Abstract
Bgp, one of the surface-localized glycosaminoglycan-binding proteins of the Lyme disease spirochete, Borrelia burgdorferi, exhibited nucleosidase activity. Infection of SCID mice with B. burgdorferi strain N40 mutants harboring a targeted insertion in bgp and apparently retaining all endogenous plasmids revealed that Bgp is not essential for colonization of immunocompromised mice.
Lyme disease is a tick-borne multisystemic infection caused by three spirochete species, Borrelia burgdorferi sensu stricto, Borrelia garinii, and Borrelia afzelii, which have been collectively termed B. burgdorferi sensu lato. These spirochetes have a complex genetic structure, with a relatively small linear chromosome and an elaborate set of linear and circular plasmids, ranging in size from 5 kb to 56 kb, which encode critical genetic information (12, 19, 43, 51). B. burgdorferi can infect multiple tissues, and the ability of this spirochete to attach to a wide variety of mammalian cells could promote bacterial colonization of the mammalian host (52). Indeed, B. burgdorferi has been shown to encode several cell binding pathways. One pathway is mediated by glycosaminoglycans (GAGs), which are long, linear highly modified repeating disaccharides present on the mammalian cell surface and in the extracellular matrix (23, 26). GAG binding by B. burgdorferi is associated with hemagglutination activity, and we previously purified Bgp (Borrelia GAG-binding protein), a hemagglutinin, from lysates of B. burgdorferi strain N40 clone D10/E9. Bgp is localized on the surface of strain N40, and recombinant Bgp inhibits GAG binding by this strain (37). Bgp, which is encoded on the linear chromosome, is not the only GAG-binding protein, since at least three other surface proteins, DbpA, DbpB, and BBK32, each encoded on linear plasmids, are capable of promoting bacterial attachment to GAGs (17, 18, 36).
In addition to its GAG-binding properties, Bgp is a homolog of the highly conserved dual substrate 5′-methylthioadenosine/S-adenosyl homocysteine (MTA/SAH) nucleosidase (Pfs), found in at least 23 genera of bacteria (14, 53). These enzymes are thought to be important in elimination of MTA and SAH, toxic by-products of metabolic pathways, in the salvage of purines and/or methionine, and in the production of a precursor of the quorum-sensing molecule, autoinducer 2 (AI-2) (2, 6, 9, 42, 45, 53). Unlike Bgp, Pfs in most bacteria is localized to the bacterial cytoplasm. In fact, B. burgdorferi possesses three genes homologous to pfs (19): BB0588 (bgp/pfs-2), BB0375 (pfs-1), and BBI06. BBI06 encodes an apparent signal peptide, suggesting that, like Bgp, it is secreted, whereas BB0375 contains no obvious signal sequence and is likely to reside in the cytoplasm.
Bgp displays MTA/SAH nucleosidase activity.
To determine whether the homology of Bgp to MTA/SAH nucleosidases indeed reflects a biological activity of this protein, we assessed the nucleosidase activity of Bgp, as well as its chromosomally encoded homolog, Pfs-1. The pfs-1 gene was amplified from strain N40 DNA using primers 5PFS1 and 3PFS1 (Table 1; annealing temperature, 48°C), cleaved with BamHI and SalI, and inserted into pET30a. His-Pfs-1 and His-Bgp proteins were purified as described previously (36). Because His-Bgp precipitated at a high concentration when stored at 4°C, aliquots of His-Bgp and His-Pfs were stored in 10% glycerol at −80°C.
TABLE 1.
Primers used in this study for various PCRs
| Primer | Sequence (5′ to 3′)a | Descriptionb |
|---|---|---|
| 5Pfs1 | CGCGGATCCATGATTTTGATAATATCAGCTATGCAA | Chr. 385449-385475 |
| 3pfs1 | ACGCGTCGACTTATATTAGCCTAATAAGTTCCTTTG | Chr. 386137-386162 |
| 5BgpL | CTATTAGCATTCCTTCAGAT | Chr. 605042-605061 |
| 3BgpL | ACAGCAAAAACTATTCTTCAT | Chr. 609410-609390 |
| 5KanPacI | CCTTAATTAAAATCATGAGCCGGCT | Amplifies kan |
| 3KanPacI | CCTTAATTAAGGCGAATGAGCTA | Amplifies kan |
| 5Bgp | CGGGATCCATGAATAATTGTTTAA | Chr. 607138-607153 |
| 3Bgp | AACTGCAGTTAAAGTTTTCTTAAA | Chr. 607920-607935 |
Restriction enzyme sites are in boldface.
