Abstract
Gene expression analysis by microarray assay revealed that when exposed to stress, Entamoeba histolytica exhibits a specific heat shock response, together with a dramatic overall reduction in gene transcription as well as differential allelic expression of key genes participating in virulence, such as the galactose/N-acetylgalactosamine (Gal/GalNAc) lectin.
Amoebiasis is a disease caused by the enteric protozoan parasite Entamoeba histolytica. Following invasion of human tissue by E. histolytica, the two major clinical manifestations are hemorrhagic colitis and liver abscesses (18). For infection to succeed, invading trophozoites must produce an adaptive response that ensures their protection against the host response and survival. Hence, E. histolytica proteins whose production is triggered or modulated by environmental stress are of great interest, since characterization of these species should help us understand the mechanisms which sustain pathogenesis and could lead to new treatments for amoebiasis.
Microbial pathogens have evolved a number of strategies for protecting themselves from their hosts. One of these is the so-called heat shock response, which is elicited by a sudden increase in ambient temperature (13) and induces the synthesis of a limited set of proteins (called heat shock proteins [HSPs] or molecular chaperones). Homologues of known HSPs have been identified and partially characterized in E. histolytica (1, 11). With the aim of determining gene expression changes during E. histolytica's adaptive response during infection, we developed an oligonucleotide-based microarray with transcript information randomly obtained from a cultured virulent strain of the pathogen. Array analysis revealed that gene transcription in E. histolytica exposed to heat shock is dramatically reduced, since 471 of 1,131 unique genes were down regulated, whereas specific HSP-encoding genes were up regulated. In conjunction with real-time PCR results, these genetic information data reveal for the first time a very interesting differential allelic expression of key genes participating in virulence, such as the immunodominant antigen Gal/GalNAc lectin, certain cysteine proteinases, and the so-called 20-kDa antigen.
The aims of this study were (i) to establish a highly discriminating method for monitoring gene expression changes in E. histolytica and (ii) to determine the mRNA expression profile of E. histolytica cells growing in a drastically modified environment. We decided to construct an oligonucleotide-based microarray, using information obtained directly from sequence analysis of E. histolytica transcripts, a strategy that is generally thought to overcome problems due to gene redundancy and the presence of introns. A cDNA library of the virulent E. histolytica strain HM-1:IMSS and a liver-specific cDNA subtraction library were prepared and sequenced. The bioinformatic analysis of sequenced clones enabled us to define 1,300 bona fide transcripts, all from parasites growing in vitro and enriched with randomly chosen transcripts from parasites growing in the liver. A comparison of our file with the recent publication of the E. histolytica genome sequence (10) allowed us to detect and reference 96 transcripts that have not yet been annotated in the genome (referenced as gDNA in our public database at http://genoscript.pasteur.fr). Additionally, of the 1,131 individual genes in the list, 342 corresponded to coding sequences of unknown function. Finally, E. histolytica genes known to be involved in virulence or in cytoskeleton activities were also added to the file. Due to the high AT base content of the E. histolytica genome and in order to achieve a high specificity and limit background noise in the hybridization procedure, we constrained the design of our 70-mer oligonucleotides with the following parameters: a 32% GC content, AT stretches of no longer than 30 nucleotides, and the absence of degenerate bases. All experimental procedures for microarray setting and statistical analysis of data are compiled at http://genoscript.pasteur.fr and summarized in the supplemental material.
