic analysis that can be used to reveal dynamic system-wide changes in protein phosphorylation. Recent studies have introduced two new innovations, namely, immobilized metal affinity chromatography (IMAC) (1–3) and liquid chromatography-mass spectrometry (LC-MS)3 neutral loss scanning (1–4), to increase the efficiency of phosphopeptide identification. Quantification of phosphopeptides in this setting has been challenging and has been successful so far in cultured cells (5) and yeast (6) but not in native mammalian cells and tissues. Here, we introduce a hybrid approach using IMAC for phosphopeptide enrichment, LC-MS multisequential (LC-MSn) neutral loss scanning to identify phosphorylated residues in the peptides, and label-free quantification using numerical integration of pseudochromatograms constructed from MS peak heights.
The method is used here for analysis of protein phosphorylation in vasopressin-sensitive inner medullary collecting duct (IMCD) cells freshly isolated from rat kidneys. Water and urea transport across this epithelium is regulated by the peptide hormone vasopressin via V2-type receptors. Ligand binding triggers an incompletely understood signaling response that includes activation of at least two adenylyl cyclase isoforms via the heterotrimeric G protein Gs (7–9), an increase in intracellular cAMP (10), an increase in cytosolic Ca2+ concentration (11), calmodulin-mediated activation of myosin light-chain kinase (12), and phosphorylation of the water channel aquaporin-2 (AQP2) (13) as well as the urea transporter isoforms A1 and A3 (UT-A1/3) (14). The work presented here identifies a large number of previously unknown phosphorylation sites in IMCD proteins, including sites in AQP2 and UT-A1/3. Furthermore, phosphorylation at several of these residues was found to be altered in response to vasopressin, including S256 and S261 of AQP2.
Results
Validation of the LC-MSn Approach.
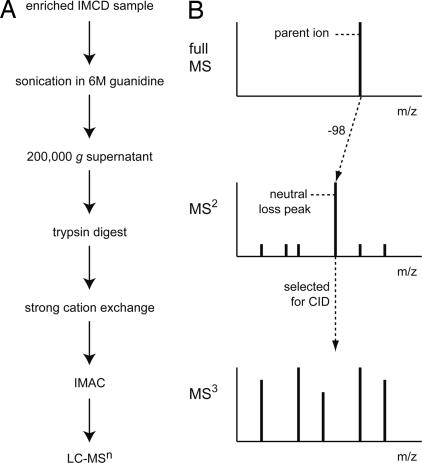
One goal of this study was to develop a robust method for isolation and identification of phosphopeptides from rat IMCD. The general experimental approach is indicated in Fig. 1A. Identification of phosphopeptides was accomplished by using LC-MSn neutral loss scanning (Fig. 1B). This method takes advantage of the poor fragmentation of phosphopeptides in the MS2 spectrum because of a predominant loss of phosphoric acid, H3PO4 (−98 Da), from the parent ion in the full MS spectrum (neutral loss). Peaks in a particular MS2 spectrum that differed from the associated parent ion by −98 were selected by the mass spectrometer for further fragmentation, followed by peptide identification using the MS3 spectrum.
Fig. 1.
Experimental approach. (A) Rat IMCD protein samples were digested with trypsin, followed by separation by strong cation exchange (SCX) chromatography. Phosphorylated peptides were enriched from total peptides by using IMAC. (B) Phosphopeptide samples were analyzed by LC-MSn neutral loss scanning in which fragmentation of the phosphopeptide resulted in loss of phosphoric acid (−98 Da; neutral loss peak) from the full MS (parent ion) scan. This neutral loss peak was selected for further fragmentation and peptide identification in the MS3 spectrum.
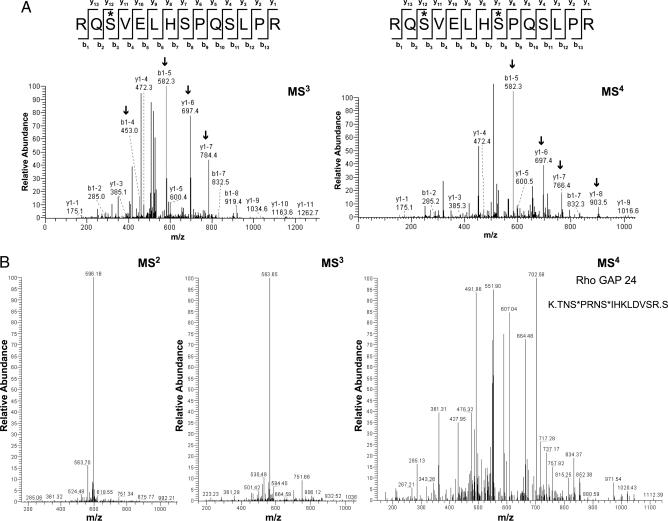
LC-MS3 analysis was often sufficient to identify the site of phosphorylation for singly phosphorylated peptides; however, multiply phosphorylated peptides usually required a further round of fragmentation and identification using MS4 spectra, as shown in the AQP2 analysis (Fig. 2). For the singly phosphorylated AQP2 peptide, the MS3 spectrum provided enough information to unambiguously identify phosphorylation at S256 (Fig. 2A Left), a previously known site. The peaks that are labeled with arrows matched correctly with phosphorylation at S256 but not with phosphorylation at any other serine residue in the peptide. In a doubly phosphorylated AQP2 peptide (S256 plus an additional site, S261), unambiguous identification of phosphorylation at both residues required LC-MS4 analysis (Fig. 2A Right). An abundant S261 monophosphorylated AQP2 peptide was also identified.
