Abstract
We investigated genome dynamics at a chromosome end in the model plant Arabidopsis thaliana through a study of natural variation in 35 wild accessions. We focused on the single-copy subtelomeric region of chromosome 1 north (∼3.5 kb), which represents the relatively simple organization of subtelomeric regions in this species. PCR fragment-length variation across the subtelomeric region indicated that the 1.4-kb distal region showed elevated structural variation relative to the centromere-proximal region. Examination of nucleotide sequences from this 1.4-kb region revealed diverse DNA rearrangements, including an inversion, several deletions, and an insertion of a retrotransposon LTR. The structures at the deletion and inversion breakpoints are characteristic of simple deletion-associated nonhomologous end-joining (NHEJ) events. There was strong linkage disequilibrium between the distal subtelomeric region and the proximal telomere, which contains degenerate and variant telomeric repeats. Variation in the proximal telomere was characterized by the expansion and deletion of blocks of repeats. Our sample of accessions documented two independent chromosome-healing events associated with terminal deletions of the subtelomeric region as well as the capture of a scrambled mitochondrial DNA segment in the proximal telomeric array. This natural variation study highlights the variety of genomic events that drive the fluidity of chromosome termini.
THE transitional region between telomeric repeats that cap the chromosome end and the most distal chromosome-specific sequence is termed the subtelomeric region. The organization of this genomic region varies among eukaryotic organisms (Pryde et al. 1997). However, some common features can be recognized, including an abundance of repetitive sequences (including microsatellites, blocks of larger tandem repeats, and transposons) (Levis et al. 1993; Vershinin et al. 1995; Pearce et al. 1996; Amarger et al. 1998) and the presence of duplicated sequences and/or paralogous genes shared among nonhomologous chromosomal ends (Carlson et al. 1985; Louis 1995; Thompson et al. 1997; 2002). The complex and extensive sequence similarity exhibited among subtelomeric regions suggests that frequent sequence exchange occurs between nonhomologous chromosome ends. A recent study in humans inferred two major processes that generate the patchwork of sequence blocks shared extensively among nonhomologous chromosome ends: chromosomal translocations through nonhomologous end-joining (NHEJ) DNA repair and subsequent homologous recombination among the duplicated segments on different chromosomes (Linardopoulou et al. 2005). The role of ectopic recombination between nonhomologous chromosomes has also been shown to underlie the complex organization of the subtelomeric regions in other organisms (Louis and Haber 1990; Freitas-Junior et al. 2000). As expected for a plastic region of the genome subject to reshuffling through recombination events, subtelomeric regions are highly polymorphic and evolutionarily dynamic (Broun et al. 1992; Royle et al. 1994; Baird and Royle 1997; Mefford and Trask 2002; Eichler and Sankoff 2003).
The role of subtelomeric regions in chromosome stability and function remains elusive. Subtelomeric regions in most organisms generally contain nonfunctional repetitive sequences, and in cases where subtelomeric sequences have been lost or omitted, cell viability and chromosome stability were not affected (see review by Mefford and Trask 2002). However, subtelomeric regions can provide a backup mechanism for acquisition of a new telomeric end through ectopic recombination with shared subtelomeric sequences on nonhomologous chromosome ends (Wang and Zakian 1990). In extreme cases where conventional telomere repeat addition is compromised in budding yeast, amplification of subtelomeric repeats can ensure chromosome maintenance through a so-called ALT alternative telomere lengthening mechanism (Lundblad and Blackburn 1993). In addition to processes that might directly affect chromosome stability and telomere function, subtelomeric plasticity may also play a role as a platform for elaborating novel gene expression programs (e.g., VSG gene expression and host immune response avoidance in Trypanosoma) (Borst et al. 1996; Barry et al. 2003). Another potential function of the subtelomeric region is to insulate the distal genes from the suppressive effects imposed by telomeric chromatin (telomere positional effect) (Gottschling et al. 1990; Baur et al. 2001; Garcia-Cao et al. 2004). Alternatively, the subtelomeric region can provide a location for genes to be modulated by chromatin level regulation, as has been demonstrated in Plasmodium (Duraisingh et al. 2005; Freitas-Junior et al. 2005). However, the plastic nature of subtelomeric regions can also cause harm to an organism, as seen in human diseases associated with subtelomeric gene rearrangements (see review by Mefford and Trask 2002). These considerations suggest that subtelomeric regions are under a range of functional constraints and that different evolutionary processes are shaping the boundary and organization of the subtelomeric regions in diverse organisms.
