Abstract
Small heat-shock proteins (sHsps) are widespread molecular chaperones for which a peroxisomal localization has not yet been reported. The Arabidopsis (Arabidopsis thaliana) genome encodes two sHsps with putative peroxisomal targeting signals type 1 or 2 (PTS1 or PTS2). As demonstrated by double-labeling experiments using full-length fusion proteins with enhanced yellow fluorescent protein and deletion constructs lacking the putative targeting domains, AtHsp15.7 (At5g37670) and AtAcd31.2 (At1g06460) are targeted to the peroxisome matrix by a functional PTS1 (SKL>) and a functional PTS2 (RLx5HF), respectively. The peroxisomal localization of AtAcd31.2 was further confirmed by isolation of leaf peroxisomes from Arabidopsis by two successive sucrose density gradients, protein separation by one- and two-dimensional gel electrophoresis, and mass spectrometric protein identification. When AtHsp15.7 and AtAcd31.2 were heterologously expressed in yeast (Saccharomyces cerevisiae) and directed to the cytosol by deletion of the PTSs, both sHsps were able to complement the morphological phenotype of yeast mutants deficient in the cytosolic homologs ScHsp42 or ScHsp26. According to expression studies by reverse transcription-PCR, AtAcd31.2 is constitutively expressed, whereas AtHsp15.7 is hardly expressed under normal conditions but strongly induced by heat and oxidative stress, the latter of which was triggered by the catalase inhibitor 3-aminotriazole or the herbicide methyl viologen applied by watering of whole plants or infiltration of rosette leaves. Thus, plants are exceptional among eukaryotes in employing sHsps in the peroxisome matrix to prevent unspecific aggregation of partially denatured proteins under both physiological and stress conditions.
In eukaryotes, many hydrogen peroxide (H2O2)-generating oxidases have been compartmentalized in peroxisomes, where the toxic by-product H2O2 can immediately be degraded by catalase (CAT) at the site of production. Further antioxidative enzymes of plant peroxisomes, such as superoxide dismutase, membrane-bound ascorbate peroxidase, and monodehydroascorbate reductase, play auxiliary roles in the detoxification of reactive oxygen species (ROS; Lisenbee et al., 2005; for review, see del Rio et al., 2002). Depending on their tissue specificity and specialization on further metabolic pathways, several variants of microbodies occur in higher plants, including the two main types, leaf peroxisomes of mesophyll cells involved in the recycling of P-glycolate formed during photosynthesis (Reumann, 2002), and glyoxysomes of triacylglyceride-storing tissues like endosperm and cotyledons, which mediate fatty acid β-oxidation during seed germination (Beevers, 1979).
Apart from the enzymes of these well-known metabolic pathways, few peroxisomal matrix proteins have been characterized to date at the molecular level, mainly because biochemical methods are generally not suitable for the identification of low-abundance proteins and polypeptides encoded by inducible genes. Recently, however, some evidence has emerged for the existence of nonenzymatic proteins like molecular chaperones in peroxisomes (Wimmer et al., 1997; Diefenbach and Kindl, 2000). Heat-shock proteins (Hsps) are expressed in response to increased temperature and other forms of abiotic stress (Young et al., 2003; Wang et al., 2004) and facilitate as chaperones the folding of newly synthesized or the refolding of partially denatured polypeptides. Two homologs of different Hsp classes have been associated with plant peroxisomes. An Hsp70 homolog from Citrullus vulgaris was shown to be targeted to both chloroplasts and peroxisomes (Wimmer et al., 1997), and a DnaJ (Hsp40) homolog from Cucumis sativus was found to be attached to the glyoxysomal membrane (Diefenbach and Kindl, 2000).
Small Hsps (sHsps) are widespread and powerful molecular chaperones that prevent the aggregation of nascent and stress-accumulated misfolded proteins (Narberhaus, 2002; Haslbeck et al., 2005). These chaperones are characterized by a small polypeptide chain (16–42 kD) and contain a conserved C-terminal α-crystallin domain of about 90 amino acid residues, which is homologous to α-crystallin proteins of the vertebrate eye lens (DeJong et al., 1998). The subunits assemble into oligomeric structures with varying degrees of order and a substantial divergence in size (4–50 subunits; Haslbeck et al., 1999, 2004; van Montfort et al., 2001; Sun et al., 2002). As exemplified by Arabidopsis (Arabidopsis thaliana), plants house an exceptionally large family of 19 closely related sHsp homologs plus 25 more distantly related proteins containing the same α-crystallin domain (Vierling, 1991; Scharf et al., 2001). Homologs of sHsps have been localized to several subcellular compartments, including the cytosol, plastids, mitochondria, and the endoplasmic reticulum (Banzet et al., 1998; Härndahl et al., 1999; Scharf et al., 2001); however, experimental evidence for targeting of sHsp paralogs to peroxisomes has not been provided for any organism previously, to our knowledge.
Peroxisomal matrix proteins are nuclear-encoded, synthesized on free cytoplasmic ribosomes, and directed to their destination by peroxisome targeting signals (PTS) in a conserved protein targeting pathway. Most of the known peroxisomal matrix proteins contain a PTS1, the C-terminal so-called SKL motif, or a PTS2, which is an N-terminal cleavable nonapeptide of the prototype RLx5HL (for review, see Johnson and Olsen, 2001). Both targeting motifs have been specified for plant peroxisomes by experimental and bioinformatics-based strategies (Reumann, 2004 and refs. therein). To identify novel low-abundance proteins of plant peroxisomes, we screened the Arabidopsis genome for genes encoding proteins with putative PTSs (Arabidopsis Genome Initiative, 2000; Reumann et al., 2004) and identified two putative sHsps, namely AtHsp15.7 and AtAcd31.2 (At5g37670 and At1g06460, respectively; Scharf et al., 2001). In the course of this study, the corresponding cDNAs were cloned, and subcellular protein targeting, gene expression, and functional complementation of yeast (Saccharomyces cerevisiae) sHsp knockout mutants were analyzed to gain first insights into the function of the putative chaperones in plant peroxisomes.
RESULTS
Subcellular Localization Studies of Putative Peroxisomal sHsps
The Arabidopsis genome encodes two predicted proteins possessing both an α-crystallin domain (Hsp20, Pfam00011) and putative targeting signals for plant peroxisomes (Reumann et al., 2004). The smaller homolog, referred to as AtHsp15.7 throughout this study (At5g37670, previously AtHsp15.7-CI for cytosolic class I; Scharf et al., 2001), carries the putative major PTS1 SKL> with high peroxisome targeting probability and a predicted mitochondrial presequence (Reumann, 2004; Fig. 1A). The second sHsp homolog AtAcd31.2 (At1g06460) contains a longer N-terminal extension and an α-crystallin domain of lower E value (E value = 5.8 × 10−6) compared to AtHsp15.7 (E = 3.2 × 10−24), indicating a higher sequence divergence of AtAcd31.2 from the defined Hsp20 motif (HMMER, hmmer.wustl.edu; Fig. 1, A and B). Phylogenetic analysis of sHsp homologs from diverse eukaryotes shows that several subclades of Arabidopsis sHsps and proteins containing one or several α-crystallin domains (Acd proteins) diverged early in evolution (Scharf et al., 2001; Supplemental Fig. 1). The homolog AtAcd31.2 carries both a predicted PTS2 nonapeptide (RLx5HF) and a predicted PTS1 tripeptide (PKL>; Fig. 1A), both of which have been defined as minor PTS peptides and presumably indicate peroxisome targeting with moderate probability (Reumann, 2004). A search of expressed sequence tag (EST) databases for plant homologs that share high sequence similarity with these proteins demonstrated that the putative PTS1 of AtHsp15.7 and the putative PTS2 of AtAcd31.2 in particular are largely conserved, suggesting that these proteins represent orthologs and are generally targeted to peroxisomes in higher plants (Supplemental Fig. 2).
Figure 1.
Domain structure and predicted targeting signals of AtHsp15.7 and AtAcd31.2. A, For both putative sHsps from plant peroxisomes, the presence of a conserved Hsp20/α-crystallin domain (pfam00011) and several subcellular targeting signals were predicted. B, For representative members of the Arabidopsis family of sHsps and Acd proteins a multiple sequence alignment of the most conserved regions of the α-crystallin domain, consensus I and II, and the E values of the Hsp20 domain are presented (HMMER, hmmer.wustl.edu). The Hsp20/α-crystallin domain is located in the C-terminal end of AtHsp15.7 (previously AtHsp15.7-CI, residues 18–134, score 105.0, E value = 3.2 × 10−24) and AtAcd31.2 (residues 182–285, score 26.0, E = 5.8 × 10−6). Notably, the E value of the Hsp20 domain of some sHsps like AtHsp18.5-CI and AtHsp14.7-P is higher, reflecting lower sequence conservation, than that of some Acd proteins including AtAcd31.2 (see also Supplemental Table II). For AtHsp15.7, a mitochondrial presequence (TargetP, 0.82; DBSubLoc, 91%; Predotar, 0.565; Mitoprot, 0.569) and a putative PTS1 are predicted. AtAcd31.2 contains both a putative PTS2 and PTS1 (Reumann et al., 2004; AraPerox, www.araperox.uni-goettingen.de). The PTS1 and the PTS2 of AtHsp15.7 and AtAcd31.2, respectively, were experimentally shown to be functional, but not the mitochondrial presequence of AtHsp15.7 nor the predicted PTS1 of AtAcd31.2 (see Fig. 2). Light and dark gray shading indicate the localization of the α-crystallin domain and the putative mitochondrial presequence, respectively.