Fig. 2.
Identification and confirmation of phosphorylation sites using LC-MSn neutral loss scanning. (A) Confirmation of AQP2 phosphorylation on S256 (∗) from an MS3 spectrum (Left). Arrows indicate specific peaks in the mass spectrum that aided in distinguishing the actual site of phosphorylation from other potential sites. Confirmation of phosphorylation on S256 as well as identification of a new site, S261, for a doubly phosphorylated AQP2 peptide from an MS4 spectrum (Right). (B) Comparison of different MS levels for Rho GTPase activating protein 24, a phosphoprotein that was identified solely on the quality of its MS4 spectrum. In both A and B, asterisks indicate residues that were phosphorylated in the full MS scan but subsequently underwent neutral loss leaving a modified original residue mass without water (−18 Da).
MS4 spectra also identified unique peptides that were not found in either MS2 or MS3 analyses, as was the case for RhoGAP 24 (Fig. 2B). In this instance, the peptide contained two phosphate groups and gave poor-quality MS2 and MS3 spectra because of “sequential neutral loss” (i.e., only one phosphate lost during each successive round of fragmentation). Once both phosphates were removed, the peptide fragmented well in MS4 and was identified.
Large-Scale Identification of Phosphoproteins from IMCD by LC-MSn Neutral Loss Scanning.
In the initial experiment, IMCD samples were treated with calyculin A, a potent serine/threonine phosphatase inhibitor, to maximize the chances of identifying phosphopeptides. In subsequent experiments, IMCD samples were incubated in the presence or absence of (deamino-Cys1, d-Arg8)vasopressin (dDAVP) (10−9 M), a V2-selective vasopressin analog, for 10 min, providing a physiologically relevant perturbation for the study of protein phosphorylation. Two different mass analyzers [Fourier transform–ion cyclotron resonance (FT-ICR) and LTQ] were used for the full (parent ion) scan in two separate MS runs for each of the above conditions. For IMCDs treated with calyculin A, we identified and manually confirmed 58 phosphoproteins by using the higher mass-resolution instrument, the FT-ICR (Table 3, which is published as supporting information on the PNAS web site). Using the higher sensitivity instrument, the LTQ on this same sample, we identified 800 potential phosphoproteins that passed a medium stringency cross-correlation (XCorr) filter. Of these proteins, 143 were found at more than one MS level (e.g., MS2 and MS3) or contained more than one unique peptide (Table 4, which is published as supporting information on the PNAS web site), thus providing a high degree of certainty to these identifications. For samples with or without dDAVP treatment, we identified and manually confirmed 46 phosphoproteins by using the FT-ICR (Table 5, which is published as supporting information on the PNAS web site). Using the LTQ on these samples, we identified 928 potential phosphoproteins that passed a medium stringency XCorr filter, 94 of which were found at more than one MS level or contained more than one unique peptide (Table 6, which is published as supporting information on the PNAS web site). A summary of the overlap between calyculin A and dDAVP data sets for separate FT-ICR and LTQ runs is provided in Fig. 5, which is published as supporting information on the PNAS web site. In total, 223 unique IMCD phosphoproteins (with 714 identified sites) were found among all MS runs, with only 50 in common between calyculin A and dDAVP-treated samples. Thus, the neutral loss scanning LC-MSn approach is capable of revealing a distinct phosphoproteomic profile in response to a physiological stimulus such as vasopressin in renal collecting duct cells.
Fig. 6, which is published as supporting information on the PNAS web site, shows a classification of the 92 phosphoproteins identified from FT-ICR spectra. This list contains proteins with a wide variety of functions, the largest groups being cytoskeletal/motor proteins, transcription factors, and other nuclear proteins. All identifications from FT-ICR and LTQ runs are listed in the Collecting Duct Phosphoprotein Database (CDPD) (http://dir.nhlbi.nih.gov/papers/lkem/cdpd/index.htm). A subset of these results highlighting proteins that are of immediate interest with regard to vasopressin signaling and AQP2 trafficking is shown in Table 1. Of the 45 phosphorylation sites listed in Table 1, only 17 had been previously identified according to searches of three phosphoprotein databases, PhosphoSite (Cell Signaling Technology, Beverly, MA), Phospho.ELM (15), and the Human Protein Reference Database (16), as well as the current scientific literature. Included in this table are previously unknown phosphorylation sites in the C terminus of AQP2 (S261, S264, and S269), the C terminus of AQP4 (S321), and the N-terminal tail, as well as the intracellular loop regions of the vasopressin-regulated urea transporter UT-A1/3 (S35, S62, S63, and S486). These results demonstrate the ability of the method to detect phosphorylation sites in integral membrane proteins using guanidine HCl alone as a membrane-disrupting agent (Fig. 7, which is published as supporting information on the PNAS web site).
Table 1.
