Abstract
Escherichia coli RecA protein catalyzes the central DNA strand-exchange step of homologous recombination, which is essential for the repair of double-stranded DNA breaks. In this reaction, RecA first polymerizes on single-stranded DNA (ssDNA) to form a right-handed helical filament with one monomer per 3 nt of ssDNA. RecA generally binds to any sequence of ssDNA but has a preference for GT-rich sequences, as found in the recombination hot spot Chi (5′-GCTGGTGG-3′). When this sequence is located within an oligonucleotide, binding of RecA is phased relative to it, with a periodicity of three nucleotides. This implies that there are three separate nucleotide-binding sites within a RecA monomer that may exhibit preferences for the four different nucleotides. Here we have used a RecA coprotease assay to further probe the ssDNA sequence specificity of E.coli RecA protein. The extent of self-cleavage of a λ repressor fragment in the presence of RecA, ADP-AlF4 and 64 different trinucleotide-repeating 15mer oligonucleotides was determined. The coprotease activity of RecA is strongly dependent on the ssDNA sequence, with TGG-repeating sequences giving by far the highest coprotease activity, and GC and AT-rich sequences the lowest. For selected trinucleotide-repeating sequences, the DNA-dependent ATPase and DNA-binding activities of RecA were also determined. The DNA-binding and coprotease activities of RecA have the same sequence dependence, which is essentially opposite to that of the ATPase activity of RecA. The implications with regard to the biological mechanism of RecA are discussed.
INTRODUCTION
RecA is a DNA-dependent ATPase that promotes a DNA strand-exchange reaction that is the central step in the repair of double-stranded DNA breaks by homologous recombination (1–3). In this reaction, RecA first polymerizes on single-stranded DNA (ssDNA) formed at the lesion to form a right-handed, helical nucleoprotein filament, within which the ssDNA is bound in a highly extended conformation (4). Each RecA monomer binds to 3 nt of DNA and there are approximately six RecA monomers per turn of the helical filament (5–7). The RecA–ssDNA–ATP filament associates with duplex DNA and after a homology search the ssDNA is paired with the complementary strand of a homologous region of duplex DNA. In a side reaction in which RecA acts as a ‘coprotease,’ the RecA–ssDNA–ATP complex binds to and activates the self-cleavage of LexA and related phage repressors (8–10), which initiate the SOS response and a switch to lytic growth, respectively.
RecA binds to all sequences of ssDNA, primarily along the sugar phosphate backbone with the bases exposed for homology recognition (11). Nonetheless, RecA does bind preferentially to certain types of DNA sequences. In early studies it was noted that RecA binds preferentially to poly(dT), owing to its lack of secondary structure (12,13). In vitro selection for sequences of DNA that bind optimally to Escherichia coli RecA (14) pulled out GT-rich sequences that bear a striking resemblance to the recombination hotspot, Chi (5′-GCTGGTGG-3′). The same in vitro selection experiment using yeast Rad51, a eukaryotic homolog of RecA, pulled out a very similar set of GT-rich sequences (15), indicating that the sequence specificity is conserved and possibly inherent to some aspect of DNA structure such as its ability to be extended. Interestingly, similar GT-rich sequences are present in highly recombinogenic regions of DNA in higher eukaryotes, including microsatellites, Alu repeat elements and the constant regions of immunoglobulin heavy chains (14,15).
The binding of RecA to different sequences of ssDNA has been examined directly by various biophysical techniques. Using isothermal titration calorimetry to compare the binding of RecA with poly(dT), poly(dA) and poly(dC) revealed that RecA binds with a substantially more favorable enthalpy to poly(dT) (16). Using surface plasmon resonance to compare the binding of RecA and Rad51 with all possible dinucleotide-repeating 39mers revealed that both proteins bind strongly to CT, GT and CA-repeating sequences and weakly to GA, AT and GC-repeats (17,18). These differences were attributed largely to secondary structure of the ssDNA. Using fluorescence anisotropy to study the binding of RecA to different trinucleotide-repeating 39mers revealed that RecA binds tightly to TTT, CCC, TCC and TAC-repeating sequences and weakly to GAA, AAA, CGG and CAG repeats (19). It was concluded that unstacking of purines is an energetic barrier to RecA binding such that pyrimidine-rich sequences bind tightly to RecA. The differences in binding could be correlated with the calculated folding energies of the ssDNA (20).
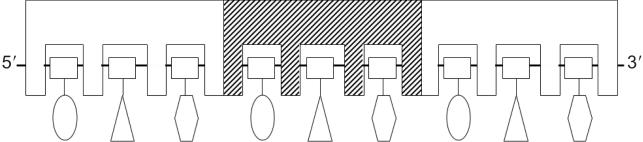
Two lines of evidence suggest that in the active state of the filament each RecA monomer binds exactly 3 nt of DNA. First, alignment of naturally occurring sequences of E.coli DNA that surround Chi sites reveals a striking TGG-repeating consensus sequence extending to at least 230 bp on either side of Chi (21). Second, DMS reactivity of Chi-containing oligonucleotides bound by RecA–ATPγS exhibits a phasing of precisely three nucleotides (22). These observations suggest that each monomer of RecA has three separate nucleotide-binding sites, each formed by a different constellation of amino acid residues (Figure 1). This gives rise to distinct preferences for different nucleotides at each of the three binding sites.
Figure 1.
Model for the binding of RecA to ssDNA. The figure is adapted from Ref. (22). Three RecA monomers are shown, and the ssDNA (9 nt) is bound along the sugar phosphate backbone, which is represented by the solid line and squares. Each RecA monomer has three separate nucleotide-binding sites. Although the bases are primarily exposed for homology recognition (11), in the model they form subtle interactions with the binding sites, giving rise to slight preferences for the different nucleotides and optimal binding to certain trinucleotide-repeating sequences.
To systematically test the ssDNA sequence specificity of E.coli RecA protein, we have used a coprotease assay to compare the binding of RecA with all 64 possible trinucleotide-repeating 15mer oligonucleotides. Since λ repressor cleavage is specific for the RecA–ssDNA–ATP complex, the extent of binding of RecA to a particular DNA sequence can be inferred from the rate of repressor cleavage. This assay is simple and rapid enough to compare large numbers of different sequences, and sensitive enough to show significant differences among them. By using 15mers as opposed to longer oligonucleotides, the inhibition of RecA–DNA binding due to secondary structure formation or intermolecular pairing of the DNA was likely minimized. We reasoned that this would allow the effects of the direct interaction between RecA and the ssDNA to be accentuated.