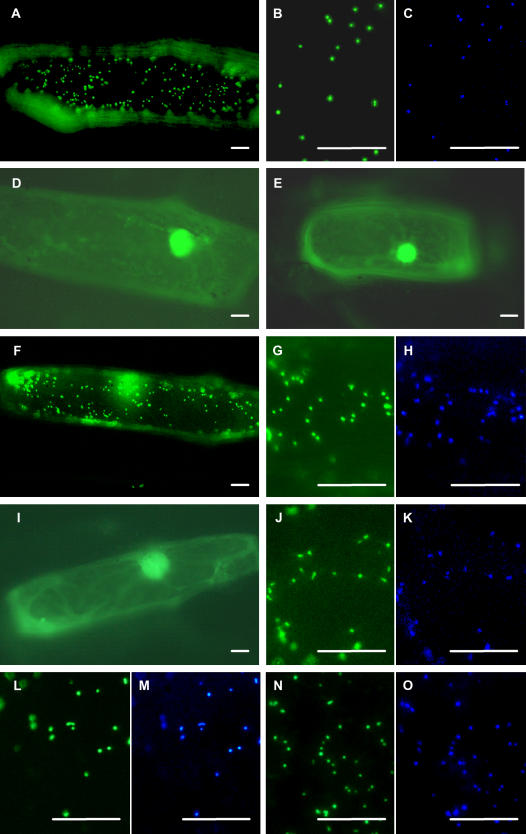
The cDNAs of AtHsp15.7 and AtAcd31.2 were cloned by reverse transcription (RT)-PCR from flowers and cold-stressed rosette leaves, respectively, and fused in frame to either of both ends of enhanced yellow fluorescent protein (EYFP) in a plant expression vector under the control of a 2-fold 35S promoter of the Cauliflower mosaic virus (CaMV; Fulda et al., 2002). Onion (Allium cepa) epidermal cells were transformed biolistically, and subcellular protein targeting of the fusion proteins was analyzed upon transient gene expression by fluorescence microscopy. The fusion protein EYFP:AtHsp15.7 with accessible C-terminal tripeptide SKL> was targeted to small punctate structures that moved quickly along cytoplasmic strands in living cells in single transformants (Fig. 2A). These punctate structures coincided with peroxisomes labeled with a control fusion protein between the PTS2 domain of glyoxysomal malate dehydrogenase from C. sativus (CsgMDH) and enhanced cyan fluorescent protein (CsgMDH:ECFP; Fulda et al., 2002), as shown for double transformants expressing simultaneously EYFP:AtHsp15.7 and CsgMDH:ECFP (Fig. 2, B and C). By contrast, mitochondria labeled with a fusion protein comprising the presequence of mitochondrial cytochrome c oxidase from yeast in frame to the N-terminal end of ECFP (ScCOX:ECFP; Fulda et al., 2002) differed from the organelles labeled with EYFP:AtHsp15.7 (data not shown).
Figure 2.
Subcellular targeting analysis of AtHsp15.7 and AtAcd31.2 in onion epidermal cells. The cDNAs of AtHsp15.7 and AtAcd31.2 were fused at the N- or the C-terminal end to EYFP under the control of a double 35S CaMV promoter, and subcellular protein targeting was analyzed in onion epidermal cells by fluorescence microscopy. In single transformants, the fusion protein EYFP:AtHsp15.7 was targeted to punctate cell structures (A) that coincided with peroxisomes, as shown by double labeling with the peroxisomal ECFP fusion protein CsgMDH:ECFP in double transformants using appropriate filter sets (B and C). The deletion construct EYFP:AtHsp15.7ΔPTS1 (D) and the inverse fusion protein AtHsp15.7:EYFP with accessible predicted mitochondrial presequence (E) remained in the cytosol. The fusion protein AtAcd31.2:EYFP with accessible PTS2 (RLx5HF) was targeted to peroxisomes (F–H), whereas site-directed mutagenesis of the PTS2 from RLx5HF to RLx5DF abolished peroxisome targeting (I). The inverse fusion protein EYFP:AtAcd31.2 was targeted to peroxisomes as well (J and K). In this case, however, deletion (EYFP:AtAcd31.2ΔPTS1; L and M) or mutagenesis of the C-terminal tripeptide (PKL→PEL; N and O) did not abolish peroxisome targeting. For imaging, either EYFP- (A, B, D–G, I, J, L, and N) or ECFP-specific filters were used (C, H, K, M, and O). The bar represents 20 μm.
Upon deletion of the putative C-terminal targeting domain from AtHsp15.7, the shortened fusion protein EYFP:AtHsp15.7ΔPTS1 remained in the cytosol (Fig. 2D; Supplemental Table I), demonstrating that the C-terminal domain is necessary for targeting of AtHsp15.7 to peroxisomes and that SKL> is the PTS1 of the protein. The inversely arranged fusion protein AtHsp15.7:EYFP with accessible N-terminal end was detected in the cytosol, strongly suggesting that AtHsp15.7 lacks a functional mitochondrial presequence (Fig. 2E).
The C-terminal fusion protein of AtAcd31.2, referred to as AtAcd31.2:EYFP, with accessible N-terminal PTS2 was also targeted to peroxisomes, as shown by double labeling (Fig. 2, F–H). The deletion construct AtAcd31.2ΔPTS2:EYFP lacking the 29 most N-terminal residues including the putative PTS2 (RLx5HF, position 11–19) was no longer targeted to peroxisomes. In line with a moderately confident prediction of a transit peptide in the N-terminal end of this deletion construct (e.g. TargetP: 0.70), however, this fusion protein entered plastids, as indicated by the larger size of the organelles and their characteristic stromuli extensions (data not shown). Conclusive evidence that the N-terminal domain of AtAcd31.2 possesses a functional PTS2 was obtained by site-directed mutagenesis of the putative PTS2 nonapeptide. When the strictly conserved His residue at position 8 of the putative PTS2 (Kato et al., 1998; Reumann, 2004) was mutated to Asp (RLx5HF→RLx5DF), the full-length fusion protein remained cytosolic (Fig. 2I).
To investigate if AtAcd31.2 contains in addition to the PTS2 a functional PTS1, EYFP was fused to its N-terminal end allowing the C-terminal tripeptide PKL> of AtAcd31.2 to be recognized by the cytosolic PTS1 receptor, Pex5p. The full-length fusion protein EYFP:AtAcd31.2 was likewise targeted to peroxisomes labeled with ECFP (Fig. 2, J and K). However, deletion of the C-terminal tripeptide (EYFP:AtAcd31.2ΔPTS1) did not abolish peroxisome targeting (Fig. 2, L and M). Likewise, a mutated construct, in which the essential basic residue of the putative PTS1 was exchanged to Glu (PKL→PEL), remained peroxisomal (Fig. 2, N and O). In summary, these results demonstrated that AtHsp15.7 and AtAcd31.2 are both peroxisomal proteins that are targeted to the matrix by a functional PTS1 and PTS2, respectively, and that the putative PTS1 of AtAcd31.2 is not required for peroxisome targeting.
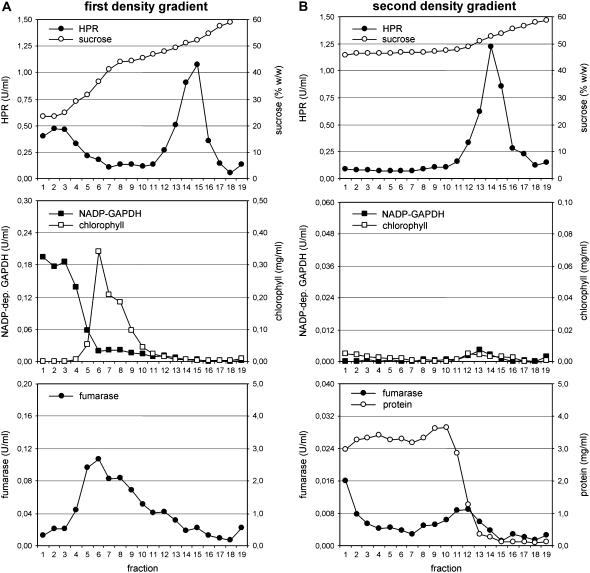
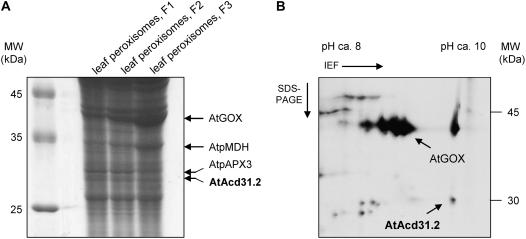
To provide a second independent line of evidence for targeting of these novel sHsps to plant peroxisomes, a method was established to isolate leaf peroxisomes from Arabidopsis. In a first Suc density gradient a significant quantity of intact leaf peroxisomes was efficiently separated from chloroplasts, thylakoids, and mitochondria and enriched near the bottom of the gradient, as indicated by the activities of appropriate organelle marker enzymes (Fig. 3A). A second Suc density gradient was added to achieve flotation of residual contaminating plastids and mitochondria to the upper fractions and concentration of leaf peroxisomes near the bottom of the gradient (Fig. 3B). Leaf peroxisomal proteins isolated from standard Arabidopsis plants were separated by one- or two-dimensional gel electrophoresis. Candidate protein bands and spots in the lower Mr and basic isoelectric point (IEP) range corresponding to the predicted size and IEP of AtAcd31.2 (AtAcd31.2: MW = 31.2, IEP = 9.82) were in-gel digested by trypsin and subsequently identified by mass spectrometry. In line with the constitutive expression of AtAcd31.2 (see below), a protein band of an apparent molecular mass of about 30 kD, as well as a distinct protein spot of the same approximate molecular mass and an IEP of about 9.5, was identified as AtAcd31.2 from at least two independent one- and two-dimensional gels, respectively (see Fig. 4, A and B, for representative gel images). On the basis of the peptide mass fingerprinting data, the protein was identified as At1g06460 (gi 15221505) with sequence coverages greater than 40%. The protein identification was further confirmed by tandem mass spectrometry (MS/MS) and sequence analysis of at least two tryptic peptides that had been assigned to AtAcd31.2 in the fingerprint analysis. In an analogous manner, leaf peroxisomal control proteins (Fig. 4, A and B) were identified by a combination of peptide mass fingerprint and MS/MS data.