In contrast to subtelomeric regions, the telomere plays a direct and essential role in maintaining genomic integrity and cell vitality (Muller 1938; McClintock 1941; Blackburn 2000). In most eukaryotes that have been examined, the extreme chromosomal termini are formed by the interaction of specialized telomere-binding proteins and tandem arrays of short G-rich repeats. These telomeric repeat arrays are synthesized by a ribonucleoprotein complex, called telomerase, using an RNA template. The telomeric structure protects the chromosome end from shortening through rounds of incomplete replication or degradation and also from being recognized as broken ends subject to DNA repair (see review by Blackburn 2001; Bucholc et al. 2001). Telomeric repeat arrays are dynamic structures that undergo expansion and contraction throughout development and possibly in response to physiological stresses (Blackburn 2001; Epel et al. 2004). As a consequence of this equilibrium, telomeric sequences added at the end of the chromosomal molecule are turned over at a higher rate than those sequences located in the centromere-proximal portion of the telomeric repeat arrays. This imbalance in turnover rate can account for the higher frequency of degenerate and variant telomeric repeats in the centromere-proximal domain of the telomere that has been observed in various organisms (Allshire et al. 1989; Richards et al. 1992; Kirk and Blackburn 1995).
Studies using the flowering plant Arabidopsis thaliana have provided basic information on both telomere DNA structure and its maintenance (Riha and Shippen 2003), but relatively little is known about the organization, function, and dynamics of A. thaliana subtelomeric regions. In most higher plant species studied, a variety of tandemly repeated sequences and transposons are associated with subtelomeric regions (Roder et al. 1993; Wu and Tanksley 1993; Vershinin et al. 1995; Pearce et al. 1996; Ohmido et al. 2001; Alkhimova et al. 2004). In contrast, A. thaliana subtelomeric regions are remarkably small and simple, in accordance with this species' small genome size and paucity of repetitive sequences. The two chromosome ends (chromosome 2 north and 4 north) that constitute the nucleolus organizer regions have telomeric repeats adjoined to the tandem array of rRNA-coding sequences (Copenhaver and Pikaard 1996). The remaining eight chromosome ends contain short subtelomeric regions (<5 kb) that are devoid of highly repetitive sequences or transposons (Arabidopsis Genome Initiative 2000; Heacock et al. 2004). Although some subtelomeric regions in Arabidopsis do share a few blocks of similarity of low-copy sequences among nonhomologous chromosomes (Kotani et al. 1999; Heacock et al. 2004), subtelomeric regions in Arabidopsis do not share extensive similarity among most nonhomologous chromosomes, such as that seen in yeast and humans (Louis 1995; Mefford and Trask 2002; Linardopoulou et al. 2005).
We examined natural variation in the nucleotide sequence structure of the single-copy subtelomeric region and adjacent proximal region of the telomere from chromosome 1 north (1N) in A. thaliana to determine whether this unique region of the genome is evolving differently from other genomic loci. In addition, we examined the pattern of variation at the chromosome 1N end to infer the molecular mechanisms that shape the unusually simple genomic organization present at chromosomal termini in Arabidopsis.
MATERIALS AND METHODS
Plant materials:
Thirty-five wild accessions of A. thaliana originating from diverse geographic locations were chosen for this study (Table 1). Seeds were obtained from the Arabidopsis Biological Resource Center (Columbus, OH), as were seven accessions of A. suecica (CS22511–CS22517). The A. suecica accessions were originally collected in the village of Tjörnarp in southern Sweden by S. Anderson (L. Comai and M. Nordborg, personal communication). An additional wild accession of A. suecica was obtained from the Helsinki Botanical Garden (seed exchange). A. suecica laboratory strains LC1 and 9502 were provided by C. Pikaard (Pontes et al. 2003). A. suecica LC1 is derived from Sue-1 (Comai et al. 2000). A. suecica 9502 is derived from accession 90-10-085-10 (originating from Finland, collected by S. O'Kane), and A. arenosa accession 3651 originated in Poland (also collected by S. O'Kane) (O'Kane et al. 1996).
TABLE 1.