MATERIALS AND METHODS
Materials
All oligonucleotides were purchased from Integrated DNA Technologies and dissolved in ddH2O. The 48mer oligonucleotides were purified by ion-exchange high-performance liquid chromatography. Oligonucleotide concentrations are expressed in nucleotides and were measured by OD260 using extinction coefficients calculated from the sequences. M13 ssDNA was from New England Biolabs. ADP, ATP, NADH, phosphoenolpyruvate (PEP), pyruvate kinase type VII and lactic dehydrogenase type XXXIX were from Sigma–Aldrich. E.coli single-stranded DNA-binding (SSB) protein was from USB. All other chemicals were Fisher ACS certified grade.
Protein expression and purification
E.coli RecA was expressed and purified as described previously (23). Briefly, RecA was expressed as a His6 fusion protein from pET14b (Novagen) in E.coli BL21(DE3)pLysS cells. The protein was purified by Ni2+-affinity and anion exchange chromatography. The final RecA protein has the extra sequence Gly-Ser-His-Met at the N-terminus, but the protein has ssDNA-dependent coprotease and ATPase activities that are essentially indistinguishable from native RecA protein based on experiments with several different oligonucleotide sequences (data not shown). Residues 93–236 of λ repressor (cI93–236) were expressed and purified by a similar procedure, as described previously for cI132–236 (24). A hypercleavable form of λ repressor (cIhc) consisting of residues 93–229 and bearing the mutations P158T and A152T, was constructed using the QuickChange (Stratagene) procedure, and purified using the same procedure as for cI93–236. Concentrations of all proteins were determined by OD280 using extinction coefficients calculated from their amino acid sequences: 20 340 M−1cm−1 for RecA, 21 095 M−1cm−1 for cI93–236, and 15 220 M−1cm−1 for cIhc.
RecA coprotease assay
The self-cleavage of a fragment of λ repressor (cI93–236) was measured in the presence of RecA, ADP-AlF4 and 64 different trinucleotide-repeating 15mer oligonucleotides. Reactions (50 µl) included 10 µM RecA, 30 µM oligonucleotide, 1 mM ADP, 2 mM aluminum nitrate, 10 mM NaF, 20 mM Tris, pH 7.4, 2 mM MgCl2 and 50 mM NaCl. The above components were mixed and incubated at 25°C for 30 min, after which 10 µM cI93–236 was added and the mixture was incubated at 25°C for 20 min. The reaction was quenched by adding 0.25 vol of 5× SDS–PAGE loading buffer and immediately heating to 95°C for 5 min. Samples were run on a 13.5% SDS–PAGE gel and stained with coomassie brilliant blue. Dried gels were digitally scanned and the intensity of each band was integrated using Kodak Digital Science 1D image analysis software. The percentage cleavage was calculated from the net intensities of the bands corresponding to cleaved and uncleaved cI93–236 (residues 112–236 and 93–236, respectively). The net intensity of cleaved cI93–236 was multiplied by 1.16 (145/125) to account for the smaller size of the cleaved product relative to the uncleaved substrate. Reactions were done in triplicate and the mean and standard deviation are reported.
ATPase assay
The DNA-dependant ATPase activity of RecA in the presence of selected 48mer trinucleotide-repeating sequences was measured by a coupled spectrophotometric assay (25). The 500 µl reaction mixture contained 25 mM Tris, pH 7.4, 10 mM MgCl2, 10 mM DTT, 10 mM ATP, 20 mM PEP, 0.5 mM NADH, 30 U/ml each of pyruvate kinase and lactic dehydrogenase, 0.8 µM RecA and 3 µM oligonucleotide. The reactions were carried out at 37°C with an electrically heated cell holder. All reaction components except RecA were pre-mixed, and reactions were initiated by mixing in RecA. The OD340 was monitored at 1 min intervals using an Ultrospec 2100 pro UV/VIS spectrophotometer (Amersham Biosciences). In the reaction with 3 µM M13 ssDNA, 0.8 µM E.coli SSB protein was added after RecA. The ATPase rate (min−1) was calculated using the formula: (−dA/dt)(1/6300 M−1cm−1)(reaction volume)/[RecA], where dA/dt is the slope in the linear region of the plot of OD340 versus time (min). Each reaction was done in triplicate and the mean and standard deviation are reported.
DNA-binding assay
Binding of RecA to 48mer oligonucleotides was measured with the double filter-binding method (26). Binding reactions (50 µl) were in buffer B (25 mM Tris–acetate, pH 7.5, 4 mM Mg-acetate, 10 mM KCl and 1 mM DTT) and contained 1 mM ATPγS, ∼10 nM 32P-end-labeled 48mer oligonucleotide and 0–2 µM RecA. The reactions were equilibrated at 37°C for 30 min and then loaded onto a Minifold I 48-well slot-blot apparatus (Schleicher & Schuell) containing nitrocellulose (Bio-Rad) and DEAE (Whatman) filters, which were pre-treated as described previously (14,26) and equilibrated in buffer B prior to use. Samples were loaded into the wells, pulled through under vacuum, and washed with 1 ml of buffer B. The radioactivity on the filters was measured using a Storm 860 phosphorimager (Amersham Biosciences) and Image Quant 5.2 software (Molecular Dynamics), and the percentage bound for each sample was calculated from the net intensities of the bound (nitrocellulose) and unbound oligonucleotide (DEAE).
RESULTS
Dependence of RecA coptrotease activity on ssDNA sequence
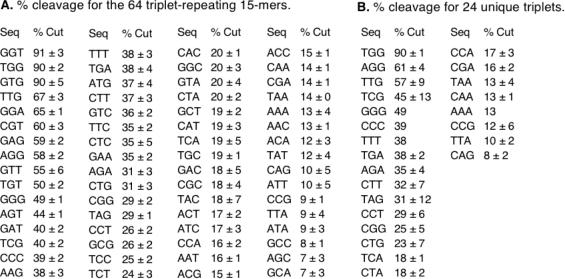
The cleavage of a λ repressor fragment (residues 93–236; cI93–236) in the presence of RecA, ADP-AlF4, and 64 different trinucleotide-repeating 15mer oligonucleotides was measured by SDS–PAGE. This fragment of λ repressor undergoes RecA-mediated cleavage in a similar manner as the full-length protein (27). The temperature of 25°C and time of 20 min were chosen in order to maximally distinguish the cleavage efficiencies in the presence of the 64 different trinucleotide-repeating 15mers. Figure 2 shows an example of the coprotease reaction for five different sequences, and Figure 3 shows the percentage cleavage in the presence of all 64 trinucleotide-repeating 15mers. The observed coprotease activity is highly dependent on the sequence of DNA. Under the conditions of the assay, the percentage cleavage values ranged from 91 to 7%. The values for 15mers containing different permutations of the same trinucleotide repeat (i.e. TGG, GTG and GGT) were observed to be very similar. Accordingly, mean values for each such group are shown in the set of 24 non-redundant trinucleotides in Figure 3B.