Figure 3.
Purification of leaf peroxisomes from Arabidopsis. Leaf peroxisomes from Arabidopsis were enriched by differential centrifugation and purified by a first (A), followed by a second Suc density gradient (B). The concentrations of protein and chlorophyll and the activities of appropriate marker enzymes for different cell compartments were analyzed to determine the purity of leaf peroxisomes (leaf peroxisomes, hydroxypyruvate reductase, HPR; chloroplast stroma, NADP-dep. glyceraldehyde dehydrogenase, NADP-GAPDH; mitochondria, fumarase). Note that the scale between the first and the second gradient is identical for HPR but reduced by a factor of five for NADP-dep. GAPDH, fumarase, and chlorophyll. Due to the high BSA concentration, the protein concentration was not determined in the first gradient.
Figure 4.
Identification of AtAcd31.2 in isolated Arabidopsis leaf peroxisomes. Leaf peroxisomes were isolated from Arabidopsis by two successive Suc density gradients and the proteins separated by SDS-PAGE (A) or by isoelectric focusing (nonlinear IPG strip, pH 3–10) followed by denaturing SDS-PAGE (B). In one-dimensional electrophoresis (A), the protein profile of leaf peroxisomes enriched by one Suc density gradient (F1) was compared to that of the upper (F2, about fractions 12 and 13, Fig. 4) and lower fractions (F3, about fractions 14 and 15, Fig. 4) of the second density gradient of lower and higher specific HPR activity, respectively, loading 200 μg protein per lane. For two-dimensional electrophoresis, about 50 μg protein of leaf peroxisomes enriched by two Suc density gradients (F3) were analyzed. Candidate protein bands and spots in the lower Mr and basic IEP range corresponding to the predicted size and pI (IEP) of AtAcd31.2 (MW = 31.2, IEP = 9.82) and several leaf peroxisomal control proteins (GOX, glycolate oxidase, At3g14420; pMDH, peroxisomal MDH, At5g09660; pAPX3, peroxisomal ascorbate peroxidase, At4g35000) were identified by mass spectrometry. One protein band (A) and a single protein spot (B) were identified as AtAcd31.2 (At1g06460, gi 15221505).
Yeast Complementation Studies
Activity analyses of putative sHsps generally rely on in vitro refolding assays using recombinant sHsps and denatured citrate synthase as model substrate (Lee, 1995; Buchner et al., 1998). Haslbeck et al. (2004) recently reported a morphological phenotype of heat-stressed yeast knockout mutants that are deficient in cytosolic sHsps and that now allow functional complementation studies of heterologously expressed sHsp genes from other organisms. Yeast expresses two sHsps, referred to as ScHsp26 and ScHsp42 (Supplemental Fig. 1), both of which are localized in the cytosol and show a broad and unspecific substrate specificity (Petko and Lindquist, 1986; Wotton et al., 1996; Haslbeck et al., 1999, 2004). At late logarithmic phase and when subjected to a heat shock, the morphology of the deletion strains changes dramatically compared to the wild type, resembling wrinkled cells undergoing dehydration or aging, as observed by scanning electron microscopy (SEM). Based on further lines of evidence including increased protein aggregation in the deletion strains (see Haslbeck et al., 2004), the altered cell morphology is thought to reflect a general disturbance in proteome homeostasis in the sHsp-deficient mutants.
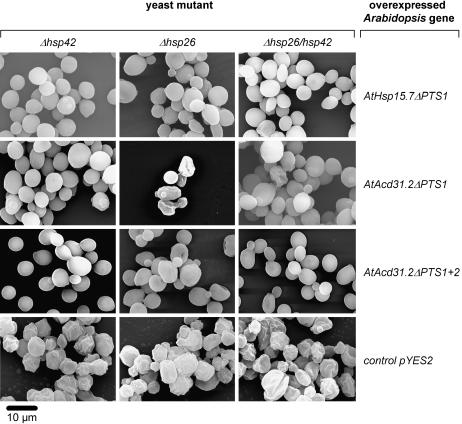
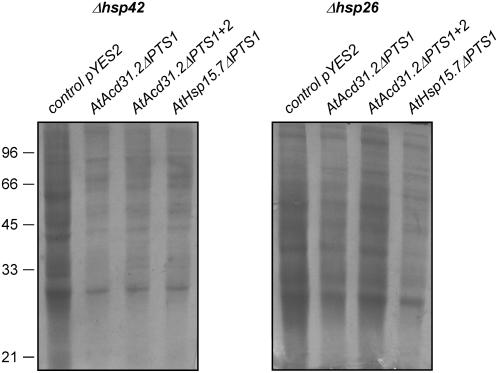
To investigate whether yeast and plant peroxisomal sHsps have a conserved chaperone function and can mutually complement a functional defect, the cDNAs of AtHsp15.7 and AtAcd31.2 were expressed by deletion of the PTSs as presumably cytosolic proteins from a Gal-inducible promoter in single or double deletion mutants, and the change in cell morphology was analyzed by SEM. Except for AtAcd31.2ΔPTS1 expressed in Δhsp26, both plant peroxisomal sHsps were able to complement the morphological defects of the single mutants deficient in ΔScHsp42 or ΔScHsp26 and the double mutant lacking both sHsps (Fig. 5). In addition, expression of AtHsp15.7ΔPTS1 and AtAcd31.2 ΔPTS1+2 led to a reduction of unspecific protein aggregation in the complemented yeast deletion strains after heat stress (Fig. 6). These results demonstrate that not only AtHsp15.7, but also AtAcd31.2, which notably belongs to a novel family of largely unknown Acd proteins with a weakly conserved α-crystallin domain (Fig. 1B; Supplemental Table II), is capable of suppressing the aggregation of a broad variety of cytosolic substrate proteins under heat stress conditions in yeast. It was concluded that both Arabidopsis proteins play a role as chaperones in the peroxisome matrix similar to yeast sHsps (Haslbeck et al., 2004; Cashikar et al., 2005).
Figure 5.
Complementation studies of single and double deletion strains of yeast deficient in ScHsp42 and ScHsp26 with plant peroxisomal sHsps. The yeast deletion strains Δhsp42, Δhsp26, and Δhsp42/26 were transformed with AtHsp15.7ΔPTS1, AtAcd31.2ΔPTS1, or AtAcd31.2ΔPTS1+PTS2 under the control of a GAL1 promoter, and yeast cells were imaged by SEM. After induction (Gal) for the expression of the Arabidopsis sHsps, the yeast cells were heat shocked for 1 h at 43°C prior to SEM analysis. As a negative control, yeast cells of equally treated deletion strains were transformed with the empty vector pYES2 and exhibited the wrinkly phenotype typical of the mutant. Yeast deletion strains and complementing Arabidopsis sHsps are indicated. The bar represents 10 μm.
Figure 6.
Reduction of protein aggregation in Δhsp42 and Δhsp42 deletion strains of yeast by plant peroxisomal sHsps. Yeast deletion strains Δhsp42 and Δhsp26 were transformed with the empty vector pYES 2 (lane 1) or the vector containing AtAcd31.2ΔPTS1, AtAcd31.2ΔPTS1+2, or AtHsp15.7ΔPTS1 (lanes 2–4), gene expression induced by galatose, and insoluble protein aggregates in lysates were analyzed by Coomassie-stained SDS-PAGE. Yeast cells were grown and heat stressed as for SEM analysis. An equal number of cells were lysed and insoluble proteins enriched by centrifugation (Haslbeck et al., 2004).