Shown are the positions on the chromosome (Chr.) (18).
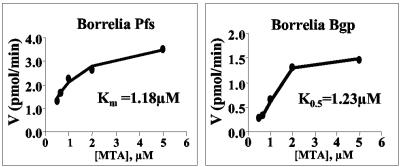
Both purified His-tagged Bgp and Pfs-1 showed nucleosidase activity when assayed as previously described (15, 28) using [3H]MTA prepared from S-adenosyl-l-[methyl-3H]methionine (ARC, St. Louis, MO) (45). The specific activities for Bgp and Pfs-1 were calculated to be 13 μmol/min/mg and 7 μmol/min/mg, respectively (Fig. 1). A Hill coefficient for Bgp of 3.5 was derived from its substrate velocity plot (Fig. 1) (10), suggesting strong cooperative behavior: i.e., binding of MTA by one Bgp subunit further enhances the MTA affinity for the second subunit of the dimer. In contrast, Pfs activity yielded a parabolic curve (Fig. 1) and displayed only weak evidence for cooperative behavior, with a calculated Hill coefficient of 1.2. The cooperative behavior of Bgp indicates that strict Michaelis-Menten kinetics were not obeyed, but kinetic constants, calculated from substrate saturation plots fitted to the equation V0 = Vmax/[1 + (S/Km)n] yielded an estimated K0.5[MTA] of 1.23 μM for Bgp and a Km[MTA] of 1.18 μM for Pfs-1. Thus, although the divergent Hill coefficients suggest that Bgp and Pfs-1 recognize substrate significantly differently at low concentrations, their calculated kinetic constants are very close to the Km[MTA] values (0.45 to 1 μM) reported for other bacterial MTA nucleosidases (14, 16). This, in combination with the high specific activities, indicates that both enzymes efficiently break down MTA and may be important in elimination of toxic metabolic products, purine and/or methionine salvage, and the production of the quorum-sensing molecule AI-2.
FIG. 1.
Bgp is an MTA nucleosidase. The conversion of [3H]MTA to [3H]methylthioribose plus adenine by His-tagged Pfs and Bgp was measured utilizing radioactive 5′ [methyl-3H]methylthioadenosine ([3H]MTA) as a substrate, and kinetic constants were calculated from nonlinear regression analysis of substrate velocity data.
Generation of bgp mutants of strain N40 clone D10/E9 that retain the detectable endogenous plasmids.
Given the potential role of Bgp in bacterial metabolism and/or in promoting bacterial attachment to GAGs, analysis of a B. burgdorferi bgp mutant would greatly facilitate delineation of the potential function(s) of this protein. To generate bgp-deficient mutants of infectious B. burgdorferi strain N40 clone D10/E9 (13), we amplified a 4.3-kb DNA fragment that contained bgp (BB0588) and flanking sequence using the primer pair 5BgpL and 3BgpL (Table 1) and the following PCR conditions: 30 cycles of 95°C for 1 min, 48°C for 1 min, and 65°C for 2 min. The PCR product was cloned into the plasmid TopoXL, which cannot replicate in B. burgdorferi, and a kanamycin resistance (Kanr) cassette driven by the B. burgdorferi flaB promoter (8) was inserted into the bgp gene at a PacI restriction site after codon 159 (see Fig. 2A). Two identical clones, pNP1 and pNP2, were generated and sequenced. Electrocompetent B. burgdorferi strain N40 clone D10/E9 was transformed with 30 μg of either plasmid, and bacteria were allowed to recover overnight in Barbour-Stoenner-Kelly H (BSK-H) medium in the absence of selection. The bacteria were then plated on BSK agarose medium containing 200 μg/ml kanamycin, as previously described (8).