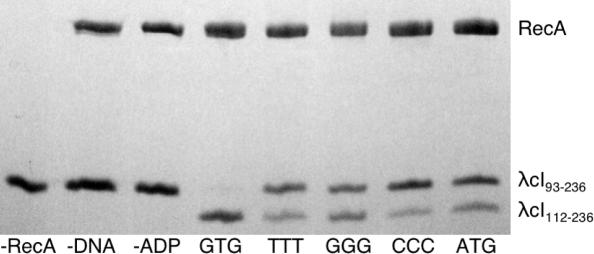
Figure 2.
RecA coprotease activity is highly dependent on the ssDNA sequence. The SDS–PAGE gel shows the self-cleavage of λcI93–236 to produce λcI112–236 in the presence of RecA, ADP-AlF4 and different trinucleotide-repeating 15mers, for a 20 min reaction as described in Materials and Methods. The first three lanes show that no cleavage is detected if RecA, ssDNA or ADP is omitted from the reaction. Notice that cleavage is particularly high for GTG.
Figure 3.
RecA coprotease activity in the presence of 64 trinucleotide-repeating 15mers. The value in the table is the percentage of λcI93–236 cleaved in a 20 min reaction at 25°C, as described in Materials and Methods. Each reaction was performed in triplicate, and the mean and standard deviation are reported. (A) Percentage cleavage values for all 64 possible trinucleotide-repeating 15mers, listed in order from high to low. (B) Mean percentage cleavage for the set of 24 non-redundant trinucleotides. The value in (B) is the average of those for the three different permutations (i.e. TGG, GTG and GGT) of each trinucleotide from (A). Notice that the TGG-repeating sequences give by far the highest coprotease activity.
By far the highest coprotease activity was observed with TGG-repeating sequences, which gave 90% cleavage in the 20 min reaction. The next best trinucleotides were AGG and TTG, which gave ∼60% cleavage. These are similar to TGG, but with the substitutions of T to A and G to T, respectively. Trinucleotide-repeating sequences that gave the lowest coprotease activity (∼10% cleavage) tended to have combinations of GC or AT. Among the polynucleotides, cleavage was highest for GGG (49%), followed by CCC (39%), TTT (38%) and AAA (13%).
From examination of Figure 3, it is clear that it is the sequence of nucleotides within a trinucleotide, and not simply the composition, that determines the ability to promote RecA coprotease activity. This is most evident in comparing the cleavage for TCG (45%) and CTG (23%), which differ in sequence but not in composition. Similarly, it is not simply the pattern of pyrimidines and purines within a trinucleotide that is important, since TGG gives the highest cleavage (90%), whereas CAG (grouped with AGC and GCA) gives the lowest cleavage (8%). Thus, individual bases have specific properties that give rise to a strong nucleotide sequence dependence of RecA coprotease activity.
Dependence of RecA coprotease activity on oligonucleotide length
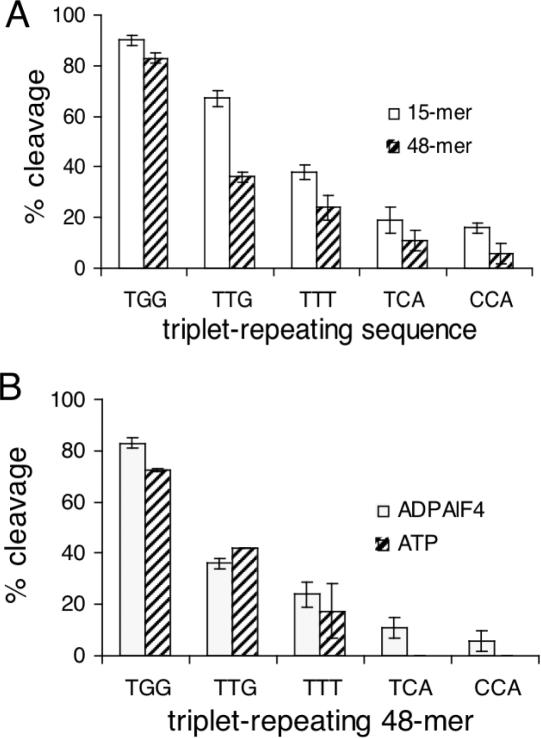
For the reasons described above, 15mer trinucleotide-repeating oligonucleotides were chosen for an exhaustive comparison. To test whether the same sequence dependence of RecA coprotease activity is observed for longer oligonucleotides, cleavage reactions for five selected trinucleotide-repeats (TGG, TTG, TTT, TCA and CCA) were compared for 15mer and 48mer oligonucleotides. For these experiments the ratio of RecA to DNA was fixed at 3 nt per RecA monomer so that the reactions with 48mer oligonucleotides contained fewer molecules of ssDNA, but the same number of nucleotides. The results show definitively that the length of the oligonucleotide does not change the sequence specificity of RecA coprotease activity (Figure 4A). Interestingly, slightly but significantly lower coprotease activity was observed for all of the 48mer sequences, which is somewhat counter-intuitive since the binding of RecA to ssDNA is known to be highly cooperative (28,29).
Figure 4.
DNA sequence specificity of RecA coprotease activity is not dependent on oligonucleotide length or type of ATP cofactor. (A) The percentage cleavage of cI93–236 in the presence of five trinucleotide-repeating sequences was determined for 15mer and 48mer oligonucleotides under otherwise identical conditions (as described in Figure 2). In all reactions there are 3 nt of ssDNA per monomer of RecA. The error bars show the standard deviation from three separate reactions. The 15mers (open bars) give slightly but significantly higher coprotease activity than the 48mers (closed bars), but the sequence dependence of RecA coprotease activity is the same. (B) The RecA coprotease activity is compared for reactions in the presence of ADP-AlF4 (open bars) and ATP (closed bars). All reactions are with 48mer oligonucleotides. Since the cleavage is normally dramatically slower with ATP, reactions in the presence of ATP used a hypercleavable form of λ repressor (see text), but under otherwise identical conditions as reactions with ADP-AlF4.