Expression Analysis of Peroxisomal sHsps
Small Hsps play an important role in plant stress tolerance because they assist in refolding of proteins that have been denatured under abiotic stress conditions such as high temperature, drought, high salt concentration, or elevated light intensity (Lee et al., 1995; Härndahl et al., 1999; Sun et al., 2002). Because sHsps are generally regulated at the transcriptional level (Scharf et al., 2001), isoform-specific expression data may indicate their involvement in stress tolerance. According to publicly available expression data retrieved using GENEVESTIGATOR (www.genevestigator.ethz.ch; Zimmermann et al., 2004), AtHsp15.7 is rather weakly expressed at most stages of plant development and in different organs, showing highest mRNA levels in roots, seeds, and suspension-cultured cells (Supplemental Fig. 3, A and B). In contrast, AtAcd31.2 is overall highly expressed at levels that exceed those of AtHsp15.7 in seedlings, leaves, flowers, and siliques about 5- to 20-fold (Supplemental Fig. 3, A and B), suggesting a constitutive expression of AtAcd31.2 under physiological conditions.
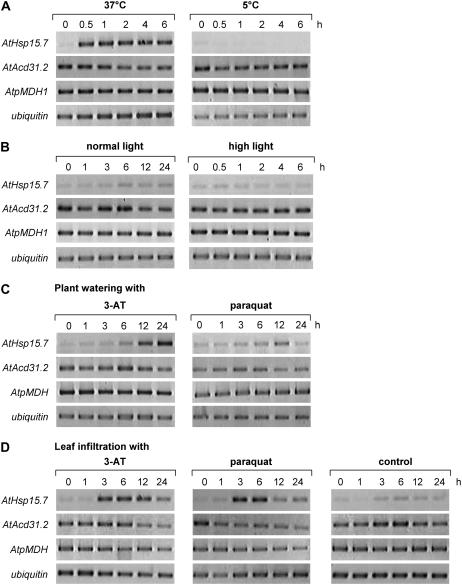
To investigate the expression of both peroxisomal sHsps in more detail, the effects of various abiotic stress conditions on sHsp expression in leaves were analyzed by RT-PCR using gene-specific oligonucleotide primers. When soil-grown Arabidopsis plants were transferred from ambient (22°C) to elevated (37°C) or cold temperature (5°C), the expression of AtHsp15.7 was hardly detectable under normal growth conditions or upon cold treatment but was quickly induced after 30 min heat shock (Fig. 7A). The pronounced induction of AtHsp15.7 expression was not due to stress-induced peroxisome proliferation and overall enhanced gene expression of peroxisomal proteins (Lopez-Huertas et al., 2000), because the expression level of house-keeping genes of leaf peroxisomal enzymes, such as peroxisomal malate dehydrogenase 1 (AtpMDH1, At5g09660; Fig. 7A), was not altered by increased temperature, demonstrating that AtHsp15.7 was specifically induced by heat stress among plant peroxisomal matrix proteins. By contrast, AtAcd31.2 was constitutively expressed under standard growth conditions and not further induced by heat (Fig. 7A).
Figure 7.
Semiquantitative expression analysis of AtHsp15.7 and AtAcd31.2 by RT-PCR. Expression analysis of AtHsp15.7 and AtAcd31.2 by heat and cold stress (A), standard and high light intensity (B), and oxidative stress applied by plant watering with inhibitor solution (C) or leaf infiltration (D). For oxidative stress analysis, soil-grown plants were either watered with 5 mm 3-AT or 100 μm methyl viologen (paraquat; C) or rosette leaves were infiltrated with 100 μm 3-AT or 10 μm methyl viologen (paraquat; D) and further incubated under standard light intensity (100 μE m−2 s−1). Gene expression was analyzed in leaves by RT-PCR using appropriate oligonucleotide primers and ubiquitin as a control for equal cDNA concentration. All experiments were performed three times and showed similar changes in transcript levels in each case.
Under elevated light and temperature conditions, the ratio of oxygenation-to-carboxylation increases (Sharkey, 1988), and ROS production is enhanced both in chloroplasts (mainly O2−• and H2O2) and peroxisomes (mainly H2O2), possibly stimulating expression of AtHsp15.7. During the long-day light period at either standard (about 100 μE m−2 s−1) or a maximum light intensity of 450 μE m−2 s−1, at which the temperature could still be controlled at about 23°C, no obvious induction of AtHsp15.7 was observed (Fig. 7B), suggesting that elevated ROS levels were efficiently reduced by CAT and the auxiliary antioxidative enzymes. To investigate whether a stronger ROS production affected the expression of peroxisomal sHsps, the CAT inhibitor 3-amino-1,2,4-triazole (3-AT) and the herbicide methyl viologen, which inhibits the D1 protein of PSII and leads to the production of O2−• in chloroplasts, were applied to soil-grown plants by watering to induce ROS production within the peroxisomal matrix or in chloroplasts, respectively. After 12 h of 3-AT application, AtHsp15.7 was strongly induced (Fig. 7C). Interestingly, expression of AtAcd31.2 seemed to decline concomitantly to AtHsp15.7 induction (Fig. 7C; see also Fig. 7, A and D). As for the temperature dependence of gene expression, the drastic increase of AtHsp15.7 expression was gene specific and not found for AtpMDH1 (Fig. 7C). Application of 100 μm methyl viologen had a comparable but minor effect on AtHsp15.7 expression with maximum levels toward the end of the light period (12 h; Fig. 7C).
Because uptake of ROS-producing agents from the soil by the plants is difficult to control, gene expression of peroxisomal sHsps was confirmed in an alternative experimental system, i.e. by infiltrating Arabidopsis leaves with the same chemicals. Leaf infiltration with 100 μm 3-AT induced expression of AtHsp15.7 after 3 h with constant levels up to the end of the light period (12 h) and lower levels in the beginning of the light period of the second day (24 h; Fig. 7D). Control plants infiltrated with solution lacking any inhibitor showed a minor induction of AtHsp15.7 expression similar to standard plants (Fig. 7B), suggesting that leaf wounding itself did not alter AtHsp15.7 expression to a considerable extent. Likewise, infiltration with 10 μm methyl viologen induced expression of AtHsp15.7 within about 3 h (Fig. 7D). In both experimental systems, the expression of AtAcd31.2 was not induced by any of the effectors applied. Degenerative symptoms such as leaf bleaching were not observed during the time period of analysis (data not shown).
In summary, the inducible gene expression of AtHsp15.7 by elevated temperature and ROS-generating chemicals compared to the constant mRNA levels of AtAcd31.2 supported a stress-inducible and general house-keeping function of AtHsp15.7 and AtAcd31.2 in plant peroxisomes, respectively.
DISCUSSION
Experimental Validation of Peroxisome Targeting
We identified two members of the expanded superfamily of sHsp-related proteins with putative targeting signals for peroxisomes in the Arabidopsis genome. The presence of a PTS peptide is a strong but not conclusive indication for peroxisome targeting of unknown proteins, as outlined previously (Neuberger et al., 2004; Reumann, 2004; Reumann et al., 2004). Experimental subcellular targeting analyses are thus required to verify predicted peroxisome targeting of unknown proteins. As shown by fluorescence microscopy using full-length and deletion constructs with EYFP, AtHsp15.7 was targeted to peroxisomes in onion epidermal cells by the C-terminal tripeptide SKL>. The double-labeling results excluded the possibility that the fluorescent punctate structures represented mitochondria or heat-shock granules, i.e. aggregates of sHsps, reported to occur in plants (Löw et al., 2000). Because a mitochondrial presequence was also predicted for AtHsp15.7 (Fig. 1A) and putative PTS1 signals can be overruled in vivo by N-terminal nonperoxisomal targeting signals (Neuberger et al., 2004), subcellular targeting of the inversely arranged fusion protein with accessible N-terminal end was investigated as well. Cytosolic targeting of AtHsp15.7:EYFP provided strong evidence for an exclusively peroxisomal localization of AtHsp15.7 in vivo. The PTS1 of AtHsp15.7 was conserved as major PTS1 in AtHsp15.7 homologs assembled from overlapping ESTs of various plant species including monocotyledons (Supplemental Fig. 2A), allowing the general conclusion that AtHsp15.7 orthologs are widespread in higher plants and targeted to peroxisomes as well.
The second sHsp AtAcd31.2 was rather unusual in containing two predicted minor PTSs (RLx5HF and PKL>; Reumann, 2004). Thorough sequence analysis pointed toward the PTS2 representing the functional PTS. First, additional Arg and Pro residues surrounded the PTS2 peptide (PTS2, RRRLAAFAAHFPA; PTS1, GILRIVIPKL>), as often observed in plant PTS2 proteins (Reumann, 2004). Second, plant ESTs homologous to the PTS2 of AtAcd31.2 showed a high degree of sequence variation, most of which were defined as PTS2 peptides (RLx5HF, RIx5H[LVF], RTx5HL, and R[MV]x5HF; Reumann, 2004; Supplemental Fig. 2B). By contrast, the C-terminal tripeptide of AtAcd31.2 homologs was mutated to PTS1-related peptides (e.g. PKI>, PKV>, PFI>, or AHM>; Supplemental Fig. 2B) that have not (yet) been defined as PTS1 peptides and the peroxisome targeting function of which is questionable (Reumann, 2004). Third, the PTS2 is located in the long presumably flexible N-terminal domain of AtAcd31.2 (Fig. 1), whereas the close proximity of PKL> to the tightly folded β-sheet sandwich of the α-crystallin domain (van Montfort et al., 2001) is likely to prohibit interaction with Pex5p and PTS1 tripeptide optimization by point mutations (Supplemental Fig. 2).