We exposed E. histolytica trophozoites growing exponentially at 37°C to a heat shock by moving them to a temperature of 42°C for 4 hours. Cell viability was monitored by using the trypan blue exclusion test, which indicated that 96% of trophozoites were still alive after the treatment. The 3.4-fold increase of Hsp70 indicated that a heat shock response was obtained under our experimental conditions (data not shown). Given that the array represented a relatively small number of genes and that major changes in gene expression are likely during changes in the parasite's milieu, we chose to compare two normalization methods. The first takes into account the overall mean value of E. histolytica gene expression. In the second method, normalization incorporates the use of external controls by spotting oligonucleotides (spikes) prepared from an unrelated source (Arabidopsis thaliana). In both cases, statistical analysis was performed using a paired Student t test. After drawing locally weighted regression normalization curves, we noted that normalization with spikes fitted the asymmetric data set well, with linear correlation coefficients between biological replicates and between technical replicates being >0.9 and >0.95, respectively; the analysis allowed us to conclude that the method using spikes showed a greater specificity for changes in gene expression observed after heat shock treatment, together with a more asymmetric data set (few induced genes and numerous repressed genes).
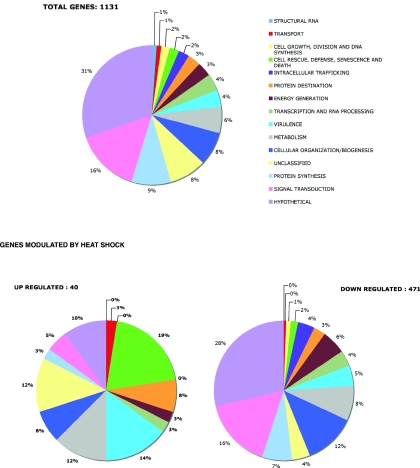
Overview of gene expression modulated by heat shock in Entamoeba histolytica.
To obtain an overview of our microarray results, we classified genes according to their genomic annotations and then tabulated the numbers of differentially regulated genes in selected functional categories. The most striking finding was our observation of massive gene down regulation after heat shock. Of the 1,131 unique genes probed by the microarray (most of which were picked randomly from cDNA sequences), almost 471 (42%) were significantly repressed during heat treatment. One hundred thirty-five (29%) of the 471 down-regulated genes potentially encode proteins of unknown function. Using genome annotation and BLAST, we identified genes involved in growth-related processes, DNA metabolism and repair, fatty acid metabolism, RNA dynamics, signaling, cytoskeletal activity, vesicular trafficking, metabolic processes, and virulence. An interesting group of genes stood out due to its involvement in protein translation (principally the synthesis of ribosomal proteins). The repression of genes encoding ribosomal proteins has been observed during stress in Saccharomyces cerevisiae (20). A smaller number of genes were up regulated by heat shock; these included the usual chaperones as well as a small set of hypothetical proteins. A comparison of the gene categories present on the microarray with the gene categories modulated by heat shock (Fig. 1) prompted us to conclude that gene down regulation is rather indiscriminate and may correspond to a general response for adaptation to new environmental conditions. These observations support the notion that signals leading to gene down regulation during the heat shock response converge on common pathways resulting from an imbalance in cell homeostasis.
FIG. 1.
Categories of genes whose transcription is modulated by heat shock treatment. The data were organized according to a number of different categories suggested for the analysis of transcripts from protozoan parasites (http://www.ebi.ac.uk/parasites/_old/FalcipProteome/geneclass.html). Note that a homogenous color code (appearing in a clockwise representation) is associated with each category. The distribution of total genes was obtained by the percentages of different categories of genes carried on the microarray (these genes were basically randomly chosen after the sequencing of cDNA libraries). Of the genes up regulated by heat shock, 19% encode factors involved in cell rescue. Interestingly, 14% are related to pathogenesis, and very few correspond to hypothetical proteins. The class distribution of transcripts down regulated by heat shock more or less matches the distribution of genes represented on the microarray.
The most up and down-regulated genes under heat shock conditions were grouped together according to function (Tables 1 and 2). Some of these sequences are discussed further below, and the complete list of clones is presented in our public, MIAME-compliant database (http://genoscript.pasteur.fr). The functional annotation of genes modulated by heat shock was organized according to (i) the gene's role in the heat shock response, (ii) potential regulatory factors, and (iii) virulence-related genes.