Dependence of RecA coprotease activity on type of ATP cofactor
The coprotease assays described above were carried out in the presence of ADP-AlF4, which gives the highest coprotease activity of any ATP analog tested. In order to determine whether the DNA sequence dependence of RecA coprotease activity was the same under conditions of ATP hydrolysis, reactions in the presence of ATP were attempted. However, with ATP as the cofactor cleavage is much less efficient, and there was no detectable cleavage of cI93–236 for reactions with 15mer or 48mer oligonucleotides, even after several hours at 37°C. In order to overcome this problem, coprotease reactions with ATP were performed with a hypercleavable form of λ repressor consisting of residues 93–229 and bearing the mutations P158T and A152T (hereafter referred to as cIhc). In Figure 4B, the coprotease activity of RecA in the presence of ADP-AlF4 (with cI93–236) and ATP (with cIhc) is compared for five selected 48mer oligonucleotides under otherwise identical conditions. The DNA sequence dependences of RecA coprotease activity in the presence of ADP-AlF4 and ATP are essentially the same. With TCA and CCA-repeating 48mers in the presence of ATP, there was no detectable cleavage of cIhc in the 20 min reactions. Thus we conclude that although the repressor cleavage reaction is dramatically slower under conditions of ATP hydrolysis, the dependence of the reaction on the DNA sequence is the same as with ADP-AlF4.
Detailed comparison of TGG, GGT and GTG
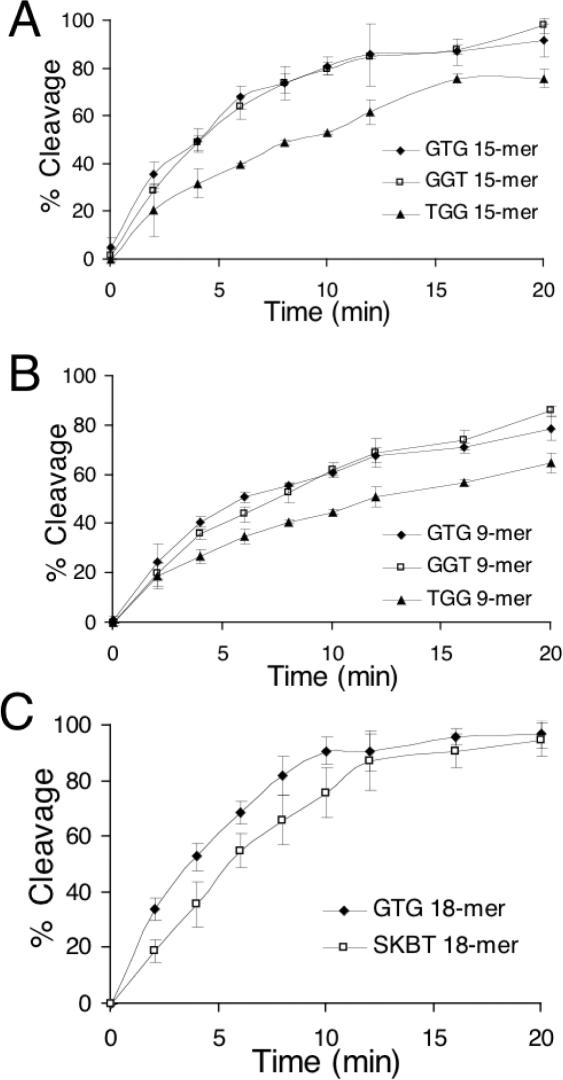
The initial experiments showed that GGT, GTG and TGG-repeating 15mers gave by far the highest RecA coprotease activity. These three oligonucleotides differ only in the point at which they start. In order to determine whether there is a single best trinucleotide repeat, these three 15mers were examined more closely by doing a time course for the cleavage reactions (Figure 5). The rationale for doing this experiment is as follows. If, for example, the optimal trinucleotide is TGG, then a TGG-repeating 15mer will have five ideal sites, whereas GTG and GGT-repeating sequences will only have four. One might expect this effect to be accentuated for 9mer sequences as compared with 15mers since the difference in the number of ideal sites would be more significant (2 versus 3 as compared with 4 versus 5).
Figure 5.
Time course of RecA coprotease activity with three different TGG-repeating sequences and the top in vitro-selected sequence. The percentage cleavage of cI93–236 in the presence of TGG, GTG and GGT-repeating sequences is plotted versus time for (A) 15mer oligonucleotides, and (B) 9mer oligonucleotides. GTG and GGT give significantly higher coprotease activity than TGG. At early time points, GTG gives slightly higher coprotease activity than GGT. (C) Comparison of a GTG-repeating 18mer and the top in vitro-selected sequence, 5′-GCGTGTGTGGTGGTGTGC-3′ (SKBT) (14,15). The value at each time-point is the average of three separate reactions and the error bars show the standard deviations.
The results of the time course show that GTG and GGT-repeating sequences give significantly higher rates of cleavage than TGG-repeats, both for the 15mers and for the 9mers. At lower time points GTG is consistently better than GGT, although the differences are not statistically significant at any single time-point. The differences do not appear to be accentuated for 9mers as compared with 15mers, as was hypothesized above. These results indicate that the ideal trinucleotide for RecA coprotease activity is GTG, with GGT, a very close second and TGG, a more distant third. As a final test, a GTG-repeating 18mer and the top 18mer sequence from in vitro selection, 5′-GCGTGTGTGGTGGTGTGC-3′ (14), were compared in a time course for the coprotease reaction (Figure 5C). The GTG-repeating sequence gives significantly higher coprotease activity, particularly at the earlier time points.
Dependence of RecA ATPase activity on DNA sequence
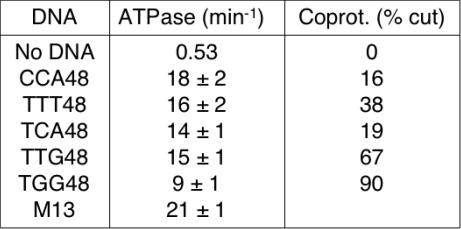
As RecA is a DNA-dependent ATPase, measurements of ATPase rate have commonly been used to indirectly assess the binding of RecA to DNA. We sought to determine whether the dependence of RecA coprotease activity on DNA sequence that we observed above was also observed for RecA ATPase activity. A coupled spectrophotometric ATPase assay was used to compare the RecA ATPase activity in the presence of five different trinucleotide-repeating sequences that gave a wide range of percentage cleavage values (TGG, TTG, TTT, TCA and CCA). Whereas 15mer sequences did not give any detectable ATPase activity (data not shown), 48mer sequences gave ATPase rates approaching that of M13 ssDNA (Figure 6), as has been observed previously (30). Surprisingly, TGG produced by far the lowest ATPase rate (9 min−1) of the five trinucleotide-repeating sequences examined, while CCA gave the highest rate (18 min−1). Thus, the sequence with the highest coprotease activity gives the lowest ATPase activity, and vice versa. In general, such an inverse correlation was observed, but did not hold exactly for all of the five sequences tested; TCA gives very low coprotease activity, but only moderate ATPase activity. We conclude that both the ATPase and coprotease activities of RecA are highly dependent on the sequence of DNA to which RecA is bound, but that the preferred sequences for the two activities are remarkably different.