Our experimental analyses demonstrated that both fusion proteins AtAcd31.2:EYFP and EYFP:AtAcd31.2 were targeted to peroxisomes. Nonperoxisomal targeting of the deletion construct AtAcd31.2ΔN:EYFP and the version with mutated PTS2 (RLx5HF→RLx5DF) demonstrated that RLx5HF is a functional PTS2 and argued against the presence of an additional internal PTS similar to those described for CAT and a few other proteins (Kamigaki et al., 2003). Subcellular targeting of AtAcd31.2ΔN:EYFP to plastids was in line with computer prediction for this fusion protein but is not thought to be of physiological significance, because a second putative alternative translational start codon is lacking in front of the predicted transit peptide and because dual targeting of the full-length fusion protein AtAcd31.2:EYFP to both peroxisomes and plastids was not observed. Peroxisome targeting of EYFP:AtAcd31.2 argued in favor of the presence of a second PTS, i.e. the putative PTS1 PKL>. Deletion of this C-terminal tripeptide or exchange of the essential basic residue Lys to Glu (PKL→PEL), however, did not abolish peroxisome targeting of the fusion protein nor increased noticeably cytosolic EYFP fluorescence. We interpret these results that the putative PTS1 is not essential for peroxisome targeting. Whether an internal PTS targets EYFP:AtAcd31.2 to peroxisomes needs to be investigated in more detail in future studies.
The N-terminal targeting domains of several plant PTS2 proteins are proteolytically removed by a specific yet unknown processing peptidase upon import into the peroxisome matrix (Kato et al., 1998). A peptide with a mass-to-charge ratio (m/z) corresponding to the N-terminal tryptic peptide of the full-length open reading frame of AtAcd31.2 (MEHESITARR, amino acids 1–10, Mcalc = 1228.61) was indeed detected in the protein band/spot of AtAcd31.2 by peptide mass fingerprinting (Fig. 4, A and B; data not shown). However, sequence analysis by MS/MS revealed that this peptide did not represent the N terminus but an internal tryptic peptide of AtAcd31.2 (QASSAQGFFMR, amino acids 99–109, Mcalc = 1228.57). Taken together with the finding that none of the other most probable N-terminal tryptic peptides (MEHESITAR, amino acids 1–9, Mcalc = 1072.50; MEHESITARRR, amino acids 1–11, Mcalc = 1384.71) was detected by peptide mass fingerprinting, these data may indicate that the PTS2 of AtAcd31.2 is proteolytically removed in vivo.
To provide independent support for the localization of both sHsps in plant peroxisomes, we established a method to isolate peroxisomes from mature leaves of Arabidopsis. Previous methods published for Arabidopsis cotyledons or leaves from spinach (Spinacia oleracea; Yu and Huang, 1986; Fukao et al., 2002, 2003) were not suitable due to an extreme fragility of peroxisomes isolated from mature Arabidopsis rosette leaves, which is probably caused by the high concentration of secondary metabolites and proteases in this tissue. Moreover, we noticed a pronounced adherence between peroxisomes, mitochondria, and plastids in Brassicaceae. Gentle sedimentation of the leaf peroxisomes onto a Suc cushion during differential centrifugation and addition of a second Suc density gradient allowed a significant organelle enrichment. Even though marker enzyme activities of contaminating organelles were hardly detectable in the final peroxisome fraction (Fig. 3), partial comigration of residual plastids and mitochondria was observed, and the purity of Arabidopsis leaf peroxisomes remained lower as compared to that of model plants like spinach (data not shown). As a result, dominant proteins of plastids and mitochondria, but notably no cytosolic proteins, were still detected in minor concentration in the fraction of enriched Arabidopsis leaf peroxisomes (data not shown). Nevertheless, AtAcd31.2 was successfully identified in several independent leaf peroxisomal protein preparations from Arabidopsis by using one- and two-dimensional gels in combination with different mass spectrometric techniques. Considering the absence of a predicted mitochondrial presequence or plastidic transit peptide in AtAcd31.2 (Fig. 1), these data provide further support for targeting of this chaperone to Arabidopsis peroxisomes in vivo.
To provide supplementary support for targeting of AtHsp15.7 to peroxisomes, we tried to isolate leaf peroxisomes from Arabidopsis plants that had been subjected to heat stress. Although leaf peroxisomes could indeed be isolated in a quantity and quality comparable to that obtained for control plants (S. Reumann, unpublished data), we did not succeed in identifying this chaperone from polyacrylamide gels so far. Whether this failure in protein identification is due to biological effects such as a high rate of protein turnover or technical limitations often observed when small and basic proteins (MW = 15.7, IEP = 7.9) are analyzed by gel-based proteomics approaches remains to be determined.
Toward an Elucidation of the Function of Peroxisomal sHsps
To investigate whether AtHsp15.7 and AtAcd31.2 play a physiological role as sHsps, we tried to complement the morphological phenotype of knockout mutants of yeast that are deficient in one of two cytosolic sHsps (Haslbeck et al., 2004). The PTSs were deleted from AtHsp15.7 and AtAcd31.2 to redirect the sHsps from peroxisomes to the cytosol in yeast. Except for AtAcd31.2ΔPTS1 expressed in Δhsp26, both peroxisomal plant Hsps were able to complement the single and the Δhsp26/42 double mutant (Fig. 5). Complementation by AtAcd31.2ΔPTS1 was generally less effective than that of the fusion protein lacking also the PTS2; it was observed in only about 70% of the experiments (data not shown). Therefore, insufficient complementation of AtAcd31.2ΔPTS1 was most likely caused by protein targeting to yeast peroxisomes by the PTS2 rather than the cytosol and not due to the higher sequence divergence of AtAcd31.2 from the consensus pattern of the α-crystallin domain (Fig. 1B). The analysis of cytosolic yeast extracts upon heat stress (Fig. 6) demonstrated that overexpression of AtHsp15.7ΔPTS1 and AtAcd31.2ΔPTS1+2 reduced the extent of unspecific protein aggregation in the deletion strains of yeast. From these data it can be concluded that AtHsp15.7 and AtAcd31.2 are involved in protein homeostasis and presumably exhibit a conserved sHsp function in vivo.
Because AtHsp15.7 clusters with Arabidopsis sHsps of the cytosolic class I and those of the endoplasmic reticulum class, which comprise some well-characterized heat-inducible sHsps (Supplemental Fig. 1; Scharf et al., 2001), a conserved chaperone function was expected for AtHsp15.7. By contrast, AtAcd31.2 belongs to a novel family of Acd proteins, only a few members of which have been analyzed experimentally to date and the function of none of which has been associated with molecular chaperones (Scharf et al., 2001 and refs. therein). Several clades of Arabidopsis Acd proteins branch deeply in the phylogenetic tree of various eukaryotic sHsps (Supplemental Fig. 1), suggesting a diverse evolutionary origin. It needs to be stressed that the previous classification of Arabidopsis proteins into sHsps and Acd proteins (Scharf et al., 2001) is not based on functional studies or Acd conservation (see Supplemental Table II), but generally refers to sequence similarity with known heat-inducible sHsp, whereas the clades of Acd proteins represent largely unknown proteins. Hence, AtAcd31.2 is the first Arabidopsis Acd protein shown to have a chaperone function in vivo, strongly suggesting that other yet unknown Acd proteins represent molecular chaperones as well.
To gain first insights into the role of both peroxisomal sHsps in plant peroxisomes, we analyzed their expression patterns in rosette leaves of plants that had been subjected to various forms of abiotic or biotic stress conditions. Leaf peroxisomes play a major role not only in photosynthesis but also in fatty acid β-oxidation. Our expression analyses at the mRNA and the protein level indicate that AtAcd31.2 is constitutively expressed at significant levels, whereas AtHsp15.7 expression is not detectable under standard conditions and strongly induced by heat and oxidative stress conditions. The heat inducibility of AtHsp15.7 is further supported by an extended cluster of heat-shock element motifs in the gene's promoter (Scharf et al., 2001). In two different experimental systems, i.e. inhibitor application by watering or infiltration, and using two alternative ROS-producing effectors (3-AT and methyl viologen), an induction of AtHsp15.7 expression by ROS was observed. For a subset of sHsps, including two located in the cytosol and one each in mitochondria and plastids, a similar induction by oxidative stress has previously been determined (Banzet et al., 1998; Lee and Vierling, 2000; for review, see Sun et al., 2002).
The induction of AtHsp15.7 by oxidative stress conditions is of particular interest because peroxisomes are one of the two major subcellular compartments in which ROS are produced during photosynthesis. At elevated temperature, the ratio of carboxylation to oxygenation reaches a value of 1:0.5 (Sharkey, 1988). Thus, fixation of 2 mol of CO2 is accompanied by the production of 1 mol of glycolate and, by the activity of glycolate oxidase, 1 mol of H2O2. Under standard conditions, the high concentration of CAT is thought to detoxify most H2O2 produced during photorespiration. However, CAT is easily inactivated by light and has a rather low turnover rate (Feierabend and Engel, 1986; Grotjohann et al., 1997). Under CAT-inactivating conditions, the ROS concentration in the peroxisome matrix may reach such a high level that matrix proteins are oxidized, hydrophobic polypeptide patches exposed, and enzymes inactivated. The induction of AtHsp15.7 by oxidative stress conditions may therefore indicate that AtHsp15.7 plays an important role in minimizing oxidative damage on leaf peroxisomal metabolism.