FIG. 2.
Insertional mutagenesis of bgp in strain N40 clone D10/E9. (A, top) Structure of the wild-type N40 bgp locus. The open arrow represents bgp (nucleotides 607138 to 607935, as indicated below map), which can be specifically amplified using primers 5Bgp and 3Bgp (small arrows). Flanking shaded or solid arrows represent neighboring open reading frames. The region between nucleotides 605042 and 609410 (indicated above map) that encompasses bgp was amplified using the primers 5BgpL and 3BgpL (small arrows), cloned, and subjected to kan insertion mutagenesis. (A, bottom) Structure of the bgp locus of mutants NP1.3, NP1.4, and NP2.1, which carry an insertion of the kan gene in bgp. Vertical open arrowheads indicate HindIII sites (see panel C). (B) PCR analysis of NP1.3, NP1.4, and NP2.1, three kanamycin-resistant B. burgdorferi colonies. (Top agarose gel) PCR using primers that specifically amplify kan resulted in the expected 1.2-kb product (Kanr→) from each Kanr clone and from plasmid pNP1, which carries bgp::kan and was used as a positive control. No product was amplified from plasmid pbgpL, which carries wild-type bgp. (Bottom agarose gel) Amplification using primers 5Bgp and 3Bgp that flank bgp (see panel A) resulted in the expected 2.0-kb bgp::kan product (bgp::Kanr→) from each Kanr clone and from plasmid pNP1. In contrast, an 800-bp bgp product (bgp→) was amplified from plasmid pbgpL, which carries wild-type bgp. The leftmost and rightmost lanes of the ethidium-stained gels contain λ HindIII and 1-kb ladder markers, respectively. (C) Southern blot analysis of wild-type N40 D10/E9, NP1.3, NP1.4, and NP2.1. HindIII-digested genomic DNA from the parental strain and each Kanr clone was subjected to Southern hybridization with a bgp probe. HindIII cleavage of the wild-type N40 bgp locus results in the predicted 800-bp bgp-containing fragment (bgp→), whereas HindIII cleavage of NP1.3, NP1.4, or NP2.1 results in the predicted 2.0-kb bgp::kan-containing fragment (bgp::Kanr→).
After 4 weeks at 33°C, eight colonies were obtained and grown in BSK-H complete medium containing 200 μg/ml kanamycin. Total genomic DNA was prepared from broth cultures of these clones as previously described (37). Three clones, termed NP1.3, NP1.4, and NP2.1, were selected, and PCR analysis using primers specific for kan (primers 5KanPacI/3KanPacI in Table 1; annealing temperature, 55°C) or bgp (primers 5Bgp/3Bgp; annealing temperature, 48°C) suggested they each carried the kan cassette and an approximately 1.2-kb insertion in bgp (Fig. 2B). Southern blots of total genomic DNA digested with HindIII were hybridized with a nonradioactively labeled bgp probe generated by PCR (Roche Diagnostics GmbH, Mannheim, Germany) using the primers 5Bgp and 3Bgp, and no wild-type bgp gene was detected in NP1.3, NP1.4, and NP2.1 (Fig. 2C), indicating that the mutants arose by allelic exchange involving recombination events, rather than a single crossover event that would result in the insertion of the entire plasmid into the chromosome.
Due to the complex structure of the B. burgdorferi genome and the sporadic loss of plasmids essential for infectivity associated with in vitro culture (20, 34, 39, 40, 46, 55), genetic studies of this organism are extremely challenging. To date, only a few B. burgdorferi mutants have been generated in infectious strain backgrounds, and most of these studies have utilized derivatives of strain B31 MI, whose genome sequence has been determined, thus facilitating a PCR-based analysis of plasmid content of mutants (12, 19, 43, 51). However, B. burgdorferi strain N40, the strain used for previous work on Bgp, has been widely studied, in part because it is capable of recapitulating many of the clinical manifestations of human Lyme disease in several animal models (1, 3-5, 11, 30, 32, 33, 56). In addition, a draft DNA sequence of the strain N40 linear chromosome indicates low (∼0.5%) sequence divergence from B31 M1 (41), and analysis of cloned and uncloned versions of N40 revealed plasmids corresponding to many of those found in strain B31 M1 (35, 50). Therefore, we attempted to amplify sequences from strain N40 clone D10/E9, using primer pairs previously used to detect plasmids in other strains (40, 50). Total plasmid DNA from this strain, as well as from the control strain B31 5A4 (40) (generously provided by Steve Norris), was isolated using a Promega Wizard-Plus Miniprep kit.