Figure 6.
Dependence of RecA ATPase activity on the sequence of ssDNA. The rate of RecA ATP hydrolysis was determined in the presence of five different trinucleotide-repeating 48mer oligonucleotides using a coupled spetrophotometric assay, as described in Materials and Methods. The ATPase rate is shown together with the coprotease activity from Figure 3. Notice that the coprotease activity is highest for TGG and lowest for CCA, while the opposite is true for the ATPase activity.
Dependence of RecA DNA-binding on DNA sequence
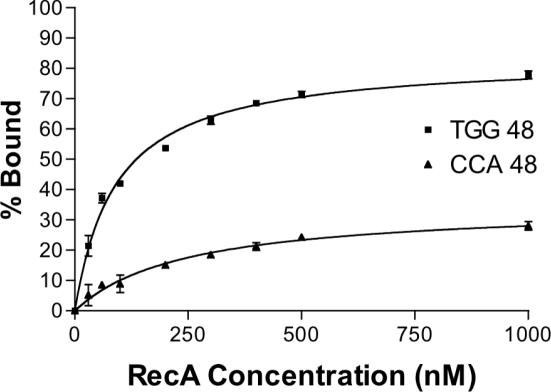
In order to directly examine the binding of RecA to different sequences of ssDNA, a double filter-binding assay was employed (26). Briefly, RecA and 32P-end-labeled oligonucleotide were incubated and passed through two filters. First a nitrocellulose filter retains protein and protein-bound oligonucleotide. The unbound oligonucleotide that passes through is then trapped on a DEAE filter. The oligonucleotide on each filter is quantified by phosphorimaging, and the percentage of DNA bound can be determined. 48mer oligonucleotides of TGG and CCA repeats were selected for this experiment due to their contrasting effects on the coprotease and ATPase activities of RecA. The binding experiments were done at ∼10 nM oligonucleotide and 0–2 µM RecA. The resulting binding curves (Figure 7) show that RecA bound substantially better to the TGG-repeating 48mer than to CCA. Whereas a maximum of ∼83% of the TGG repeat was retained on the nitrocellulose filter, only ∼35% of the CCA repeat was maximally retained. Moreover, the apparent KD estimated from these measurements is 91 nM for the TGG-repeating 48mer and 246 nM for the CCA-repeating 48mer. We attempted to obtain similar binding curves for TTT, TCA and TTG-repeating 48mers, but a high level of binding of these oligonucleotides to the nitrocellulose filter in the absence of RecA complicated these experiments.
Figure 7.
Binding of RecA to TGG and CCA-repeating 48mer oligonucleotides. The amount of 32P-end-labeled 48mer bound to RecA as a function of RecA concentration was determined by the double filter-binding method (26). The percentage bound values reported are the average of three experiments and error bars indicate the standard deviation. Notice that RecA binds much more tightly to the TGG repeat than to the CCA repeat.
In conclusion, the double filter-binding assay shows that RecA binds much more tightly to TGG-repeating sequences than to CCA repeats. Thus, based on this limited comparison, the sequence dependence of RecA–DNA binding is apparently similar to that of RecA coprotease activity, while that of RecA ATPase activity is different.
DISCUSSION
Here we have used a coprotease assay to systematically compare the binding of RecA with all possible trinucleotide-repeating 15mer oligonucleotides. The coprotease activity of RecA is highly dependent on the DNA sequence: the percentage cleavage for each sequence was reproducible and varied from 90% to <10%. Thus, the coprotease assay is a highly efficient and sensitive method for comparing the binding of RecA to different sequences of ssDNA. The most significant result from this analysis is that TGG-repeating sequences (including GGT and GTG) stand out as giving by far the highest coprotease activity.
Basis of sequence specificity of coprotease
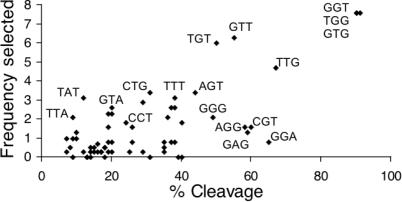
What is the physical basis for the strong preference for TGG-repeating sequences in promoting RecA coprotease activity? We consider two possibilities. First, TGG-repeating sequences could induce a particular conformation of the RecA filament, such as a precise degree of extension, that is just right for binding of the repressor and promoting the self-cleavage reaction. Alternatively, the higher coprotease activity may simply reflect the tighter binding of RecA to TGG-repeating sequences than to other sequences, giving rise to a higher percentage of RecA bound under the conditions of the assay. While the first possibility is intriguing, there is very little information regarding how different sequences of ssDNA affect the structure of RecA. On the other hand, there is evidence supporting the second possibility. First, in the binding experiment of Figure 7, the TGG-repeating 48mer, which gave the highest coprotease activity, clearly binds to RecA better than the CCA repeat, which gave low coprotease activity. Second, based on an in vitro selection experiment (14), TGG, GTG and GGT were among the most frequently occurring trinucleotides within the selected sequences. In fact, for all trinucleotides there is a general correlation between the ability to activate RecA coprotease activity and the frequency of occurrence in the selected sequences (Figure 8). Since the in vitro selection experiment is based on binding, we therefore conclude that the observed differences in coprotease activity seen for the sequences are very likely to be largely, if not entirely, due to differences in extent of RecA binding.
Figure 8.
Correlation between RecA–DNA binding and coprotease activity for the 64 trinucleotides. ‘Frequency Selected’ is the number of times each trinucleotide occurs within 24 in vitro-selected 18mer sequences, divided by the total number of trinucleotide occurrences [taken from Table 3 of Ref. (14)]. Percentage of cleavage is the value for each trinucleotide-repeating 15mer reported in Figure 3. Each point in the plot is a single trinucleotide, and the trinucleotides with highest percentage cleavage or frequency selected are labeled. Notice that there is a general correlation between trinucleotides that give high coprotease activity and those that are frequently selected.
Based on the time course of the coprotease reactions for the three different permutations of TGG-repeats, it was determined that the best trinucleotide is GTG, closely followed by GGT and more distantly TGG. Interestingly, in the in vitro selection experiments, GTG was found to be the most frequently selected trinucleotide, both for E.coli RecA (14), in which case it was tied with TGG, and especially in the case of yeast Rad51, in which case it was significantly higher than GGT and TGG (15). Thus, together the three studies point to GTG as being the most preferred trinucleotide.