A constitutive expression as determined for AtAcd31.2 is atypical for sHsps and has, to the best of our knowledge, not been described yet for any Arabidopsis or tomato (Lycopersicon esculentum) sHsp. In a previous study, AtAcd31.2 was shown to be negatively regulated by floral induction and gibberellins (Chandler and Melzer, 2004). Interestingly, in contrast to the typical heat inducibility of sHsps, several yet unknown Arabidopsis Acd genes are not heat inducible (Zimmermann et al., 2004; Supplemental Fig. 4). The constitutive and high expression level of AtAcd31.2 (Fig. 7; Supplemental Figs. 3 and 4) suggests that this chaperone and possibly other Acd proteins play an important role even under normal physiological conditions. The analysis of Arabidopsis knockout mutants deficient in AtHsp15.7 or AtAcd31.2 may reveal the precise function of both sHsps in plant peroxisomal metabolism and their mode of action in the future.
Hsps Acting in Concert with Peroxisomal sHsps
The localization of two distinct sHsps to the matrix of plant peroxisomes raises new questions and hypotheses. Because sHsps lack ATP-hydrolyzing activity, the chaperone function of sHsps appears to be limited to binding and maintaining the solubility of unfolded proteins, without promoting their refolding directly and actively in an ATP-dependent manner. Therefore, substrate renaturation by sHsps is generally thought to require their interaction with other Hsps (Forreiter et al., 1997; Lee and Vierling, 2000). Targeting of additional Hsps, mainly a Hsp70 or Hsp100 homolog (Mogk et al., 2003; Cashikar et al., 2005), needs to be postulated to act in concert with AtHsp15.7 and AtAcd31.2. Because of its membrane topology facing the cytosolic side, the DnaJ ATPase chaperone of the glyoxysomal membrane of C. sativus (Diefenbach and Kindl, 2000) is not expected to mediate protein folding in the peroxisome matrix. An Hsp70 homolog from Citrullus lanatus was shown to be targeted to both chloroplasts and the peroxisome matrix by the use of two alternative translation start codons, leading to the translation of either a transit peptide or a PTS2 at the N-terminal end (Wimmer et al., 1997). Two Hsp70 isoforms have also been detected in highly purified glyoxysomes from C. sativus (Diefenbach and Kindl, 2000). Members of the Hsp70 family with putative PTSs, however, have not been detected in the Arabidopsis genome to date (Reumann et al., 2004). In one of two Arabidopsis Hsp70 homologs, the second Met of the peroxisomal Hsp70 homolog from Cucumis is indeed conserved but not obviously followed by a putative PTS2. More research is required to investigate dual targeting of Arabidopsis Hsp70 homologs to plastids and peroxisomes. Considering the small number of cloned cDNAs of plant peroxisomal proteins (Reumann, 2004) and the current limits in predicting alternative splice and translational variants, currently unknown PTS are likely to be unraveled in the ongoing postgenomic era and further peroxisomal Arabidopsis proteins to be identified, possibly including peroxisomal Hsp70 and Hsp100 homologs. If neither a matrix-targeted Hsp70 nor an Hsp100 homolog is involved in protein refolding in plant peroxisomes, an alternative mechanism can be envisioned, in which sHsp-substrate complexes are transported from the peroxisome matrix back to the cytosol for proper refolding and the renatured polypeptides subsequently reimported into the peroxisome matrix.
CONCLUSION
This study shows that low-abundance regulatory proteins from plant peroxisomes can indeed be identified by screening the Arabidopsis genome for genes encoding proteins with putative PTSs. Arabidopsis is thus the first organism shown to contain sHsps in the matrix of peroxisomes. In addition to the previously defined six classes of plant sHsps (Scharf et al., 2001; Sun et al., 2002), we identified a seventh class for peroxisomes and suggest to change the acronyms of these proteins from AtHsp15.7-CI (cytosolic class I; Scharf et al., 2001) to AtHsp15.7-Px and AtAcd31.2-Px (Px, peroxisome). The characterization of AtAcd31.2 as a constitutively expressed sHsp suggests that plant sHsps function not only under stress conditions but also assist in protein refolding under standard physiological conditions. The localization of sHsps to plant peroxisomes and the detection of Acd homologs with putative PTS1s in other organisms (Supplemental Fig. 1) indicate that homologs of larger sHsp families may be targeted to peroxisomes in other eukaryotes as well. Regarding higher eukaryotes, however, plants may be the prototypical organisms that require peroxisomal sHsps due to their sessile nature in combination with their permanent subjection to quickly and drastically changing environmental conditions.
MATERIALS AND METHODS
Plant Growth
Standard Arabidopsis (Arabidopsis thaliana) ecotype Columbia plants were grown for about 4 weeks in a 16-h-light/8-h-dark cycle at 22°C under a light intensity of 100 to 150 μE m−2 s−1. All the stress treatments were initiated after 3 h of light. For heat and cold stress experiments, plants were incubated in the dark at 37°C and 5°C, respectively, whereas the control plants were incubated at 22°C in the dark. For high light stress, the light intensity was raised to 450 μE m−2 s−1 while keeping the temperature constant at about 23°C. Control plants grown at the same temperature, but under normal light, were analyzed in parallel. For the oxidative stress experiments, soil-grown plants were either watered with 5 mm 3-AT or 100 μm methyl viologen (about 50 mL/9-cm pot and day), or rosette leaves were infiltrated with 100 μm 3-AT or 10 μm methyl viologen (in water) using a syringe and floated on inhibitor solution. Rosette leaves infiltrated with water were used as a mock control.
Gene Cloning and Semiquantitative RT-PCR
Total RNA was isolated from different tissues of Arabidopsis ecotype Columbia using the Invisorb Spin plant mini kit (Invitek). Full-length cDNAs for AtHsp15.7 (At5g37670) and AtAcd31.2 (At1g06460) were isolated from flowers and cold-treated rosette leaves, respectively, using appropriate oligonucleotide primers (Supplemental Table I). Total RNA was converted to single-strand cDNA by reverse transcriptase (Superscript III, Invitrogen) and used as template for PCR using a proof-reading DNA polymerase (Thermozyme, Invitrogen). Amplified products were subcloned into pGEMT using the pGEM-T Easy Vector system (Promega) and sequenced. Amplification errors that resulted in amino acid exchanges were not observed. Semiquantitative RT-PCR was performed using a First-Strand cDNA Synthesis kit (MBI, Fermentas) according to the manufacturer's instruction. For PCR, standard parameter and an appropriate number of cycles were used (AtHsp15.7 and ubiquitin, 30 cycles; AtAcd31.2, 26 cycles; and AtpMDH1, 24 cycles). Complete removal of residual genomic DNA by enzymatic digestion was verified for the intronless gene of AtHsp15.7 using control samples lacking reverse transcriptase. The specificity of AtHsp15.7 amplification was confirmed by restriction endonuclease digest of the RT-PCR products. All RT-PCR experiments were repeated at least three times using independent plant material.
Subcellular Localization Studies
Targeting prediction was performed as described earlier (Reumann et al., 2004). Fusion proteins with N- or C-terminally located EYFP were generated by PCR (Supplemental Table I) to investigate the function of C-terminal and N-terminal targeting signals, respectively, and subcloned in frame into the plant expression vectors pCAT-EYFP-Nfus and pCAT-EYFP-Cfus (Fulda et al., 2002) under control of a double 35S CaMV promoter. The C-terminal deletion construct EYFP:AtHsp15.7ΔPTS1 lacked the C-terminal 14 residues including the putative PTS1 SKL>. The deletion constructs AtAcd31.2ΔPTS2:EYFP and EYFP:AtAcd31.2ΔPTS1 lacked the N-terminal 29 residues including the putative PTS2 (RLx5HF, residues 11–19) or the C-terminal tripeptide PKL>, respectively (Supplemental Table I). Site-directed mutagenesis (PTS2 of AtAcd31.2, RLx5HF to RLx5DF; PTS1, PKL> to PEL>) was performed using PfuUltra high-fidelity DNA polymerase for mutagenic primer-directed replication of both plasmid strands of AtAcd31.2:EYFP in pCAT-EYFP-Cfus and EYFP:AtAcd31.2 in pCAT-EYFP-Nfus using the Quick-Change II site-directed mutagenesis kit (Stratagene; Supplemental Table I). Onion (Allium cepa) epidermal cells were transformed biolistically as described (Biolistic PDS 1000/He Biolistic Particle Delivery system, Bio-Rad; Fulda et al., 2002) using 1,100 psi rupture discs and a vacuum of 0.1 bar. All subcellular analyses were reproduced at least three times in independent experiments.