Control amplifications of strain B31 5A4 using each of the 22 primer pairs that amplify sequences from plasmids present in this strain generated PCR products of the predicted sizes. Fifteen of the 22 primer pairs also amplified products of identical sizes from strain N40 DNA (see Table S1 in the supplemental material). (The primers specific for lp56 and cp9 gave rise to detectable products, but with lower apparent molecular weights than the corresponding strain B31 amplicons [data not shown].) In addition, a primer pair that detects cp32-12, which is not present in strain B31 but is found in an independent clone of strain N40 (50), amplified a product of the predicted size from strain N40 D10/E9, but not from strain B31 5A4 (see Table S1 in the supplemental material). Although amplification by itself provides no information regarding the physical location of the detected sequences on the main chromosome or linear or circular plasmids, this result, as well as analysis of plasmids from diverse strains of B. burgdorferi (25), including strain N40 (35), makes it reasonable to postulate that the primer pairs utilized amplified plasmid sequences from strain N40 D10/E9.
The plasmid content of the three N40 D10/E9 bgp::kan mutants was then assessed using the 16 primer pairs that amplify (presumed) plasmid sequences in the parental strain. Amplification of DNA of the clones NP1.3 and NP1.4 resulted in products of the predicted size for all 16 primer pairs (not shown). Amplification of NP2.1 did not give rise to any product when done with primers specific for lp25, suggesting that this plasmid, which is somewhat unstable when B. burgdorferi is propagated in vitro and encodes a nicotinamidase that is required for survival in mice (39), is not present in this mutant.
Bgp is not essential for infection of immunodeficient mice.
Bgp-mediated bacterial attachment to extracellular matrix or host cells might promote the establishment of an initial infectious niche in host tissues, or Bgp-mediated MTA nucleosidase activity could be required to inactivate toxic metabolic products during growth in the mammalian host. Antibodies directed against B. burgdorferi have been shown to induce dramatic changes in the expression of several spirochetal surface proteins (29), and an adaptive immune response might alter the expression of bgp during infection. To examine the role of Bgp in early tissue colonization without the potentially confounding effect of an adaptive immune response on its production, we infected severe combined immunodeficient (SCID) mice and assessed colonization at a relatively early time point. Groups of three C3H/HeJ SCID mice were inoculated subcutaneously with 1 × 104 wild-type N40 D10/E9, NP1.3, NP1.4, or NP2.1 colonies, and at 2 weeks after infection, skin samples from the injection site, ear, heart, and bladder were cultured in BSK-H complete medium in the absence or presence of 200 μg/ml kanamycin for the wild-type strain and bgp mutants, respectively. NP2.1, the bgp mutant clone that apparently lacks lp25, a plasmid required for infectivity (39), was not recovered from any tissue. Wild-type N40 D10/E9, as well as NP1.3 and NP1.4, the two bgp mutants that apparently retained the 16 plasmids, was isolated from multiple sites (Table 2).
TABLE 2.
Bgp is not required for infection of immunodeficient micea
| Infecting strain | No. of positive cultures/total from:
|
||||
|---|---|---|---|---|---|
| Injection site | Blood | Skin | Joint | Bladder | |
| N40 D10/E9 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 |
| NP1.3 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 |
| NP1.4 | 3/3 | 1/3 | 1/3 | 3/3 | 3/3 |
| NP2.1 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 |
Groups of three C3H SCID mice were infected intradermally or subcutaneously with wild-type B. burgdorferi N40 clone D10/E9 or one of the bgp mutant derivatives NP1.3, NP1.4, and NP2.1. After 2 weeks of infection, the indicated sites were cultured for 1 week in BSK-H medium, at which time positive cultures were determined by microscopic examination.