IMCD phosphoproteins of potential relevance to vasopressin signaling/AQP2 trafficking
| ID | Site (putative kinase) | Swiss-Prot | Spectrum | MS quant ratio | Comment (Ref.) |
|---|---|---|---|---|---|
| AQP2 | S256* (PKA), S261* (p38), S264 (PKC), S269 (PKA) | P41181 | MS2, MS3, MS4 | 2.67 ± 0.84** | Associated with insertion into the apical membrane (13) |
| AQP4 | S321 (PKC) | P55087 | MS2, MS3 | PDZ binding motif (22) | |
| Arrestin, β1 | S412(ERK1/2) | P49407 | MS2, MS3 | 1.46 ± 0.34 | Role in V2 receptor endocytosis (29) |
| Bcl-2-associated transcription factor (Bclaf1) | S177 (Cdk5) | Q9NYF8 | MS2, MS3 | 0.33 ± 0.07** | Transcriptional repressor |
| Dynein, cytoplasmic, light intermediate chain 1 | S510 (ERK1), S516(GSK3) | Q9Y6G9 | MS2 | Associated with AQP2-containing intracellular vesicles (30) | |
| EGFR pathway substrate 8-like protein 2 (Eps8L2) | T306 (PKC) | Q9H6S3 | MS2, MS3 | Activates Rac-GEF activity of Sos-1; actin remodeling | |
| Epsin 3† | S175 (PKC), S177 (GSK3), S178 (PKC) | Q9H201 | MS2, MS3 | Clathrin-binding adaptor protein | |
| Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | T182 (DNAPK) | P04406 | MS2, MS3 | Interacts with Rab2; ER to Golgi transport (31) | |
| Heat shock 27 kDa protein 1 | S13 (ERK1), S15*(PKC), S85(PKC),S86(PKC) | P04792 | MS2, MS3 | 0.94 ± 0.08 | Induced by AVP in smooth muscle (32) |
| Heat shock 90 kDa protein 1, beta | S255(CK2) | P08238 | MS2, MS3 | 0.64 ± 0.32 | Upregulated by osmotic stress (33) |
| Leucine-rich repeat containing protein 47 (LRRC47) | S293 (PKA) | Q8N1G4 | MS2 | 2.46 ± 0.54** | Protein synthesis |
| MAP kinase 1 (ERK2) | T183(MEK1), Y185(MEK1) | P28482 | MS2, MS3 | Role in insulin-induced AQP2 expression (34) | |
| MAP kinase 3 (ERK1) | T203 (MEK1), Y205(MEK1) | P27361 | MS2, MS3 | ||
| Myosin heavy chain 10 (IIb) | S1935 (PKC), S1956 (CK2) | P35580 | MS2, MS3 | Potential role in AQP2 trafficking (12) | |
| Myosin heavy chain 9 (IIa) | S1944(CK2) | P35579 | MS2, MS3 | 1.14 ± 0.04 | |
| p21-activated kinase 2 (PAK2) | S141(CAMKII) | Q13177 | MS2, MS3 | Effector of Rac/cdc42-induced actin reorganization (35) | |
| PKA, regulatory, type II alpha | S97 (PKA) | P13861 | MS2, MS3 | Associated with AQP2 via AKAP (36) | |
| PKA, regulatory, type II beta | S112(PKA) | P31323 | MS2, MS3 | ||
| Protein phosphatase I, regulatory subunit 12A | S507* (PKA), S941 (PKA) | O14974 | MS2, MS3 | 0.98 ± 0.25 | Regulates the interaction of actin and myosin downstream of Rho (37) |
| Protein phosphatase I, regulatory subunit 12B | S566 (PKA) | O60237 | MS2, MS3 | Regulates the interaction of actin and myosin downstream of Rho (37) | |
| RalGDS-related effector protein of M-Ras (Rgl3) | S561, S562 (cdc2) | Q3MIN7 | MS2 | 0.69 ± 0.11** | Interacts with Ral, Rap1, and Ras; vesicle trafficking (38) |
| Reticulon 4 (Nogo) | S107(MAPKAPK2) | Q9NQC3 | MS3 | 1.03 ± 0.15 | Vesicle trafficking (39) |
| Rho GTPase activating protein 15 | S620 (PKA) | Q86WP1 | MS2, MS3 | Rho inhibits cAMP-induced translocation of AQP2 (40) | |
| Rho GTPase activating protein 21 | S1400 (CK2), S1406 | Q5T5U3 | MS2, MS3 | ||
| Rho GTPase activating protein 24 | S311 (PKC), S315 (cdc2) | Q4KMG1 | MS2, MS3, MS4 | ||
| Scaffold-associated factor B2 (SAFB2) | S366(ERK1) | Q14151 | MS2, MS3 | 0.009 ± 0.003** | DNA binding/transcription |
| Septin 2 | S218 (CK2) | Q15019 | MS2, MS3 | 1.20 ± 0.32 | Vesicle exocytosis and actin filament dynamics (41) |
| Spectrin, αII | S1217 (PKA) | Q13813 | MS2, MS3 | 1.74 ± 0.31 | Ca2+/calmodulin-dependent movement of the cytoskeleton (42) |
| Transcytosis associated protein | S940(CK2) | O60763 | MS2, MS3 | Vesicle trafficking (43) | |
| Urea transporter UT-A1/3 | S35 (Cdk5), S62 (CK2), S63 (CK2), S486 (PKA) | Q96PH5 | MS2, MS3, MS4 | AVP increases UT-A1 phosphorylation at unknown site (14) |
Sites in bold italics have been previously identified. Abundance ratios are expressed as mean ± SE (dDAVP/control), n= 3.