Yeast Complementation Studies
Deletion constructs of AtHsp15.7 or AtAcd31.2 lacking the C-terminal three residues and/or the N-terminal PTS2 (residues 1–29), referred to as AtHsp15.7ΔPTS1, AtAcd31.2ΔPTS1, and AtAcd31.2ΔPTS1+2, were generated to target the proteins to the yeast (Saccharomyces cerevisiae) cytosol and subcloned without any terminal tags into the pYES2.1-topo cloning vector under the control of a GAL1 promoter and containing the URA3 gene for selection of transformants (pYES2.1 TOPO TA expression kit, Invitrogen). After transformation, yeast deletion strains deficient in ScHsp42 and/or ScHsp26 (Haslbeck et al., 2004) and expressing Arabidopsis sHsps were first selected on complete supplement media lacking uracil (CSM-URA) and containing Glc. Prior to induction of the expression of the peroxisomal sHsps, mid logarithmic phase cells cultivated at 30°C were transferred to CSM-URA media containing raffinose for 2 h. Next, the cells were transferred to CSM-URA media containing Gal for induction. After 4 h of induction, the cultures were heat shocked for 1 h at 43°C and subsequently analyzed by SEM. As negative control, equally treated cells transformed with the empty vector pYES2 were used. To determine the total amount of aggregated protein in the complemented yeast strains, 2 × 108 yeast cells were collected subsequently after heat shock and lysed using a Basic Z cell disrupter at 2.5 kbar (Constant Systems). After separation of cellular fragments by gentle centrifugation at 500g for 10 min, insoluble protein aggregates were sedimented by centrifugation (10 min, 13,000g at 4°C) and analyzed by SDS-PAGE.
Microscopy
Analysis of onion epidermal cells was performed using a fluorescence microscope (Olympus BX51) with the following filter sets: EYFP (F41-028; excitation filter HQ500/20, barrier HQ535/30), and ECFP (F31-044; excitation filter D436/20, barrier D480/40). Digital images were captured using a CCD camera (ColorViewII) with analySIS3.1 Imaging software (Soft imagine system GMDH). For analysis of yeast morphology, cells were fixed and prepared as described by Spector et al. (1998). SEM was performed with a JEOL 5900 LV microscope. Pictures were taken at a constant voltage of 20 kV and a spot size of 20 nm at a magnification of 5,500×.
Isolation of Leaf Peroxisomes
Leaves were ground in grinding buffer (170 mm Tricine-KOH, pH 7.5, 1.0 m Suc, 1% (w/v) bovine serum albumin (BSA), 2 mm EDTA, 5 mm dithiothreitol, 10 mm KCl, and 1 mm MgCl2) in the presence of protease inhibitors using a mortar and a pestle, the suspension filtered, and chloroplasts were sedimented at 5,000g (1 min). Leaf peroxisomes were sedimented onto a Suc cushion of 50% (w/w) by centrifugation at 20,000g for 5 min. The resuspended organelles were homogenized using a Potter-Elvehjem homogenizer, loaded onto a Suc density gradient prepared in TE buffer (10 mm Tricine-KOH, pH 7.5, 1 mm EDTA) supplemented with 0.5% (w/v) BSA (from the top to the bottom: 2 mL 30% [w/w], 3 mL 35% [w/w], linear gradient of 2× 7.5 mL 40% to 52% [w/w], 3 mL 52% [w/w], 5 mL 60% [w/w]), and centrifuged for 2 h at 80,000g (Beckman SW28 rotor). For analytical purpose, the gradient was fractionated in 2-mL fractions. For preparative purposes, the peroxisome fraction located at the interface between 52% and 60% (w/w) Suc was harvested, combined from several gradients, diluted to 48% (w/w), and loaded onto a second Suc density gradient (linear part of 6 mL each 48% and 60% [w/w], 3 mL 60% [w/w] in TE buffer lacking BSA) by diluting the peroxisome fraction in a gradient mixer with 40% (w/w) in TE buffer. For two-dimensional gel electrophoresis, BSA was omitted in all Suc solutions of a density higher than 40% (w/w). The proteins were precipitated according to Wessel and Flügge (1984), dissolved in urea buffer (7 m urea, 2 m thiourea, 4% [w/v] CHAPS, 0.5% immobilized pH gradient (IPG) buffer, and 3 mg/mL dithiothreitol) and subjected to isoelectric focusing (nonlinear IPG strip, pH 3–10). For SDS-PAGE and the second dimension, the proteins were separated on a large 7.5% to 15% acrylamide gradient gel under denaturing conditions and stained with silver or colloidal Coomassie Blue. Chlorophyll and protein were determined according to Arnon (1949) and Lowry et al. (1951), respectively. The activities of marker enzymes (hydroxypyruvate reductase [HPR] for leaf peroxisomes, fumarase for mitochondria, and NADP-GAPDH for chloroplasts) were determined as described earlier (Reumann et al., 1995).
Protein Identification
Excised spots were subjected to automated in-gel digest with prior alkylation according to standard protocols supplied with the ProTeam Advanced Digest system (Tecan). Mass spectrometry grade trypsin was purchased from Promega. For the acquisition of peptide mass fingerprints by matrix-assisted laser-desorption ionization time of flight-mass spectrometry, the peptides extracted from the gel plugs were applied to a prestructured sample support (AnchorChip target; Bruker Daltonics) coated with a thin layer of α-cyano-4-hydroxy-cinnamic acid (Gobom et al., 2001) using the same liquid handling system as for the in-gel digest. The target was inserted into a Bruker Ultraflex TOF/TOF instrument (Suckau et al., 2003) and submitted to an automated analysis loop using external calibration. Database searches in the NCBInr primary sequence database restricted to the taxonomy Arabidopsis were performed using the Mascot Software 2.0 (Matrix Science) with carboxamidomethylation of Cys as fixed and oxidation of Met as variable modification, respectively. The monoisotopic mass tolerance was set to 100 ppm and one missed cleavage was allowed. For confirmation of the peptide mass fingerprinting results, the samples were analyzed by MS/MS using the LIFT technology of the Ultraflex TOF/TOF instrument (Suckau et al., 2003) to obtain sequence information of selected peptides. Database searches using combined peptide mass fingerprint and MS/MS datasets were performed as described above with the fragment mass tolerance set to 0.7 D.
Sequence data from this article can be found in the EMBL/GenBank data libraries under accession numbers DQ403190 (AtHsp15.7, At5g37670) and DQ403189 (AtAcd31.2, At1g06460).
Supplementary Material
Acknowledgments
We are grateful for practical assistance in yeast complementation studies by B. Richter, for the initial SEM and proteome analyses performed by Dr. A. Olbrich and Dr. H. Kratzin, respectively, for stimulating discussions with Dr. K.D. Scharf and M. Siddique, and for provision of the pCAT plant expression vectors by Dr. M. Fulda. We thank K. Pawlowski, I. Heilmann, and O. Voitsekhovskaja for critical review of the manuscript, and Professor I. Feussner for provision of the infrastructure for our research. Sequencing of cDNAs by the Göttingen Genomics Center (Professor Gottschalk, Department of Microbiology) and the Department of Developmental Biochemistry (Professor Pieler) is greatly acknowledged.
This work was supported by the Deutsche Forschungsgemeinschaft (grant no. RE1304/2 to S.R.), by Fonds der Chemischen Industrie (to M.H.), and by the government of Lower Saxony (a Dorothea-Erxleben stipend to S.R.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Sigrun Reumann (sreuman@gwdg.de).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.073841.