The results obtained here indicate that Bgp is not required for infection of immunodeficient mice. It is possible that two activities of Bgp, i.e., MTA/SAH nucleosidase and GAG binding, are not required during infection. Although quorum-sensing pathways influence virulence phenotypes in several bacterial pathogens (27, 31, 44, 47, 48, 54) and AI-2 has been implicated in the production of several B. burgdorferi proteins in vitro (49), a luxS mutant of B. burgdorferi, which is incapable of producing AI-2, displays no defect in tick or mammalian infection (7, 24). It is also possible that the spirochete encodes redundant functions that mask a virulence defect due to the lack of Bgp. For example, two other Pfs homologs are encoded by this bacterium, at least one of which, Pfs-1, we show here to possess nucleosidase activity. In addition, the Lyme disease spirochete encodes multiple GAG-binding adhesins, such as DbpA and DbpB (18, 21, 22, 36) and the fibronectin binding protein BBK32 (17, 38). Rigorous testing of the roles of MTA/SAH nucleosidase and GAG binding during mammalian infection is likely to involve the generation of B. burgdorferi strains with defined lesions in multiple genes. The development of methods to easily assess the plasmid content of strain N40 should facilitate such genetic studies.
Supplementary Material
Acknowledgments
We thank S. Casjens and C. Fraser (University of Utah), as well as Brian Stevenson (University of Kentucky College of Medicine), for providing invaluable advice for detection of plasmid content.
This study was supported by American Heart Association grant AHA/0265355T to N.P. and NIH grant R01-AI37601 to J.M.L.
Editor: J. T. Barbieri
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Armstrong, A. L., S. W. Barthold, D. H. Persing, and D. S. Beck. 1992. Lyme disease susceptible and resistant strains of laboratory mice infected with Borrelia burgdorferi. Am. J. Trop. Med. Hyg. 47:249-258. [DOI] [PubMed] [Google Scholar]
- 2.Backlund, P. S., Jr., C. P. Chang, and R. A. Smith. 1982. Identification of 2-keto-4-methylthiobutyrate as an intermediate compound in methionine synthesis from 5′-methylthioadenosine. J. Biol. Chem. 257:4196-4202. [PubMed] [Google Scholar]
- 3.Bai, Y., K. Narayan, D. Dail, M. Sondey, E. Hodzic, S. W. Barthold, A. R. Pachner, and D. Cadavid. 2004. Spinal cord involvement in the nonhuman primate model of Lyme disease. Lab. Investig. 84:160-172. [DOI] [PubMed] [Google Scholar]
- 4.Barthold, S. W., D. S. Beck, G. M. Hansen, G. A. Terwilliger, and K. D. Moody. 1990. Lyme borreliosis in selected strains and ages of laboratory mice. J. Infect. Dis. 162:133-138. [DOI] [PubMed] [Google Scholar]
- 5.Barthold, S. W., M. S. de Souza, J. L. Janotka, A. L. Smith, and D. H. Persing. 1993. Chronic Lyme borreliosis in the laboratory mouse. Am. J. Pathol. 143:959-971. [PMC free article] [PubMed] [Google Scholar]
- 6.Beeston, A. L., and M. G. Surette. 2002. pfs-dependent regulation of autoinducer 2 production in Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:3450-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blevins, J. S., A. T. Revel, M. J. Caimano, X. F. Yang, J. A. Richardson, K. E. Hagman, and M. V. Norgard. 2004. The luxS gene is not required for Borrelia burgdorferi tick colonization, transmission to a mammalian host, or induction of disease. Infect. Immun. 72:4864-4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bono, J. L., A. F. Elias, J. J. Kupko III, B. Stevenson, K. Tilly, and P. Rosa. 2000. Efficient targeted mutagenesis in Borrelia burgdorferi. J. Bacteriol. 182:2445-2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borchardt, R. T. 1980. S-Adenosyl-L-methionine-dependent macromolecule methyltransferases: potential targets for the design of chemotherapeutic agents. J. Med. Chem. 23:347-357. [DOI] [PubMed] [Google Scholar]