*Indicates a site that was quantified.
†Indicates an identification containing ambiguous site(s).
**, Ratio is significant (P < 0.05).
Quantification of Protein Phosphorylation: Effects of Vasopressin.
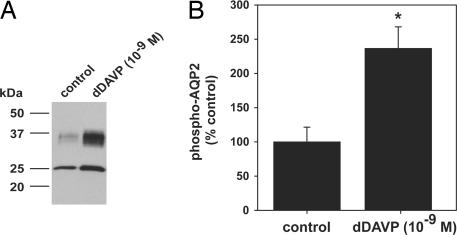
IMCD suspensions were incubated with 10−9 M dDAVP or vehicle for 10 min. Before LC-MSn analysis, samples were analyzed for AQP2 phosphorylation by immunoblotting (Fig. 3A) and dot blotting (Fig. 3B) by using a phosphospecific AQP2 antibody that recognizes phosphorylation at S256 (17). S256 phospho-AQP2 abundance was significantly increased, a hallmark of short-term dDAVP treatment (Fig. 3B). These samples were analyzed by LC-MSn neutral loss scanning by using the FT-ICR for the full ion scan to obtain the mass resolution required for accurate quantification.
Fig. 3.
Quantification of AQP2 phosphorylation in response to short-term dDAVP treatment by immunoblotting. (A) IMCD suspensions treated with dDAVP (10−9 M) or without (control) for 10 min. Immunoblots were probed by using a phosphospecific AQP2 antibody that recognizes phosphorylated S256. (B) Phosphorylated AQP2 levels were significantly increased with dDAVP treatment (dDAVP 237 ± 31.4% vs. control 100 ± 21.5%; ∗, P < 0.05).
Label-free quantification of phosphoproteins was performed using quoil (quantification without isotopic labeling) software (see Materials and Methods). This approach utilizes numerical integration of reconstructed ion chromatogram peaks from separate LC-MS runs to quantify peptides. A subset of phosphoproteins and associated quantification ratios are included in Table 1. Phosphoproteins that significantly increased in abundance with short-term dDAVP included AQP2 and LRRC47. Phosphoproteins that significantly decreased in abundance were Bclaf1, Rgl3, and SAFB2.
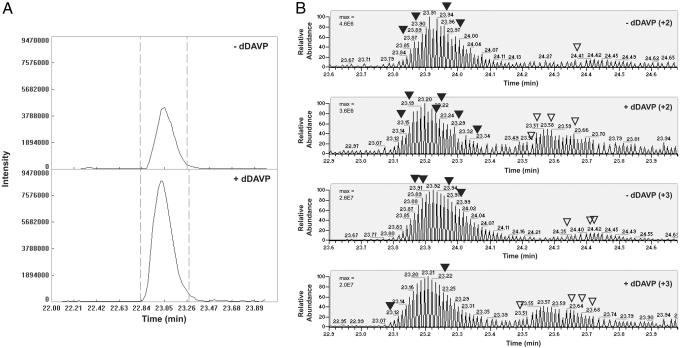
A detailed analysis of AQP2 peptide phosphorylation revealed that doubly phosphorylated AQP2 (pS256/261) appeared as a single peak in the reconstructed chromatogram. The abundance of this doubly phosphorylated form was increased by dDAVP as shown in Fig. 4A. For three pairs of samples, the average peak area ratio was 2.67 ± 0.84. A more complex profile emerged for singly phosphorylated AQP2 peptides (pS256 and pS261) (Fig. 4B). These chromatograms exhibited two distinct peak clusters: The left cluster was associated with pS261, whereas the one on the right was associated with pS256. Subsequent targeted ion selection (TIS) analysis increased the number of identifications from each of these reconstructed clusters and confirmed this association. Interestingly, these peaks underwent reciprocal changes in abundance in the presence of dDAVP, with pS261 decreasing 2.5-fold and pS256 increasing 7.2-fold. In addition, these changes were present for peptides at both +2 and +3 charge states.
Fig. 4.
MS quantification of AQP2 phosphorylation in response to dDAVP treatment. (A) Aligned chromatogram plots of overall peptide abundance for AQP2 phosphorylated at both S256 and S261. Retention time in minutes (x axis) and intensity (y axes). The doubly phosphorylated AQP2 peptide increased an average of 2.67-fold with dDAVP treatment. Values were calculated by using the “area under the curve” between the two cutoff lines (dashed lines). (B) Targeted ion selection (TIS) analysis of singly phosphorylated AQP2 peptides. Labeled peaks corresponding to either pS261 (▾) or pS256 (▿) indicate spectra that were confirmed by manual validation. Max, maximum peak intensity.
An analysis of mixtures of purified, synthetic AQP2 monophosphopeptide standards confirmed that pS256 and pS261 peptides have distinct elution profiles (Fig. 8, which is published as supporting information on the PNAS web site), demonstrating that pS256 ionizes more efficiently than pS261. Accounting for this differential ionization, it was possible to obtain normalized quantification ratios for pS261 vs. pS256 under both control and dDAVP treatment. We found that pS261 was ≈24-fold more abundant that pS256 under basal conditions, whereas pS261 and pS256 are almost equally abundant after short-term dDAVP treatment.