Validity of using trinucleotide-repeating sequences
In the coprotease assay, we focused on comparing all possible trinucleotide-repeating sequences, for the reasons described above. How appropriate was this choice? Although it seems clear that one monomer of RecA binds to precisely three nucleotides of ssDNA (21,22), it is conceivable that neighboring monomers within a filament interact with one another in such a way as to influence one another's sequence specificity, thus giving rise to sequence preferences extending beyond the context of a trinucleotide. In the in vitro selection experiments, which selected an 18 nt sequence flanked by two 18 nt fixed sequences, precisely the same sequence, 5′-GCGTGTGTGGTGGTGTGC-3′, was clearly the most frequently selected, both for E.coli RecA and yeast Rad51 (14,15). This would tend to support the idea that the specificity of the RecA–DNA interaction extends beyond the trinucleotide. It is possible, however, that the fixed 18 nt flanking regions, which are needed for PCR amplification, somehow influenced the preferred nucleotides at positions within the selected region, particularly at the ends where non-GTG trinucleotides were found. Consistent with this latter interpretation, we found that the GTG-repeating 18mer gives slightly higher coprotease activity than an 18mer corresponding to the top sequence from in vitro selection (Figure 5C), suggesting that it binds to RecA more tightly.
Another point to consider is that while it has been observed that RecA binds in a phased manner with a period of 3 nt to oligonucleotides containing the Chi sequence (22), the phasing of RecA on the sequences used in this study has not been examined. Thus, it is possible, especially for the weaker binding sequences, that RecA is not bound in homogeneous alignment for all molecules within a given sample. Nonetheless, with these caveats in mind, since the trinucleotide is the basic unit of ssDNA to which a monomer of RecA binds, it seems clear that trinucleotide-repeating sequences are the most appropriate choice for determining the sequence specificity of RecA in a comprehensive, systematic manner.
Basis of sequence specificity of binding
If we assume that the observed differences in coprotease activity are due to differences in binding, then what is the physical basis for this? The binding of RecA to a particular sequence of DNA could be influenced by (i) the intermolecular interactions between the amino acid residues within the three binding sites on RecA and the four different nucleotides of DNA and/or (ii) some intrinsic property of the DNA itself, such as its propensity to be extended, form intramolecular secondary structure (folding) or intermolecular pairing interactions. The observation from in vitro selection experiments that both E.coli RecA and yeast Rad51 exhibit remarkably similar sequence preferences would tend to suggest that they are due to some intrinsic property of the DNA itself, although it is conceivable that both proteins have evolved to form interactions that favor the same sequences.
The sequence preferences seen in Figure 3 would tend to suggest that both intermolecular interactions between RecA and ssDNA as well as intrinsic properties of the DNA are at play. On the one hand, the fact that the sequences that give the lowest coprotease activity tend to have consecutive GC or AT pairs indicates that secondary structure or intermolecular pairing does to some extent play a role in dictating binding affinity, at least for those sequences. On the other hand, the differences in coprotease activity cannot simply be accounted for by differences in secondary structure formation of the DNA or ability to be extended. For example, TTT-repeating sequences, which form minimal secondary structure and stacking interactions, are clearly not the most preferred sequences for RecA-binding and coprotease activity. In addition, purine bases are known to resist the unstacking required for forming the extended conformation. Yet, the optimal sequences are not pyrimidine-rich, but instead contain two consecutive guanosines. In fact, GGG gave significantly higher coprotease activity than TTT or CCC, indicating that clearly the penalty for unstacking the purine bases does not play a dominant role in dictating the sequence specificity. Thus, it would appear that the atomic interactions between the amino acid residues of the binding sites on RecA and the individual nucleotides of ssDNA play a significant role in dictating the sequence specificity.
Contrasting preferences for ATPase and coprotease activities
Coprotease activity is high for TGG-repeating sequences and low for CCA-repeating sequences (90 and 17% cleavage, respectively). By contrast, the ATPase rate is highest for the CCA-repeating 48mer, and lowest by far for the TGG-repeat (18 and 9 min−1, respectively). Since the coprotease activity correlates well with binding, as seen in Figures 7 and 8, it is clear that ATPase rate does not. Thus, while the ATPase activity of RecA is dependent on its being bound to DNA, apparently sequences that bind to RecA optimally actually prevent maximal ATPase activity. Conceivably, this could be due to an effect on release of the products of ATP hydrolysis (ADP and Pi) as opposed to the hydrolysis event per se. In the active state of the filament, which is required for ATP hydrolysis, ATP is bound at the interface between neighboring subunits (31–33). Release of ADP and Pi would require a local subunit reorientation similar to that seen in the inactive compressed state (34), in which the ATP site is exposed toward the central axis of the filament. Perhaps this conformational transition is somewhat restricted when RecA is bound to TGG-repeating sequences.
In addition to the differences in sequence preference, the oligonucleotide length has contrasting effects on the ATPase and coprotease activities. Whereas coprotease activity is higher on 15mers as compared with 48mers, ATPase activity is high on 48mers and undetectable on 15mers. The dependence of ATPase activity on longer sequences has been well established (30), and is generally attributed to the highly cooperative behavior of the DNA-dependent ATPase activity of RecA (35). The observation that coprotease activity is actually higher on shorter sequences is less well documented. In the coprotease assay, the ratio of RecA to nucleotides of ssDNA was fixed at 1:3. Therefore, assuming complete binding, with 15mer oligonucleotides there will be a greater number of filaments than with 48mers. If the binding of repressor to a particular site on a filament prevents binding of additional repressor molecules to neighboring subunits, as is in fact suggested by EM studies of a RecA–LexA complex (36), then a greater number of shorter filaments would therefore give rise to higher coprotease activity, as is observed. An alternative explanation, as has been noted (16), is that longer DNA sequences possess more nucleation sites for RecA, and thus have a higher potential to form discontinuous filaments. This would be especially problematic in the presence of ATPγS or ADP-AlF4, where there is less dissociation and redistribution of RecA monomers than with ATP.
Biological implications
It has been shown previously that RecA binds preferentially to TGG-rich sequences, that a TGG-repeating consensus pattern is found around Chi sites in the E.coli genome, and that TGG-repeating sequences are more recombinogenic than other sequences (14,15). The implications with regard to similar GT-rich sequences found in higher eukaryotes, such as Alu repeat elements, microsatellites, and the constant regions of immunoglobulin heavy chains have been discussed (14,15). Two new observations that come out of our study are that (i) RecA coprotease activity is highest on TGG-repeating sequences, and (ii) RecA ATPase activity is significantly lower on TGG-repeating sequences than on other sequences.