TABLE 1.
Genes up regulated during heat shocka
| Oligonucleotide and functional category | NCBI accession no.b | Identification | Increase in expression (fold) |
|---|---|---|---|
| Heat shock response | |||
| EH-IP0483 | 284.t00002 | 90-kDa heat shock protein | 7.8 |
| EH-IP0992 | 92.t00037 | 90-kDa heat shock protein | 6.2 |
| EH-IP0432 | 256.t00015 | 90-kDa heat shock protein | 2.3 |
| EH-IP0298 | 197.t00013 | Hsp70 family | 6.1 |
| EH-IP0727.2 | 45.t00004 | 70-kDa heat shock protein | 3.9 |
| EH-IP0309.1 | 2.t00027 | DnaJ | 4.7 |
| EH-IP0309.2 | 2.t00027 | DnaJ | 4.7 |
| EH-IP0260 | 18.t00071 | p23 cochaperone homologue | 2.0 |
| EH-IP0136 | 13.t00024 | BAG domain | 3.7 |
| EH-IP0958 | 84.t00012 | FKBP homologue | 5.8 |
| EH-IP0368 | 216.t00010 | Ehssp1 | 6.3 |
| EH-IP1010 | 99.t00006 | Ubiquitin-conjugating enzyme | 2.4 |
| EH-IP0753.1 | 49.t00025 | Ubiquitin-activating enzyme | 1.7 |
| EH-IP0962 | 86.t00015 | PCI domain, 26S proteasome | 2.0 |
| EH-IP1063 | X98567/Q27935 | Ubiquitin 1 | 2.1 |
| Regulatory factors | |||
| EH-IP0299 | 198.t00003 | Transcription initiation factor IIIB (BRF domain) | 2.5 |
| EH-IP0677 | 40.t00045 | Reverse transcriptase | 1.8 |
| Specific genes | |||
| EH-IP0334.2 | 202.t00007 | 20-kDa antigen-related protein | 8.2 |
| EH-IP0312 | 2.t00055 | Cysteine protease 6 | 8.9 |
| EH-IP0026 | 10.t00046 | Cysteine protease 4 | 2.1 |
| EH-IP0731.1 | 46.t00004 | Cysteine desulfurase | 2.3 |
| EH-IP0731.2 | 46.t00004 | Cysteine desulfurase | 2.4 |
| EH-IP0791 | 52.t00007 | Lysozyme | 2.0 |
| EH-IP0144 | 131.t00014 | Iron-sulfur flavoprotein | 1.7 |
| EH-IP0493 | 29.t00025 | Sugar/ion transporter | 1.5 |
| EH-IP0210.2 | 16.t00014 | Gal/GalNAc lectin Hgl-2 | 3.7 |
| Unknown proteins | |||
| EH-IP0556 | 313.t00008 | Hypothetical protein/multifamily | 8.8 |
| EH-IP0377 | 22.t00061 | Hypothetical protein | 1.8 |
The Hsp90 chaperone, together with Hsp70, helps newly synthesized proteins to fold. Identified chaperone cofactors were the following: Hsp40 (DNAJ), BAG-1, and CHIP (3, 7), which interacts with the molecular chaperones Hsp70 and Hsp90 through its TPR domain, whereas its U-box domain contains the protein’s E3 ubiquitin ligase activity. A cochaperone for Hsp90 is p23, which bears an SGS domain found in eukaryotic proteins mediating ubiquitination (22). Notice that the heat shock up regulation response also included 84.t00012, a FKBP gene encoding a protein with homology to FK506-binding proteins (FKBP or immunophilin). FKBP proteins display both peptidyl-prolyl cis-trans isomerase and chaperone activity associated with Hsp90 (9). The FKBPs also specifically bind fungal macrolides such as FK506 and rapamycin, which are not yet defined to have an anti-amoeba activity. Finally, the “313.t00008” 81-amino-acid protein sequence, with no known homologies, was encountered within at least 100 other open reading frames in E. histolytica.
http://www.ncbi.nlm.nih.gov/gquery/gquery.fcgi?term=ENTAMOEBA.