Taken together, these results indicate that differential phosphorylation can alter a peptide's affinity for the reverse phase LC column and thus its elution profile. Importantly, this phenomenon presents a potential caveat for automated quantification of phosphopeptides, demonstrating the requirement for both manual validation of identified peaks and manual adjustment of peak boundaries. A summary of quantification ratios for all singly and doubly phosphorylated AQP2 peptides is given in Table 2.
Table 2.
Summary of MS quantification of AOP2 phosphopeptides
| Site (charge state) | Sequence | Ratio ± SE (dDAVP/control) |
|---|---|---|
| S256 (+2) | R.RQS*VELHSPQSLPR.G | 9.29 ± 6.20 |
| S256 (+3) | R.RQS*VELHSPQSLPR.G | 5.06 ± 1.30** |
| S261 (+2) | R.RQSVELHS*PQSLPR.G | 0.43 ± 0.17** |
| S261 (+3) | R.RQSVELHS*PQSLPR.G | 0.37 ± 0.08** |
| S256/261 (+2) | R.RQS*VELHS*PQSLPR.G | 2.67 ± 0.84** |
*Indicates site of phosphorylation.
**, Considered significant (P< 0.05).
Discussion
This study presents a detailed phosphoproteomic analysis of IMCD using a “shotgun” approach involving four main steps: (i) isolation and in-solution digestion of IMCD proteins without prior subcellular fractionation, (ii) enrichment and fractionation of the resulting phosphopeptides by IMAC, (iii) identification of phosphopeptides through LC-MSn neutral loss scanning, and (iv) label-free quantification of phosphopeptides.
Crucial to this approach is extensive quality control to avoid false positive identifications, including stringent filtering of data sets and manual examination of the spectra. Filtering was based on several criteria, including peptide XCorr score, the presence of one or more phosphorylated residues, the number of unique peptides per protein, and the number of MS levels at which a particular identification was found. This process eliminated all nonphosphorylated peptides as well as the majority of poor-quality spectra. Moreover, spectra from smaller FT-ICR data sets were manually confirmed of the associated spectra, which often assisted in assigning exact phosphorylation sites. Consequently, all identifications presented in Tables 1 and 3–6 represent the highest quality identifications from much larger data sets, at the possible cost of losing some valid identifications.
This large-scale approach was effective in identifying a number of phosphoproteins with potential relevance to vasopressin signaling in collecting duct (Table 1), including a number of phosphorylation sites for well-known membrane transport proteins. We were able to detect phosphorylation of AQP2 at S256, a site in the C terminus previously shown to be important for vasopressin-stimulated trafficking to the apical plasma membrane (13, 18–20). In addition, three other sites in the C-terminal tail of AQP2 were also found to be phosphorylated: S261, S264, and S269. Although doubly phosphorylated AQP2 (pS256/261) and singly phosphorylated AQP2 at S256 (pS256) increased with dDAVP treatment, singly phosphorylated AQP2 at S261 (pS261) exhibited a decrease. We also demonstrated that pS261 is ≈24-fold more abundant than pS256 under basal conditions, whereas the amounts of these monophosphorylated forms are similar in the presence of dDAVP. This dramatic decrease in overall pS261 abundance in the presence of the hormone could be due to either dephosphorylation at S261 or phosphorylation at S256, which would create the doubly phosphorylated form. It will be important to address whether phosphorylation of S261 is a requirement for phosphorylation of S256 in vivo or whether these two events are independent. S261 is contained in a motif suggesting the involvement of a member of the “proline-directed” class of kinases, which includes mitogen-activated protein kinases, cdc2, and Cdk5. S269, which was not quantified in this study, is part of a potential PDZ (PSD95/Dlg/ZO-1)-binding motif that contributes to apical trafficking of AQP2 in transfected MDCK cells (21).
AQP4, another collecting duct water channel, was phosphorylated at S321 near its C terminus. This site is included in an established PDZ (PSD95/Dlg/ZO-1)-binding motif that has been implicated in both the localization and membrane stability of AQP4 in brain perivascular astrocytes (22).
This study also includes the identification of previously unknown phosphorylation sites in a urea transporter. The two isoforms, UT-A1 and UT-A3, expressed in collecting duct show increases in urea uptake in response to agents that increase intracellular cAMP (forskolin + 3-isobutyl-1-methylxanthine) when expressed in Xenopus oocytes (23). They are produced by alternative splicing from a single precursor RNA and share the same N-terminal 459 aa (24). Thus, it is impossible to assign residues phosphorylated near the N terminus (S35, S62, and S63) to either isoform. However, phosphorylation at S486 must occur in UT-A1 because this site is not present in UT-A3. Being a strong consensus site for protein kinase A (PKA), S486 may be the target for PKA-mediated phosphorylation of UT-A1 in response to short-term vasopressin treatment (14).
Quantitative phosphoproteomic approaches, such as those described in this study, offer great promise for rapid progress in the analysis of complex cell regulatory processes, especially when combined with more traditional biochemical approaches presently used for studying individual proteins. Moreover, the ability to simultaneously measure changes in phosphorylation state of many proteins in a single experiment can provide unique information needed for quantitative modeling of signaling pathways. Such an approach is likely to be especially effective when experiments are designed to reveal time courses of responses of multiple proteins, allowing the formulation of dynamical models.