Biologically, it makes sense that the coprotease activity should be highest on the GT-rich sequences found around Chi sites where RecA filaments often originate, in order to provide a rapid and robust signal for LexA destruction and initiation of the SOS response. Similarly, high coprotease activity on short RecA filaments might allow for rapid LexA destruction, even for relatively short ssDNA regions formed at single strand gaps. It also makes sense that ATPase activity should be low on the TGG-rich sequences where filaments often originate, since ATP hydrolysis is not required for filament formation and is in fact coupled to release of RecA monomers from the 5′ end of the filaments (37,38). Thus, the low ATPase activity on the TGG-rich sequences that we observe may be important for stabilizing initial filament formation at Chi sites. It has been shown previously that RecA does in fact dissociate more slowly from a GT-rich, in vitro-selected 18mer sequence than its complement, though this was not attributed to differences in ATPase rates (14).
ATP hydrolysis is necessary, however, for later stages of strand-exchange such as branch migration past regions of heterology (39,40), strand-exchange beyond ∼3000 nt or in a unidirectional 5′–3′ manner (41), and recycling of RecA monomers at the end of strand exchange (37). All of these activities presumably occur on long filaments, away from the GT-rich sites where filaments tend to originate. Thus, it appears that the sequence preferences of the DNA-binding (14), coprotease, and ATPase activities of RecA may be finely tuned, together with a sequence bias at recombination hotspots (21), to maximize efficiency of recombination in vivo.
CONCLUSIONS
RecA coprotease activity is an effective means of probing the sequence specificity of the RecA–ssDNA interaction. TGG-repeating sequences, including GTG and GGT, induce particularly high coprotease activity. Based on RecA–ssDNA binding measurements and comparison to previous in vitro selection experiments, the enhanced ability of these sequences to induce RecA coprotease activity is attributed largely to their efficient binding, as opposed to their stabilizing a particular conformational state. Contrasting sequence and length preferences are seen for RecA ATPase and coprotease activities, which can be rationalized in terms of their biological roles. Overall this study highlights properties of RecA–DNA filaments that are mechanistically informative.
Acknowledgments
We thank Dr Dieudonné Ndjonka for providing the hypercleavable fragment of lambda repressor used for measuring coprotease activity under conditions of ATP hydrolysis. This work was supported by NIH grant GM 067947 (to C.E.B.). Funding to pay the Open Access publication charges for this article was provided by NIH GM 067947.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bianco P.R., Tracy R.B., Kowalczykowski S.C. DNA strand exchange proteins: a biochemical and physical comparison. Front. Biosci. 1998;3:570–603. doi: 10.2741/a304. [DOI] [PubMed] [Google Scholar]
- 2.Cox M.M. The bacterial RecA protein as a motor protein. Annu. Rev. Microbiol. 2003;57:551–577. doi: 10.1146/annurev.micro.57.030502.090953. [DOI] [PubMed] [Google Scholar]
- 3.McGrew D.A., Knight K.L. Molecular design and functional organization of the RecA protein. Crit. Rev. Biochem. Mol. Biol. 2003;38:385–432. doi: 10.1080/10409230390242489. [DOI] [PubMed] [Google Scholar]
- 4.Stasiak A., DiCapua E., Koller T. Elongation of duplex DNA by recA protein. J. Mol. Biol. 1981;151:557–564. doi: 10.1016/0022-2836(81)90010-3. [DOI] [PubMed] [Google Scholar]
- 5.DiCapua E., Engel A., Stasiak A., Koller T. Characterization of complexes between recA protein and duplex DNA by electron microscopy. J. Mol. Biol. 1982;157:87–103. doi: 10.1016/0022-2836(82)90514-9. [DOI] [PubMed] [Google Scholar]
- 6.Egelman E.H., Stasiak A. Structure of helical RecA–DNA complexes. Complexes formed in the presence of ATP-gamma-S or ATP. J. Mol. Biol. 1986;191:677–697. doi: 10.1016/0022-2836(86)90453-5. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi M., Kubista M., Nordén B. Binding stoichiometry and structure of RecA–DNA complexes studied by flow linear dichroism and fluorescence spectroscopy. Evidence for multiple heterogeneous DNA co-ordination. J. Mol. Biol. 1989;205:137–147. doi: 10.1016/0022-2836(89)90371-9. [DOI] [PubMed] [Google Scholar]
- 8.Craig N.L., Roberts J.W. E.coli recA protein-directed cleavage of phage lambda repressor requires polynucleotide. Nature. 1980;283:26–30. doi: 10.1038/283026a0. [DOI] [PubMed] [Google Scholar]
- 9.Craig N.L., Roberts J.W. Function of nucleoside triphosphate and polynucleotide in Escherichia coli recA protein-directed cleavage of phage lambda repressor. J. Biol. Chem. 1981;256:8039–8044. [PubMed] [Google Scholar]
- 10.Little J.W. Autodigestion of lexA and phage lambda repressors. Proc. Natl Acad. Sci. USA. 1984;81:1375–1379. doi: 10.1073/pnas.81.5.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leahy M.C., Radding C.M. Topography of the interaction of recA protein with single-stranded deoxyoligonucleotides. J. Biol. Chem. 1986;261:6954–6960. [PubMed] [Google Scholar]
- 12.McEntee K., Weinstock G.M., Lehman I.R. Binding of the recA protein of Escherichia coli to single- and double-stranded DNA. J. Biol. Chem. 1981;256:8835–8844. [PubMed] [Google Scholar]
- 13.Amaratunga M., Benight A.S. DNA sequence dependence of ATP hydrolysis by RecA protein. Biochem. Biophys. Res. Commun. 1988;157:127–133. doi: 10.1016/s0006-291x(88)80022-6. [DOI] [PubMed] [Google Scholar]
- 14.Tracy R.B., Kowalczykowski S.C. In vitro selection of preferred DNA pairing sequences by the Escherichia coli RecA protein. Genes Dev. 1996;10:1890–1903. doi: 10.1101/gad.10.15.1890. [DOI] [PubMed] [Google Scholar]