TABLE 2.
Virulence-related genes down regulated during heat shocka
| Oligonucleotide | NCBI accession no.b | Identification | Inhibition (fold)c |
|---|---|---|---|
| EH-IP0234.1 | 17.t00056 | Gal/GalNAc lectin 35 subunit Lgl-1 | 7.4 |
| EH-IP0234.2 | 17.t00056 | Gal/GalNAc lectin 35 subunit Lgl-1 | 6.5 |
| EH-IP0234.3 | 17.t00056 | Gal/GalNAc lectin 35 subunit Lgl-1 | 1.9 |
| EH-IP0816 | 56.t00038 | Gal/GalNAc lectin 35 subunit Lgl-3 | 2.5 |
| EH-IP0796.1 | 53.t00006 | Gal/GalNAc lectin subunit Igl1 | 1.8 |
| EH-IP0796.2 | 53.t00006 | Gal/GalNAc lectin subunit Igl1 | 2.2 |
| EH-IP0100 | 119.t00005 | Gal/GalNAc lectin subunit Igl2 | 6.6 |
| EH-IP0491.2 | L14815d | Gal/GalNAc lectin subunit Hgl-3 | 2.4 |
| EH-IP0715 | 442.t00004 | Peroxiredoxin | 7.6 |
| EH-IP0160 | 136.t00007 | Peroxiredoxin | 18.6 |
| EH-IP1002 | 963.t00001 | Peroxiredoxin | 16.7 |
| EH-IP0928.2 | 79.t00025 | Cysteine protease 1 | 7.9 |
| EH-IP0928.1 | 79.t00025 | Cysteine protease 1 | 9.1 |
| EH-IP0409 | 242.t00001 | Cysteine protease 1 | 7.5 |
| EH-IP0343.2 | 206.t00005 | Cysteine proteinase 2 | 7.1 |
| EH-IP0343.1 | 206.t00005 | Cysteine proteinase 2 | 7.1 |
| EH-IP0343.3 | 206.t00005 | Cysteine proteinase 2 | 7.5 |
| EH-IP0343.6 | 206.t00005 | Cysteine proteinase 2 | 6.3 |
| EH-IP0343.4 | 206.t00005 | Cysteine proteinase 2 | 6.1 |
| EH-IP0343.5 | 206.t00005 | Cysteine proteinase 2 | 6.9 |
| EH-IP1066 | X87214e | Cysteine protease 3 | 1.3 |
| EH-IP0626.2 | 358.t00002 | Cysteine protease 8 | 6.0 |
| EH-IP0626.1 | 358.t00002 | Cysteine protease 8 | 7.6 |
| EH-IP1070 | AY156071f | Cysteine protease 13 | 2.6 |
| EH-IP0261 | 180.t00007 | Cysteine protease 17 | 1.7 |
| EH-IP0288.2 | 191.t00018 | Cysteine proteinase | 1.2 |
| EH-IP0254.1 | 18.t00027 | Amoebapore C | 7.5 |
| EH-IP0254.2 | 18.t00027 | Amoebapore C | 5.2 |
| EH-IP0149.2 | 132.t00016 | 20-kDa antigen | 1.7 |
| EH-IP0149.1 | 132.t00016 | 20-kDa antigen | 1.4 |
| EH-IP0216 | 160.t00008 | Surface antigen ariel1 | 17.4 |
| EH-IP0614 | 349.t00010 | Galactokinase | 16.4 |
Notice that the surface molecule ARIEL (an asparagine-rich E. histolytica antigen of unknown function) (12) was down regulated 17-fold by heat shock, whereas amoebapore C was down regulated fivefold. Galactokinase (with a 16-fold down regulation) catalyzes phosphorylation of galactose during its catabolism. The enzyme has attracted significant research attention because of its important metabolic role, notably in humans, where defects in the enzyme can result in the disease state referred to as galactosemia (6). In addition, galactokinase-like molecules have been shown to act as sensors for the intracellular galactose concentration and, under suitable conditions, to function as transcriptional regulators (17).
http://www.ncbi.nlm.nih.gov/gquery/gquery.fcgi?term=ENTAMOEBA.