Materials and Methods
Enrichment of Phosphopeptides from IMCD.
IMCD suspensions were prepared from inner medulla of rat kidney using the method of Stokes et al. (25) with some modifications (26). After isolation, IMCD suspensions were treated with 100 nM calyculin A for 30 min, 10−9 M dDAVP for 10 min, or vehicle control, followed by resuspension in 6 M guanidine·HCl/50 mM NH4HCO3. Samples were sonicated and then spun at 200,000 × g for 1 h at 4°C in an ultracentrifuge. The cleared 200,000 × g supernatant was used for subsequent analysis.
Protein samples (1–2 mg) were reduced with 50 mM DTT for 1 h at 56°C and then alkylated using 100 mM iodoacetamide for 1 h at room temperature in the dark. Samples were diluted in 50 mM NH4HCO3 to a final guanidine concentration ≤0.5 M before addition of trypsin (1:50 wt/wt). Digestion was carried out overnight at 37°C. Peptide samples were desalted using a 1-ml hydrophilic-lipophilic-balanced (HLB) cartridge (Oasis, Milford, MA), followed by volume reduction in vacuo. Strong cation exchange (SCX) was performed using a polySULFOETHYL A cartridge (Poly LC, Columbia, MD) using a gradient of 5–200 mM ammonium formate, pH 2.7, with 20% acetonitrile, followed by a final elution in 500 mM ammonium acetate, pH 6.5, with 20% acetonitrile. Eluted fractions were reduced to a minimal volume in vacuo and then resuspended in 20% acetonitrile, a process that was repeated three times.
Fractions were subsequently resuspended in 100 μl of 5% acetic acid, pH 2.5–3.0, and then loaded onto an IMAC column (Pierce) for phosphopeptide enrichment. Peptides were allowed to incubate with the Ga3+ resin for 30 min with gentle agitation every 10 min. Washing and elution conditions were performed according to the supplied protocol. Samples were then dried in vacuo, resuspended in 0.1% formic acid, and desalted with C18 Ziptips (Millipore) before analysis by MS.
LC-MSn Analysis of Phosphopeptides.
Isolated phosphopeptide samples were analyzed on an Agilent 1100 nanoflow system (Agilent Technologies, Palo Alto, CA) LC connected to a Finnigan LTQ FT mass spectrometer (Thermo Electron, San Jose, CA) equipped with a nanoelectrospray ion source. In experiments where the highest mass accuracy was required (i.e., quantification), the FT-ICR was used for the full MS (parent ion) scan, and subsequent MS2 and MS3 spectra were acquired by using the LTQ. These runs are designated “FT-ICR.” For runs where sensitivity was paramount and high mass accuracy was not critical, the LTQ was used exclusively for the full MS scan and subsequent spectra (MS2, MS3, and MS4). These runs are designated “LTQ.”
Peptide Identification, Validation, and Quantification.
MS spectra were analyzed by using bioworks software (Thermo Electron) running the sequest search algorithm for peptide identification. Peak masses were searched against the most current version of the Rat Refseq Database (National Center for Biotechnology Information) with the following parameters: fixed carbamidomethylation of Cys; variable phosphorylation of Ser, Thr, and Tyr when searching MS2; and variable phosphorylation of Ser, Thr, and Tyr as well as variable loss of water (−18 Da) from Ser and Thr when searching MS3 and MS4.
A preliminary exclusion filter was used to remove the poorest-quality spectra. This low-stringency filter removed any spectrum with the following values for the charge state of the peptide and the associated XCorr score: +1 ≤ 1.5; +2 ≤ 2.0; and +3 ≤ 2.5. XCorr takes into account the number of peaks matched between actual and theoretical spectra and is directly proportional to spectral quality (27). Phosphopeptide data sets were further filtered by using customized algorithms that removed nonphosphorylated peptides and the majority of poor-quality spectra (see Supporting Materials and Methods, which is published as supporting information on the PNAS web site).
Relative quantification of phosphopeptides was performed using quoil, an in-house software program designed for label-free protein quantification by LC-MS (28). This program is useful for quantifying a peptide's normalized chromatogram peaks among multiple LC-MS runs and is an alternative to expensive isotope-based labeling techniques (e.g., cICAT, SILAC, and iTRAQ). Briefly, a peptide's chromatogram peak in each LC-MS run is reconstructed based on its precursor m/z value. Quantitative data are obtained by comparing the peak areas against a chosen reference. The resulting ratios are then normalized, which reflects the relative quantity of a peptide (and hence the corresponding protein) in different samples. Identification of individual peaks from reconstructed ion chromatograms by targeted ion selection consisted of MS2 fragmentation of selected precursor ion masses for singly phosphorylated AQP2 peptides [m/z = 857.43 (+2), 571.95 (+3)].
Supplementary Material
Acknowledgments
We thank Dr. S. Nielsen (University of Aarhus, Denmark) for providing the p-AQP2 antibody and F. Trepiccione for help with database construction. This work was supported by the National Heart, Lung, and Blood Institute intramural budget ZO1-HL001285.