- 10.Bowen, W. P., and J. C. Jerman. 1995. Nonlinear regression using spreadsheets. Trends Pharmacol. Sci. 16:413-417. [DOI] [PubMed] [Google Scholar]
- 11.Cadavid, D., Y. Bai, E. Hodzic, K. Narayan, S. W. Barthold, and A. R. Pachner. 2004. Cardiac involvement in non-human primates infected with the Lyme disease spirochete Borrelia burgdorferi. Lab. Investig. 84:1439-1450. [DOI] [PubMed] [Google Scholar]
- 12.Casjens, S., N. Palmer, R. van Vugt, W. M. Huang, B. Stevenson, P. Rosa, R. Lathigra, G. Sutton, J. Peterson, R. J. Dodson, D. Haft, E. Hickey, M. Gwinn, O. White, and C. M. Fraser. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490-516. [DOI] [PubMed] [Google Scholar]
- 13.Coburn, J., J. Leong, and J. Erban. 1993. Integrin aIIbb3 mediates binding of the Lyme disease agent, Borrelia burgdorferi, to human platelets. Proc. Natl. Acad. Sci. USA 90:7058-7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cornell, K. A., and M. K. Riscoe. 1998. Cloning and expression of Escherichia coli 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase: identification of the pfs gene product. Biochim. Biophys. Acta 1396:8-14. [DOI] [PubMed] [Google Scholar]
- 15.Cornell, K. A., W. E. Swarts, R. D. Barry, and M. K. Riscoe. 1996. Characterization of recombinant Escherichia coli 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase: analysis of enzymatic activity and substrate specificity. Biochem. Biophys. Res. Commun. 228:724-732. [DOI] [PubMed] [Google Scholar]
- 16.Della Ragione, F., M. Porcelli, M. Carteni-Farina, V. Zappia, and A. E. Pegg. 1985. Escherichia coli S-adenosylhomocysteine/5′-methylthioadenosine nucleosidase. Purification, substrate specificity and mechanism of action. Biochem. J. 232:335-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischer, J. R., K. T. LeBlanc, and J. M. Leong. 2006. Fibronectin binding protein BBK32 of the Lyme disease spirochete promotes bacterial attachment to glycosaminoglycans. Infect. Immun. 74:435-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischer, J. R., N. Parveen, L. Magoun, and J. M. Leong. 2003. Decorin-binding proteins A and B confer distinct mammalian cell type-specific attachment by Borrelia burgdorferi, the Lyme disease spirochete. Proc. Natl. Acad. Sci. USA 100:7307-7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J. F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, J. C. Venter et al. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 20.Grimm, D., A. F. Elias, K. Tilly, and P. A. Rosa. 2003. Plasmid stability during in vitro propagation of Borrelia burgdorferi assessed at a clonal level. Infect. Immun. 71:3138-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo, B. P., E. L. Brown, D. W. Dorward, L. C. Rosenberg, and M. Hook. 1998. Decorin-binding adhesins from Borrelia burgdorferi. Mol. Microbiol. 30:711-723. [DOI] [PubMed] [Google Scholar]
- 22.Guo, B. P., S. J. Norris, L. C. Rosenberg, and M. Höök. 1995. Adherence of Borrelia burgdorferi to the proteoglycan decorin. Infect. Immun. 63:3467-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hook, M., L. Kjellen, S. Johansson, and J. Robinson. 1984. Cell-surface glycosaminoglycans. Annu. Rev. Biochem. 53:847-869. [DOI] [PubMed] [Google Scholar]
- 24.Hubner, A., A. T. Revel, D. M. Nolen, K. E. Hagman, and M. V. Norgard. 2003. Expression of a luxS gene is not required for Borrelia burgdorferi infection of mice via needle inoculation. Infect. Immun. 71:2892-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iyer, R., O. Kalu, J. Purser, S. Norris, B. Stevenson, and I. Schwartz. 2003. Linear and circular plasmid content in Borrelia burgdorferi clinical isolates. Infect. Immun. 71:3699-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson, R., S. Busch, and A. Cardin. 1991. Glycosaminoglycans: molecular properties, protein interactions, and role in physiological processes. Physiol. Rev. 71:481-539. [DOI] [PubMed] [Google Scholar]