Rate of inhibition of gene transcription after heat shock.
Hgl-3 allele published in reference 15.
CP3 sequence published in reference 4.
CP13 sequence published in reference 4.
Genes encoding heat shock proteins were among the 40 most highly expressed genes (Table 1). We identified three isoforms of Hsp90, two isoforms of Hsp70, and several well-known chaperone cofactors, such as DNAJ, a protein containing a BAG domain, a homologue of cochaperone p23, a candidate presenting a TPR domain, and a protein with structural homologies with CHIP. In E. histolytica, heat shock activates ubiquitin and a component of the 26S proteasome regulatory complex, as well as several other ubiquitination components involved in the removal of damaged proteins. A copy of the Ehssp1 stress-inducible gene (16), which has a possible role in cellular adaptation to stress conditions, was up regulated.
The BRF subunit of transcription factor IIIB (TFIIIB) from E. histolytica was overexpressed threefold during heat shock. TFIIIB recruits RNA polymerase III (Pol III) to initiate transcription of short, untranslated RNA molecules. The RNA Pol III also transcribes short interspersed nuclear elements (SINEs), which contain Pol III promoters. The E. histolytica genome contains three classes of long interspersed nuclear elements (LINEs) and SINEs (2), accounting for about 6% of the total nuclear DNA. Indeed, SINE transcription accounts for 25% of all RNA transcription in this parasite, and the transcription of these elements may be modulated during stress.
Specific virulence-related genes are modulated by heat shock.
Heat shock treatment of the parasite induced the transcription of genes previously described as cysteine proteases 4 and 6 (CP4 and CP6), which have never previously been found to be expressed during parasite growth (4). CP6 was especially up regulated by heat shock, indicating a particular activity of this protease during stress due to its potential role in the degradation of damaged proteins. In contrast, CP1, CP2, CP8, and CP17 were all down regulated (Table 2).
One of the most striking findings generated by the microarray analysis was the observation of differential expression of alleles for the heavy-chain subunit of the Gal/GalNAc lectin, which is an abundant surface protein complex involved in contact-dependent killing and phagocytosis of target cells by E. histolytica (14). The Gal/GalNAc lectin is composed of two subunits, a light (Lgl), glycosylphosphatidylinositol-anchored, 31/35-kDa subunit and a heavy (Hgl), 170-kDa subunit. The complex is associated with an intermediate, 120-kDa subunit (Igl). Each lectin subunit is represented in the E. histolytica genome by multiple alleles; however, the functional distinctions between these alleles have not yet been established. Six genes with 53 to 85% identity encode Lgl isoforms, and at least two genes encoding specific Hgl isoforms (Hgl-2 and Hgl-3) have been isolated and well studied (15, 19). Furthermore, another Hgl isoform and an incomplete sequence have been described from the genome (10). We found that the Hgl-2 allele and one Lgl allele (which we suggest calling Lgl-6) were involved in the heat shock response, whereas all other lectin family components were not. In contrast, the Hgl-3 allele and all other Lgl and Igl subunits underwent gene repression during heat shock. This was also true for other factors that have been associated with the Gal/GalNAc lectin, such as peroxiredoxin (the principal thiol-containing surface antigen in E. histolytica). Peroxiredoxin is important during the transition from the anaerobic environment of the large intestine to the aerobic environment in human tissue (5) and has been found to associate with the Gal/GalNAc lectin in two-hybrid and cellular experiments (8). Our findings suggest that there is a need to assess Hgl-3's specific activity during oxygen peaks. Quantitative reverse transcription-PCR confirmed that transcription of the Hgl-2 allele (16.t0014) and the Lgl-6 allele (168.t0024) increased roughly sixfold during heat shock, whereas all other Hgl, Igl, and Lgl alleles were down regulated (see the supplemental material). Finally, one of the previously described 20-kDa antigen (202.t00007) (21)-encoding genes was found to be up regulated, whereas the second (132.t00016) was down regulated.