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Arnon DI (1949) Copper enzymes in isolated chloroplasts: polyphenoloxidase in Beta vulgaris. Plant Physiol 24: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banzet N, Richaud C, Deveaux Y, Kazmaier M, Gagnon J, Triantaphylides C (1998) Accumulation of small heat shock proteins, including mitochondrial HSP22, induced by oxidative stress and adaptive response in tomato cells. Plant J 13: 519–527 [DOI] [PubMed] [Google Scholar]
- Beevers H (1979) Microbodies in higher plants. Annu Rev Plant Physiol 30: 159–193 [Google Scholar]
- Buchner J, Grallert H, Jakob U (1998) Analysis of chaperone function using citrate synthase as a nonnative substrate protein. Methods Enzymol 290: 323–338 [DOI] [PubMed] [Google Scholar]
- Cashikar AG, Duennwald M, Lindquist SL (2005) A chaperone pathway in protein disaggregation: Hsp26 alters the nature of protein aggregates to facilitate reactivation by Hsp104. J Biol Chem 280: 23869–23875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler JW, Melzer S (2004) An alpha-crystallin gene, ACD31.2 from Arabidopsis is negatively regulated by FPF1 overexpression, floral induction, gibberellins, and long days. J Exp Bot 55: 1433–1435 [DOI] [PubMed] [Google Scholar]
- De Jong WW, Caspers GJ, Leunissen JA (1998) Genealogy of the alpha-crystallin-small heat-shock protein superfamily. Int J Biol Macromol 22: 151–162 [DOI] [PubMed] [Google Scholar]
- del Rio LA, Corpas FJ, Sandalio LM, Palma JM, Gomez M, Barroso JB (2002) Reactive oxygen species, antioxidant systems and nitric oxide in peroxisomes. J Exp Bot 53: 1255–1272 [PubMed] [Google Scholar]
- Diefenbach J, Kindl H (2000) The membrane-bound DnaJ protein located at the cytosolic site of glyoxysomes specifically binds the cytosolic isoform 1 of Hsp70 but not other Hsp70 species. Eur J Biochem 267: 746–754 [DOI] [PubMed] [Google Scholar]
- Feierabend J, Engel S (1986) Photoinactivation of catalase in vitro and in leaves. Arch Biochem Biophys 251: 567–576 [DOI] [PubMed] [Google Scholar]
- Forreiter C, Kirschner M, Nover L (1997) Stable transformation of an Arabidopsis cell suspension culture with firefly luciferase providing a cellular system for analysis of chaperone activity in vivo. Plant Cell 9: 2171–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao Y, Hayashi M, Hara-Nishimura I, Nishimura M (2003) Novel glyoxysomal protein kinase, GPK1, identified by proteomic analysis of glyoxysomes in etiolated cotyledons of Arabidopsis thaliana. Plant Cell Physiol 44: 1002–1012 [DOI] [PubMed] [Google Scholar]
- Fukao Y, Hayashi M, Nishimura M (2002) Proteomic analysis of leaf peroxisomal proteins in greening cotyledons of Arabidopsis thaliana. Plant Cell Physiol 43: 689–696 [DOI] [PubMed] [Google Scholar]
- Fulda M, Shockey J, Werber M, Wolter FP, Heinz E (2002) Two long-chain acyl-CoA synthetases from Arabidopsis thaliana involved in peroxisomal fatty acid beta-oxidation. Plant J 32: 93–103 [DOI] [PubMed] [Google Scholar]
- Gobom J, Schuerenberg M, Mueller M, Theiss D, Lehrach H, Nordhoff E (2001) Alpha-cyano-4-hydroxycinnamic acid affinity sample preparation: a protocol for MALDI-MS peptide analysis in proteomics. Anal Chem 73: 434–438 [DOI] [PubMed] [Google Scholar]
- Grotjohann N, Janning A, Eising R (1997) In vitro photoinactivation of catalase isoforms from cotyledons of sunflower (Helianthus annuus L.). Arch Biochem Biophys 346: 208–218 [DOI] [PubMed] [Google Scholar]
- Härndahl U, Hall RB, Osteryoung KW, Vierling E, Bornman JF, Sundby C (1999) The chloroplast small heat shock protein undergoes oxidation-dependent conformational changes and may protect plants from oxidative stress. Cell Stress Chaperones 4: 129–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslbeck M, Braun N, Stromer T, Richter B, Model N, Weinkauf S, Buchner J (2004) Hsp42 is the general small heat shock protein in the cytosol of Saccharomyces cerevisiae. EMBO J 23: 638–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslbeck M, Franzmann T, Weinfurtner D, Buchner J (2005) Some like it hot: the structure and function of small heat-shock proteins. Nat Struct Mol Biol 12: 842–846 [DOI] [PubMed] [Google Scholar]
- Haslbeck M, Walke S, Stromer T, Ehrnsperger M, White HE, Chen S, Saibil HR, Buchner J (1999) Hsp26: a temperature-regulated chaperone. EMBO J 18: 6744–6751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TL, Olsen LJ (2001) Building new models for peroxisome biogenesis. Plant Physiol 127: 731–739 [PMC free article] [PubMed] [Google Scholar]
- Kamigaki A, Mano S, Terauchi K, Nishi Y, Tachibe-Kinoshita Y, Nito K, Kondo M, Hayashi M, Nishimura M, Esaka M (2003) Identification of peroxisomal targeting signal of pumpkin catalase and the binding analysis with PTS1 receptor. Plant J 33: 161–175 [DOI] [PubMed] [Google Scholar]
- Kato A, Takeda-Yoshikawa Y, Hayashi M, Kondo M, Hara-Nishimura I, Nishimura M (1998) Glyoxysomal malate dehydrogenase in pumpkin: cloning of a cDNA and functional analysis of its presequence. Plant Cell Physiol 39: 186–195 [DOI] [PubMed] [Google Scholar]
- Lee GJ (1995) Assaying proteins for molecular chaperone activity. Methods Cell Biol 50: 325–334 [DOI] [PubMed] [Google Scholar]
- Lee GJ, Vierling E (2000) A small heat shock protein cooperates with heat shock protein 70 systems to reactivate a heat-denatured protein. Plant Physiol 122: 189–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Hubel A, Schoffl F (1995) Derepression of the activity of genetically engineered heat shock factor causes constitutive synthesis of heat shock proteins and increased hermotolerance in transgenic Arabidopsis. Plant J 8: 603–612 [DOI] [PubMed] [Google Scholar]
- Lisenbee CS, Lingard MJ, Trelease RN (2005) Arabidopsis peroxisomes possess functionally redundant membrane and matrix isoforms of monodehydroascorbate reductase. Plant J 43: 900–914 [DOI] [PubMed] [Google Scholar]
- Lopez-Huertas E, Charlton WL, Johnson B, Graham IA, Baker A (2000) Stress induces peroxisome biogenesis genes. EMBO J 19: 6770–6777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löw D, Brandle K, Nover L, Forreiter C (2000) Cytosolic heat-stress proteins Hsp17.7 class I and Hsp17.3 class II of tomato act as molecular chaperones in vivo. Planta 211: 575–582 [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193: 265–275 [PubMed] [Google Scholar]
- Mogk A, Deuerling E, Vorderwulbecke S, Vierling E, Bukau B (2003) Small heat shock proteins, ClpB and the DnaK system form a functional triade in reversing protein aggregation. Mol Microbiol 50: 585–595 [DOI] [PubMed] [Google Scholar]
- Narberhaus F (2002) Alpha-crystallin-type heat shock proteins: socializing minichaperones in the context of a multichaperone network. Microbiol Mol Biol Rev 66: 64–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuberger G, Kunze M, Eisenhaber F, Berger J, Hartig A, Brocard C (2004) Hidden localization motifs: naturally occurring peroxisomal targeting signals in non-peroxisomal proteins. Genome Biol 5: R97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petko L, Lindquist S (1986) Hsp26 is not required for growth at high temperatures, nor for thermotolerance, spore development, or germination. Cell 45: 885–894 [DOI] [PubMed] [Google Scholar]
- Reumann S (2002) The photorespiratory pathway of leaf peroxisomes. In A Baker, IA Graham, eds, Plant Peroxisomes: Biochemistry, Cell Biology and Biotechnological Applications, Ed 1. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 141–189
- Reumann S (2004) Specification of the peroxisome targeting signals type 1 and type 2 of plant peroxisomes by bioinformatics analyses. Plant Physiol 135: 783–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S, Ma C, Lemke S, Babujee L (2004) AraPerox: a database of putative Arabidopsis proteins from plant peroxisomes. Plant Physiol 136: 2587–2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S, Maier E, Benz R, Heldt HW (1995) The membrane of leaf peroxisomes contains a porin-like channel. J Biol Chem 270: 17559–17565 [DOI] [PubMed] [Google Scholar]
- Scharf KD, Siddique M, Vierling E (2001) The expanding family of Arabidopsis thaliana small heat stress proteins and a new family of proteins containing alpha-crystallin domains (Acd proteins). Cell Stress Chaperones 6: 225–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharkey T (1988) Estimating the rate of photorespiration in leaves. Physiol Plant 73: 147–152 [Google Scholar]
- Spector DL, Goldmann RD, Leinwand LA (1998) Preparative methods for scanning electron microscopy. In DL Spector, RD Goldmann, LA Leinwand, eds, Cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, pp 122.1–123.27
- Suckau D, Resemann A, Schuerenberg M, Hufnagel P, Franzen J, Holle A (2003) A novel MALDI LIFT-TOF/TOF mass spectrometer for proteomics. Anal Bioanal Chem 376: 952–965 [DOI] [PubMed] [Google Scholar]
- Sun W, Van Montagu M, Verbruggen N (2002) Small heat shock proteins and stress tolerance in plants. Biochim Biophys Acta 1577: 1–9 [DOI] [PubMed] [Google Scholar]
- van Montfort RL, Basha E, Friedrich KL, Slingsby C, Vierling E (2001) Crystal structure and assembly of a eukaryotic small heat shock protein. Nat Struct Biol 8: 1025–1030 [DOI] [PubMed] [Google Scholar]
- Vierling E (1991) The roles of heat shock proteins in plants. Annu Rev Plant Physiol Plant Mol Biol 42: 579–620 [Google Scholar]
- Wang W, Vinocur B, Shoseyov O, Altman A (2004) Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci 9: 244–252 [DOI] [PubMed] [Google Scholar]
- Wessel D, Flügge UI (1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem 138: 141–143 [DOI] [PubMed] [Google Scholar]
- Wimmer B, Lottspeich F, van der Klei I, Veenhuis M, Gietl C (1997) The glyoxysomal and plastid molecular chaperones (70-kDa heat shock protein) of watermelon cotyledons are encoded by a single gene. Proc Natl Acad Sci USA 94: 13624–13629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wotton D, Freeman K, Shore D (1996) Multimerization of Hsp42p, a novel heat shock protein of Saccharomyces cerevisiae, is dependent on a conserved carboxyl-terminal sequence. J Biol Chem 271: 2717–2723 [DOI] [PubMed] [Google Scholar]
- Young JC, Barral JM, Hartl UF (2003) More than folding: localized functions of cytosolic chaperones. Trends Biochem Sci 28: 541–547 [DOI] [PubMed] [Google Scholar]
- Yu C, Huang AH (1986) Conversion of serine to glycerate in intact spinach leaf peroxisomes: role of malate dehydrogenase. Arch Biochem Biophys 245: 125–133 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.