- 15.Tracy R.B., Baumohl J.K., Kowalczykowski S.C. The preference for GT-rich DNA by the yeast Rad51 protein defines a set of universal pairing sequences. Genes Dev. 1997;11:3423–3431. doi: 10.1101/gad.11.24.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wittung P., Ellouze C., Maraboeuf F., Takahashi M., Nordén B. Thermochemical and kinetic evidence for nucleotide-sequence-dependent RecA-DNA interactions. Eur. J. Biochem. 1997;245:715–719. doi: 10.1111/j.1432-1033.1997.00715.x. [DOI] [PubMed] [Google Scholar]
- 17.Biet E., Sun J., Dutreix M. Conserved sequence preference in DNA binding among recombination proteins: an effect of ssDNA secondary structure. Nucleic Acids Res. 1999;27:596–600. doi: 10.1093/nar/27.2.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dutreix M. (GT)n repetitive tracts affect several stages of RecA-promoted recombination. J. Mol. Biol. 1997;273:105–113. doi: 10.1006/jmbi.1997.1293. [DOI] [PubMed] [Google Scholar]
- 19.Bar-Ziv R., Libchaber A. Effects of DNA sequence and structure on binding of RecA to single-stranded DNA. Proc. Natl Acad. Sci. USA. 2001;98:9068–9073. doi: 10.1073/pnas.151242898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tracy R.B., Chedin F., Kowalczykowski S.C. The recombination hot spot Chi is embedded within islands of preferred DNA pairing sequences in the E.coli genome. Cell. 1997;90:205–206. doi: 10.1016/s0092-8674(00)80328-1. [DOI] [PubMed] [Google Scholar]
- 22.Volodin A.A., Camerini-Otero R.D. Influence of DNA sequence on the positioning of RecA monomers in RecA–DNA cofilaments. J. Biol. Chem. 2002;277:1614–1618. doi: 10.1074/jbc.M108871200. [DOI] [PubMed] [Google Scholar]
- 23.Xing X., Bell C.E. Crystal structures of Escherichia coli RecA in a compressed helical filament. J. Mol. Biol. 2004;342:1471–1485. doi: 10.1016/j.jmb.2004.07.091. [DOI] [PubMed] [Google Scholar]
- 24.Bell C.E., Frescura P., Hochschild A., Lewis M. Crystal structure of the lambda repressor C-terminal domain provides a model for cooperative operator binding. Cell. 2000;101:801–811. doi: 10.1016/s0092-8674(00)80891-0. [DOI] [PubMed] [Google Scholar]
- 25.Morrical S.W., Lee J., Cox M.M. Continuous association of Escherichia coli single-stranded DNA binding protein with stable complexes of recA protein and single-stranded DNA. Biochemistry. 1986;25:1482–1494. doi: 10.1021/bi00355a003. [DOI] [PubMed] [Google Scholar]
- 26.Wong I., Lohman T.M. A double-filter method for nitrocellulose-filter binding: application to protein-nucleic acid interactions. Proc. Natl Acad. Sci. USA. 1993;90:5428–5432. doi: 10.1073/pnas.90.12.5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sauer R.T., Ross M.J., Ptashne M. Cleavage of the lambda and P22 repressors by recA protein. J. Biol. Chem. 1982;257:4458–4462. [PubMed] [Google Scholar]
- 28.Menetski J.P., Kowalczykowski S.C. Interaction of recA protein with single-stranded DNA. Quantitative aspects of binding affinity modulation by nucleotide cofactors. J. Mol. Biol. 1985;181:281–295. doi: 10.1016/0022-2836(85)90092-0. [DOI] [PubMed] [Google Scholar]
- 29.Takahashi M., Strazielle C., Pouyet J., Daune M. Co-operativity value of DNA RecA protein interaction. Influence of the protein quaternary structure on the binding analysis. J. Mol. Biol. 1986;189:711–714. doi: 10.1016/0022-2836(86)90501-2. [DOI] [PubMed] [Google Scholar]
- 30.Bianco P.R., Weinstock G.M. Interaction of the RecA protein of Escherichia coli with single-stranded oligodeoxyribonucleotides. Nucleic Acids Res. 1996;24:4933–4939. doi: 10.1093/nar/24.24.4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conway A.B., Lynch T.W., Zhang Y., Fortin G.S., Fung C.W., Symington L.S., Rice P.A. Crystal structure of a Rad51 filament. Nature Struct. Mol. Biol. 2004;11:791–796. doi: 10.1038/nsmb795. [DOI] [PubMed] [Google Scholar]
- 32.VanLoock M.S., Yu X., Yang S., Lai A.L., Low C., Campbell M.J., Egelman E.H. ATP-mediated conformational changes in the RecA filament. Structure. 2003;11:187–196. doi: 10.1016/s0969-2126(03)00003-0. [DOI] [PubMed] [Google Scholar]
- 33.Wu Y., He Y., Moya I.A., Qian X., Luo Y. Crystal structure of archaeal recombinase RADA: a snapshot of its extended conformation. Mol. Cell. 2004;15:423–435. doi: 10.1016/j.molcel.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 34.Story R.M., Weber I.T., Steitz T.A. The structure of the E.coli recA protein monomer and polymer. Nature. 1992;355:318–325. doi: 10.1038/355318a0. [DOI] [PubMed] [Google Scholar]
- 35.Weinstock G.M., McEntee K., Lehman I.R. Hydrolysis of nucleoside triphosphates catalyzed by the recA protein of Escherichia coli. Steady state kinetic analysis of ATP hydrolysis. J. Biol. Chem. 1981;256:8845–8849. [PubMed] [Google Scholar]
- 36.Yu X., Egelman E.H. The LexA repressor binds within the deep helical groove of the activated RecA filament. J. Mol. Biol. 1993;231:29–40. doi: 10.1006/jmbi.1993.1254. [DOI] [PubMed] [Google Scholar]
- 37.Rosselli W., Stasiak A. Energetics of RecA-mediated recombination reactions. Without ATP hydrolysis RecA can mediate polar strand exchange but is unable to recycle. J. Mol. Biol. 1990;216:335–352. doi: 10.1016/S0022-2836(05)80325-0. [DOI] [PubMed] [Google Scholar]
- 38.Bork J.M., Cox M.M., Imnam R.B. RecA protein filaments disassemble in the 5′ to 3′ direction on single-stranded DNA. J. Biol. Chem. 2001;276:45740–45743. doi: 10.1074/jbc.M109247200. [DOI] [PubMed] [Google Scholar]
- 39.Rosselli W., Stasiak A. The ATPase activity of RecA is needed to push the DNA strand exchange through heterologous regions. EMBO J. 1991;10:4391–4396. doi: 10.1002/j.1460-2075.1991.tb05017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim J.I., Cox M.M., Inman R.B. On the role of ATP hydrolysis in RecA protein-mediated DNA strand exchange I. Bypassing a short heterologous insert in one DNA substrate. J. Biol. Chem. 1992;267:16438–16443. [PubMed] [Google Scholar]
- 41.Jain S.K., Cox M.M., Inman R.B. On the role of ATP hydrolysis in RecA protein-mediated DNA strandexchange III. Unidirectional branch migration and extensive hybrid DNA formation. J. Biol. Chem. 1994;269:20653–20661. [PubMed] [Google Scholar]