- 27.Joyce, E. A., A. Kawale, S. Censini, C. C. Kim, A. Covacci, and S. Falkow. 2004. LuxS is required for persistent pneumococcal carriage and expression of virulence and biosynthesis genes. Infect. Immun. 72:2964-2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, J. E., K. A. Cornell, M. K. Riscoe, and P. L. Howell. 2001. Expression, purification, crystallization and preliminary X-ray analysis of Escherichia coli 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase. Acta Crystallogr. D Biol. Crystallogr. 57:150-152. [DOI] [PubMed] [Google Scholar]
- 29.Liang, F. T., J. Yan, M. L. Mbow, S. L. Sviat, R. D. Gilmore, M. Mamula, and E. Fikrig. 2004. Borrelia burgdorferi changes its surface antigenic expression in response to host immune responses. Infect. Immun. 72:5759-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma, Y., K. P. Seiler, E. J. Eichwald, J. H. Weis, C. Teuscher, and J. J. Weis. 1998. Distinct characteristics of resistance to Borrelia burgdorferi-induced arthritis in C57BL/6N mice. Infect. Immun. 66:161-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marouni, M. J., and S. Sela. 2003. The luxS gene of Streptococcus pyogenes regulates expression of genes that affect internalization by epithelial cells. Infect. Immun. 71:5633-5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moody, K. D., G. A. Terwilliger, G. M. Hansen, and S. W. Barthold. 1994. Experimental Borrelia burgdorferi infection in Peromyscus leucopus. J. Wildl. Dis. 30:155-161. [DOI] [PubMed] [Google Scholar]
- 33.Pachner, A. R., E. Delaney, T. O'Neill, and E. Major. 1995. Inoculation of nonhuman primates with the N40 strain of Borrelia burgdorferi leads to a model of Lyme neuroborreliosis faithful to the human disease. Neurology 45:165-172. [DOI] [PubMed] [Google Scholar]
- 34.Pal, U., X. Yang, M. Chen, L. K. Bockenstedt, J. F. Anderson, R. A. Flavell, M. V. Norgard, and E. Fikrig. 2004. OspC facilitates Borrelia burgdorferi invasion of Ixodes scapularis salivary glands. J. Clin. Investig. 113:220-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palmer, N., C. Fraser, and S. Casjens. 2000. Distribution of twelve linear extrachromosomal DNAs in natural isolates of Lyme disease spirochetes. J. Bacteriol. 182:2476-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parveen, N., M. Caimano, J. D. Radolf, and J. M. Leong. 2003. Adaptation of the Lyme disease spirochaete to the mammalian host environment results in enhanced glycosaminoglycan and host cell binding. Mol. Microbiol. 47:1433-1444. [DOI] [PubMed] [Google Scholar]
- 37.Parveen, N., and J. M. Leong. 2000. Identification of a candidate glycosaminoglycan-binding adhesin of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:1220-1234. [DOI] [PubMed] [Google Scholar]
- 38.Probert, W. S., and B. J. Johnson. 1998. Identification of a 47 kDa fibronectin-binding protein expressed by Borrelia burgdorferi isolate B31. Mol. Microbiol. 30:1003-1015. [DOI] [PubMed] [Google Scholar]
- 39.Purser, J. E., M. B. Lawrenz, M. J. Caimano, J. K. Howell, J. D. Radolf, and S. J. Norris. 2003. A plasmid-encoded nicotinamidase (PncA) is essential for infectivity of Borrelia burgdorferi in a mammalian host. Mol. Microbiol. 48:753-764. [DOI] [PubMed] [Google Scholar]