Abbreviations
- AQP
aquaporin
- dDAVP
(deamino-Cys1, d-Arg8)vasopressin
- FT
Fourier transform
- ICR
ion cyclotron resonance
- IMAC
immobilized metal affinity chromatography
- IMCD
inner medullary collecting duct
- LC
liquid chromatography
- MS
mass spectrometry
- n
multisequential
- UT-A1/3
urea transporter isoforms A1 and A3
- XCorr
cross-correlation.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Giorgianni F., Beranova-Giorgianni S., Desiderio D. M. Proteomics. 2004;4:587–598. doi: 10.1002/pmic.200300584. [DOI] [PubMed] [Google Scholar]
- 2.Jin W. H., Dai J., Zhou H., Xia Q. C., Zou H. F., Zeng R. Rapid Commun. Mass Spectrom. 2004;18:2169–2176. doi: 10.1002/rcm.1604. [DOI] [PubMed] [Google Scholar]
- 3.Gruhler A., Olsen J. V., Mohammed S., Mortensen P., Faergeman N. J., Mann M., Jensen O. N. Mol. Cell. Proteomics. 2005;4:310–327. doi: 10.1074/mcp.M400219-MCP200. [DOI] [PubMed] [Google Scholar]
- 4.Beausoleil S. A., Jedrychowski M., Schwartz D., Elias J. E., Villen J., Li J., Cohn M. A., Cantley L. C., Gygi S. P. Proc. Natl. Acad. Sci. USA. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amanchy R., Kalume D. E., Iwahori A., Zhong J., Pandey A. J. Proteome Res. 2005;4:1661–1671. doi: 10.1021/pr050134h. [DOI] [PubMed] [Google Scholar]
- 6.Nuhse T. S., Stensballe A., Jensen O. N., Peck S. C. Mol. Cell. Proteomics. 2003;2:1234–1243. doi: 10.1074/mcp.T300006-MCP200. [DOI] [PubMed] [Google Scholar]
- 7.Chabardes D., Firsov D., Aarab L., Clabecq A., Bellanger A. C., Siaume-Perez S., Elalouf J. M. J. Biol. Chem. 1996;271:19264–19271. doi: 10.1074/jbc.271.32.19264. [DOI] [PubMed] [Google Scholar]
- 8.Helies-Toussaint C., Aarab L., Gasc J. M., Verbavatz J. M., Chabardes D. Am. J. Physiol. 2000;279:F185–F194. doi: 10.1152/ajprenal.2000.279.1.F185. [DOI] [PubMed] [Google Scholar]
- 9.Hoffert J. D., Chou C. L., Fenton R. A., Knepper M. A. J. Biol. Chem. 2005;280:13624–13630. doi: 10.1074/jbc.M500040200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orloff J., Handler J. S. J. Am. Soc. Nephrol. 1999;10:1623–1630. [PubMed] [Google Scholar]
- 11.Star R. A., Nonoguchi H., Balaban R., Knepper M. A. J. Clin. Invest. 1988;81:1879–1888. doi: 10.1172/JCI113534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chou C. L., Christensen B. M., Frische S., Vorum H., Desai R. A., Hoffert J. D., de Lanerolle P., Nielsen S., Knepper M. A. J. Biol. Chem. 2004;279:49026–49035. doi: 10.1074/jbc.M408565200. [DOI] [PubMed] [Google Scholar]
- 13.Kuwahara M., Fushimi K., Terada Y., Bai L., Marumo F., Sasaki S. J. Biol. Chem. 1995;270:10384–10387. doi: 10.1074/jbc.270.18.10384. [DOI] [PubMed] [Google Scholar]
- 14.Zhang C., Sands J. M., Klein J. D. Am. J. Physiol. 2002;282:F85–F90. doi: 10.1152/ajprenal.0054.2001. [DOI] [PubMed] [Google Scholar]
- 15.Diella F., Cameron S., Gemund C., Linding R., Via A., Kuster B., Sicheritz-Ponten T., Blom N., Gibson T. J. BMC Bioinformatics. 2004;5:79. doi: 10.1186/1471-2105-5-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peri S., Navarro J. D., Amanchy R., Kristiansen T. Z., Jonnalagadda C. K., Surendranath V., Niranjan V., Muthusamy B., Gandhi T. K., Gronborg M., et al. Genome Res. 2003;13:2363–2371. doi: 10.1101/gr.1680803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishimoto G., Zelenina M., Li D., Yasui M., Aperia A., Nielsen S., Nairn A. C. Am. J. Physiol. 1999;276:F254–F259. doi: 10.1152/ajprenal.1999.276.2.F254. [DOI] [PubMed] [Google Scholar]
- 18.Katsura T., Gustafson C. E., Ausiello D. A., Brown D. Am. J. Physiol. 1997;272:F817–F822. [PubMed] [Google Scholar]
- 19.Fushimi K., Sasaki S., Marumo F. J. Biol. Chem. 1997;272:14800–14804. doi: 10.1074/jbc.272.23.14800. [DOI] [PubMed] [Google Scholar]
- 20.Deen P. M., van Balkom B. W., Savelkoul P. J., Kamsteeg E. J., Van Raak R. M., Jennings M. L., Muth T. R., Rajendran V., Caplan M. J. Am. J. Physiol. 2002;282:F330–F340. doi: 10.1152/ajprenal.0168.2001. [DOI] [PubMed] [Google Scholar]