In conclusion, this work is a first step toward defining E. histolytica's overall stress-triggered expression program and should help us to understand this parasite's remarkable ability to survive the host response during tissue infection.
Supplementary Material
Acknowledgments
We thank Marie-Christine Rigothier for help with animal experiments, Catherine Bourgouin for advice with the quantitative reverse transcription-PCR experiments, and Samantha Blazquez for helpful comments on the manuscript.
This work was supported by grants from the Pasteur-Weizmann Research Council and the French Ministry of National Education (via the PRFMMIP program).
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Bakatselou, C., and C. G. Clark. 2000. A mitochondrial-type hsp70 gene of Entamoeba histolytica. Arch. Med. Res. 31:S176-S177. [DOI] [PubMed] [Google Scholar]
- 2.Bakre, A. A., K. Rawal, R. Ramaswamy, A. Bhattacharya, and S. Bhattacharya. 2005. The LINEs and SINEs of Entamoeba histolytica: comparative analysis and genomic distribution. Exp. Parasitol. 110:207-213. [DOI] [PubMed] [Google Scholar]
- 3.Ballinger, C. A., P. Connell, Y. Wu, Z. Hu, L. J. Thompson, L. Y. Yin, and C. Patterson. 1999. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol. Cell. Biol. 19:4535-4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruchhaus, I., B. J. Loftus, N. Hall, and E. Tannich. 2003. The intestinal protozoan parasite Entamoeba histolytica contains 20 cysteine protease genes, of which only a small subset is expressed during in vitro cultivation. Eukaryot. Cell 2:501-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi, M. H., D. Sajed, L. Poole, K. Hirata, S. Herdman, B. E. Torian, and S. L. Reed. 2005. An unusual surface peroxiredoxin protects invasive Entamoeba histolytica from oxidant attack. Mol. Biochem. Parasitol. 143:80-89. [DOI] [PubMed] [Google Scholar]
- 6.Elsas, L. J., II, and K. Lai. 1998. The molecular biology of galactosemia. Genet. Med. 1:40-48. [DOI] [PubMed] [Google Scholar]
- 7.Frydman, J., and J. Hohfeld. 1997. Chaperones get in touch: the Hip-Hop connection. Trends Biochem. Sci. 22:87-92. [DOI] [PubMed] [Google Scholar]
- 8.Hughes, M. A., C. W. Lee, C. F. Holm, S. Ghosh, A. Mills, L. A. Lockhart, S. L. Reed, and B. J. Mann. 2003. Identification of Entamoeba histolytica thiol-specific antioxidant as a GalNAc lectin-associated protein. Mol. Biochem. Parasitol. 127:113-120. [DOI] [PubMed] [Google Scholar]
- 9.Kumar, R., B. Adams, A. Musiyenko, O. Shulyayeva, and S. Barik. 2005. The FK506-binding protein of the malaria parasite, Plasmodium falciparum, is a FK506-sensitive chaperone with FK506-independent calcineurin-inhibitory activity. Mol. Biochem. Parasitol. 141:163-173. [DOI] [PubMed] [Google Scholar]