- 40.Purser, J. E., and S. J. Norris. 2000. Correlation between plasmid content and infectivity in Borrelia burgdorferi. Proc. Natl. Acad. Sci. USA 97:13865-13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qiu, W. G., S. E. Schutzer, J. F. Bruno, O. Attie, Y. Xu, J. J. Dunn, C. M. Fraser, S. R. Casjens, and B. J. Luft. 2004. Genetic exchange and plasmid transfers in Borrelia burgdorferi sensu stricto revealed by three-way genome comparisons and multilocus sequence typing. Proc. Natl. Acad. Sci. USA 101:14150-14155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Riscoe, M. K., A. J. Ferro, and J. H. Fitchen. 1989. Methionine salvage as a target for antiprotozoal drug development. Parasitol. Today 5:330-333. [DOI] [PubMed] [Google Scholar]
- 43.Rosa, P. A., K. Tilly, and P. E. Stewart. 2005. The burgeoning molecular genetics of the Lyme disease spirochaete. Nat. Rev. Microbiol. 3:129-143. [DOI] [PubMed] [Google Scholar]
- 44.Rumbaugh, K. P., J. A. Griswold, and A. N. Hamood. 2000. The role of quorum sensing in the in vivo virulence of Pseudomonas aeruginosa. Microbes Infect. 2:1721-1731. [DOI] [PubMed] [Google Scholar]
- 45.Schlenk, F. 1983. Methylthioadenosine. Adv. Enzymol Relat. Areas Mol. Biol. 54:195-265. [DOI] [PubMed] [Google Scholar]
- 46.Schwan, T. G., W. Burgdorfer, and C. F. Garon. 1988. Changes in infectivity and plasmid profile of the Lyme disease spirochete, Borrelia burgdorferi, as a result of in vitro cultivation. Infect. Immun. 56:1831-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sircili, M. P., M. Walters, L. R. Trabulsi, and V. Sperandio. 2004. Modulation of enteropathogenic Escherichia coli virulence by quorum sensing. Infect. Immun. 72:2329-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sperandio, V., C. C. Li, and J. B. Kaper. 2002. Quorum-sensing Escherichia coli regulator A: a regulator of the LysR family involved in the regulation of the locus of enterocyte effacement pathogenicity island in enterohemorrhagic E. coli. Infect. Immun. 70:3085-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stevenson, B., and K. Babb. 2002. LuxS-mediated quorum sensing in Borrelia burgdorferi, the Lyme disease spirochete. Infect. Immun. 70:4099-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stevenson, B., and J. C. Miller. 2003. Intra- and interbacterial genetic exchange of Lyme disease spirochete erp genes generates sequence identity amidst diversity. J. Mol. Evol 57:309-324. [DOI] [PubMed] [Google Scholar]
- 51.Stewart, P. E., R. Byram, D. Grimm, K. Tilly, and P. A. Rosa. 2005. The plasmids of Borrelia burgdorferi: essential genetic elements of a pathogen. Plasmid 53:1-13. [DOI] [PubMed] [Google Scholar]
- 52.Vallance, B. A., and B. B. Finlay. 2000. Exploitation of host cells by enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 97:8799-8806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Winzer, K., K. R. Hardie, N. Burgess, N. Doherty, D. Kirke, M. T. Holden, R. Linforth, K. A. Cornell, A. J. Taylor, P. J. Hill, and P. Williams. 2002. LuxS: its role in central metabolism and the in vitro synthesis of 4-hydroxy-5-methyl-3(2H)-furanone. Microbiology 148:909-922. [DOI] [PubMed] [Google Scholar]
- 54.Winzer, K., and P. Williams. 2001. Quorum sensing and the regulation of virulence gene expression in pathogenic bacteria. Int. J. Med. Microbiol. 291:131-143. [DOI] [PubMed] [Google Scholar]
- 55.Yang, X. F., U. Pal, S. M. Alani, E. Fikrig, and M. V. Norgard. 2004. Essential role for OspA/B in the life cycle of the Lyme disease spirochete. J. Exp. Med. 199:641-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zeidner, N. S., M. S. Nuncio, B. S. Schneider, L. Gern, J. Piesman, O. Brandao, and A. R. Filipe. 2001. A Portuguese isolate of Borrelia lusitaniae induces disease in C3H/HeN mice. J. Med. Microbiol. 50:1055-1060. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.