- 21.Kuwahara M., Asai T., Terada Y., Sasaki S. Kidney Int. 2005;68:1999–2009. doi: 10.1111/j.1523-1755.2005.00654.x. [DOI] [PubMed] [Google Scholar]
- 22.Neely J. D., Amiry-Moghaddam M., Ottersen O. P., Froehner S. C., Agre P., Adams M. E. Proc. Natl. Acad. Sci. USA. 2001;98:14108–14113. doi: 10.1073/pnas.241508198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fenton R. A., Chou C. L., Stewart G. S., Smith C. P., Knepper M. A. Proc. Natl. Acad. Sci. USA. 2004;101:7469–7474. doi: 10.1073/pnas.0401704101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sands J. M. Annu. Rev. Physiol. 2003;65:543–566. doi: 10.1146/annurev.physiol.65.092101.142638. [DOI] [PubMed] [Google Scholar]
- 25.Stokes J. B., Grupp C., Kinne R. K. Am J. Physiol. 1987;253:F251–F262. doi: 10.1152/ajprenal.1987.253.2.F251. [DOI] [PubMed] [Google Scholar]
- 26.Hoffert J. D., van Balkom B. W., Chou C. L., Knepper M. A. Am. J. Physiol. 2004;286:F170–F179. doi: 10.1152/ajprenal.00223.2003. [DOI] [PubMed] [Google Scholar]
- 27.Sadygov R. G., Cociorva D., Yates J. R., III. Nat. Methods. 2004;1:195–202. doi: 10.1038/nmeth725. [DOI] [PubMed] [Google Scholar]
- 28.Wang G., Wu W. W., Zeng W., Chou C. L., Shen R.-F. J. Proteome Res. 2006 doi: 10.1021/pr050406g. in press. [DOI] [PubMed] [Google Scholar]
- 29.Lin F. T., Miller W. E., Luttrell L. M., Lefkowitz R. J. J. Biol. Chem. 1999;274:15971–15974. doi: 10.1074/jbc.274.23.15971. [DOI] [PubMed] [Google Scholar]
- 30.Marples D., Schroer T. A., Ahrens N., Taylor A., Knepper M. A., Nielsen S. Am. J. Physiol. 1998;274:F384–F394. doi: 10.1152/ajprenal.1998.274.2.F384. [DOI] [PubMed] [Google Scholar]
- 31.Tisdale E. J., Kelly C., Artalejo C. R. J. Biol. Chem. 2004;279:54046–54052. doi: 10.1074/jbc.M409472200. [DOI] [PubMed] [Google Scholar]
- 32.Kaida T., Kozawa O., Ito T., Tanabe K., Ito H., Matsuno H., Niwa M., Miyata H., Uematsu T., Kato K. Exp. Cell Res. 1999;246:327–337. doi: 10.1006/excr.1998.4277. [DOI] [PubMed] [Google Scholar]
- 33.Dihazi H., Asif A. R., Agarwal N. K., Doncheva Y., Muller G. A. Mol. Cell. Proteomics. 2005;4:1445–1458. doi: 10.1074/mcp.M400184-MCP200. [DOI] [PubMed] [Google Scholar]
- 34.Bustamante M., Hasler U., Kotova O., Chibalin A. V., Mordasini D., Rousselot M., Vandewalle A., Martin P. Y., Feraille E. Am. J. Physiol. 2005;288:F334–F344. doi: 10.1152/ajprenal.00180.2004. [DOI] [PubMed] [Google Scholar]
- 35.Zhan Q., Ge Q., Ohira T., Van D. T., Badwey J. A. J. Immunol. 2003;171:3785–3793. doi: 10.4049/jimmunol.171.7.3785. [DOI] [PubMed] [Google Scholar]
- 36.Jo I., Ward D. T., Baum M. A., Scott J. D., Coghlan V. M., Hammond T. G., Harris H. W. Am. J. Physiol. 2001;281:F958–F965. doi: 10.1152/ajprenal.2001.281.5.F958. [DOI] [PubMed] [Google Scholar]
- 37.Feng J., Ito M., Ichikawa K., Isaka N., Nishikawa M., Hartshorne D. J., Nakano T. J. Biol. Chem. 1999;274:37385–37390. doi: 10.1074/jbc.274.52.37385. [DOI] [PubMed] [Google Scholar]
- 38.Bos J. L. EMBO J. 1998;17:6776–6782. doi: 10.1093/emboj/17.23.6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oertle T., Schwab M. E. Trends Cell Biol. 2003;13:187–194. doi: 10.1016/s0962-8924(03)00035-7. [DOI] [PubMed] [Google Scholar]
- 40.Tamma G., Klussmann E., Maric K., Aktories K., Svelto M., Rosenthal W., Valenti G. Am. J. Physiol. 2001;281:F1092–F1101. doi: 10.1152/ajprenal.0091.2001. [DOI] [PubMed] [Google Scholar]
- 41.Hall P. A., Russell S. E. J. Pathol. 2004;204:489–505. doi: 10.1002/path.1654. [DOI] [PubMed] [Google Scholar]
- 42.Noda Y., Horikawa S., Katayama Y., Sasaki S. Biochem. Biophys. Res. Commun. 2005;330:1041–1047. doi: 10.1016/j.bbrc.2005.03.079. [DOI] [PubMed] [Google Scholar]
- 43.Sztul E., Colombo M., Stahl P., Samanta R. J. Biol. Chem. 1993;268:1876–1885. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.