- 10.Loftus, B., I. Anderson, R. Davies, U. C. Alsmark, J. Samuelson, P. Amedeo, P. Roncaglia, M. Berriman, R. P. Hirt, B. J. Mann, T. Nozaki, B. Suh, M. Pop, M. Duchene, J. Ackers, E. Tannich, M. Leippe, M. Hofer, I. Bruchhaus, U. Willhoeft, A. Bhattacharya, T. Chillingworth, C. Churcher, Z. Hance, B. Harris, D. Harris, K. Jagels, S. Moule, K. Mungall, D. Ormond, R. Squares, S. Whitehead, M. A. Quail, E. Rabbinowitsch, H. Norbertczak, C. Price, Z. Wang, N. Guillen, C. Gilchrist, S. E. Stroup, S. Bhattacharya, A. Lohia, P. G. Foster, T. Sicheritz-Ponten, C. Weber, U. Singh, C. Mukherjee, N. M. El-Sayed, W. A. Petri, Jr., C. G. Clark, T. M. Embley, B. Barrell, C. M. Fraser, and N. Hall. 2005. The genome of the protist parasite Entamoeba histolytica. Nature 433:865-868. [DOI] [PubMed] [Google Scholar]
- 11.Mai, Z., S. Ghosh, M. Frisardi, B. Rosenthal, R. Rogers, and J. Samuelson. 1999. Hsp60 is targeted to a cryptic mitochondrion-derived organelle (“crypton”) in the microaerophilic protozoan parasite Entamoeba histolytica. Mol. Cell. Biol. 19:2198-2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mai, Z., and J. Samuelson. 1998. A new gene family (ariel) encodes asparagine-rich Entamoeba histolytica antigens, which resemble the amebic vaccine candidate serine-rich E. histolytica protein. Infect. Immun. 66:353-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neidhardt, F. C., R. A. VanBogelen, and V. Vaughn. 1984. The genetics and regulation of heat-shock proteins. Annu. Rev. Genet. 18:295-329. [DOI] [PubMed] [Google Scholar]
- 14.Petri, W. A., Jr., R. Haque, and B. J. Mann. 2002. The bittersweet interface of parasite and host: lectin-carbohydrate interactions during human invasion by the parasite Entamoeba histolytica. Annu. Rev. Microbiol. 56:39-64. [DOI] [PubMed] [Google Scholar]
- 15.Purdy, J. E., B. J. Mann, E. C. Shugart, and W. A. Petri, Jr. 1993. Analysis of the gene family encoding the Entamoeba histolytica galactose-specific adhesin 170-kDa subunit. Mol. Biochem. Parasitol. 62:53-59. [DOI] [PubMed] [Google Scholar]
- 16.Satish, S., A. A. Bakre, S. Bhattacharya, and A. Bhattacharya. 2003. Stress-dependent expression of a polymorphic, charged antigen in the protozoan parasite Entamoeba histolytica. Infect. Immun. 71:4472-4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Slepak, T., M. Tang, F. Addo, and K. Lai. 2005. Intracellular galactose-1-phosphate accumulation leads to environmental stress response in yeast model. Mol. Genet. Metab. 86:360-371. [DOI] [PubMed] [Google Scholar]
- 18.Stanley, S. L., Jr. 2003. Amoebiasis. Lancet 361:1025-1034. [DOI] [PubMed] [Google Scholar]
- 19.Tannich, E., F. Ebert, and R. D. Horstmann. 1991. Primary structure of the 170-kDa surface lectin of pathogenic Entamoeba histolytica. Proc. Natl. Acad. Sci. USA 88:1849-1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warner, J. R. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24:437-440. [DOI] [PubMed] [Google Scholar]
- 21.Zhang, W. W., and J. Samuelson. 1993. The cDNA sequence of an abundant Entamoeba histolytica 20-kilodalton protein containing four repetitive domains. Mol. Biochem. Parasitol. 60:323-326. [DOI] [PubMed] [Google Scholar]
- 22.Zhu, S., and J. Tytgat. 2004. Evolutionary epitopes of Hsp90 and p23: implications for their interaction. FASEB J. 18:940-947. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.