Plant materials used in this study
| Species | Name codes | Origin | Source/collectors |
|---|---|---|---|
| Arabidopsis thaliana | Br-0 | Czech Republic | CS22628 |
| Bur-0 | Ireland | CS22656 | |
| C24 | Portugal | CS22620 | |
| Col | USA (Germany?) | F. Ausubel lab stock (Harvard University) | |
| Ct-1 | Italy | CS22639 | |
| Cvi-0 | Cape Verde Islands | CS22614 | |
| Edi-0 | Scotland | CS22657 | |
| Fei-0 | Portugal | CS22645 | |
| HR-5 | England | CS22596 | |
| Kas-2 | India | CS22638 | |
| Knox-10 | USA/midwest (Indiana) | CS22566 | |
| Knox-18 | USA/midwest (Indiana) | CS22567 | |
| Kondara | Tajikistan | CS22651 | |
| Kz-1 | Kazakhstan | CS22606 | |
| Ler-1 | Germany | CS22618 | |
| Lz-0 | France | CS22615 | |
| Ms-0 | Russia | CS22655 | |
| Mt-0 | Libya | CS22642 | |
| N13 | Russia | CS22621 | |
| NFA-8 | England | CS22598 | |
| Oy-0 | Norway | CS22658 | |
| Pro-0 | Spain | CS22649 | |
| Pu2-7 | Czech Republic | CS22592 | |
| Ra-0 | France | CS22632 | |
| Shakdara | Tajikistan | CS22652 | |
| Sorbo | Tajikistan | CS22653 | |
| Tamm-27 | Finland | CS22605 | |
| Ts-1 | Spain | CS22647 | |
| Tsu-1 | Japan | CS22641 | |
| Uod-1 | Austria | CS22612 | |
| Van-0 | Canada | CS22627 | |
| Wa-1 | Poland | CS22644 | |
| Wei-0 | Switzerland | CS22622 | |
| Ws-0 | Ukraine | CS22623 | |
| Ws-2 | Ukraine | CS22659 | |
| A. suecica | 9502 | Finland | C. Pikaard lab stock (Washington University) |
| LC1 | Unknown | C. Pikaard lab stock (Washington University) | |
| Finland | Helsinki Botanical Garden (seed exchange) | ||
| Sue 10 | Tjörnarp, Sweden | CS22511 | |
| Sue 11 | Tjörnarp, Sweden | CS22512 | |
| Sue 12 | Tjörnarp, Sweden | CS22513 | |
| Sue 13 | Tjörnarp, Sweden | CS22514 | |
| Sue 14 | Tjörnarp, Sweden | CS22515 | |
| Sue 15 | Tjörnarp, Sweden | CS22516 | |
| Sue 16 | Tjörnarp, Sweden | CS22517 | |
| A. arenosa | 3651 | Poland | S. O'Kane |
DNA preparation, PCR analysis, and DNA sequencing:
Seeds were germinated and grown axenically on plates containing MS germination medium [4.3 g/liter MS salt, 1× Gamborg's vitamins, 2% (w/v) sucrose, 0.8% agar, pH 5.7] (Murashige and Skoog 1962) in the growth chamber under a 16 hr light/8 hr dark, 22° growth regime. After 2 weeks, genomic DNA was extracted from whole plants following the CTAB procedure (Rogers and Bendich 1985). Primers (21–24 nucleotides in length) used for PCR analysis were designed on the basis of the published genomic sequence of the lab strain Columbia (TAIR 6.0; http://www.arabidopsis.org) and are labeled according to their chromosomal coordinates as shown in Figure 1. DNA sequences of the 1.4-kb telomere-proximal region were analyzed by direct sequencing of multiple independent PCR-amplified genomic products [except for A. suecica CS22511–22517, for which DNA sequencing analysis was based on a single amplicon from F2367 and TeloRep(R)]. In a few instances, DNA sequencing of cloned PCR products was used to resolve sequence ambiguities (TA Cloning Kit; Invitrogen). DNA sequence reactions were set up according to the protocols developed by Applied Biosystems with minor modifications, and the extension products were separated and imaged using a Prism 3130 16-capillary automated sequencer (Applied Biosystems) by the DNA Sequence core facility in the Department of Biology, Washington University, St. Louis.
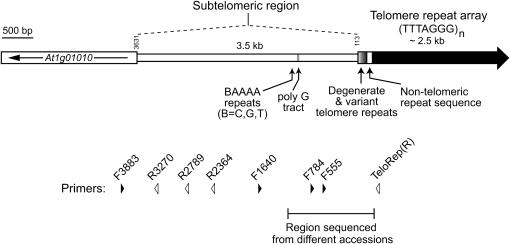
Figure 1.
Genomic organization of chromosome 1N subtelomeric region from A. thaliana accession Columbia. The terminal telomeric repeat array is depicted as a thick solid arrow oriented toward the end of the chromosome. The most distal gene, At1g01010, is shown as an open rectangle with an arrow indicating the direction of transcription. Vertical numbers marked above the chromosome are coordinates in the most current version of the Col genomic sequence (TAIR 6.0) and denote the boundaries of the subtelomeric region defined in this study. Various sequence features are shown below the chromosome map. The position and orientation of primers used in this study are shown by arrowheads below; primer names correspond to nucleotide coordinates.
Data analysis:
For the subtelomeric region, contiguous sequences were assembled using DNASTAR-SeqMan 5.0 software (DNASTAR) and contigs from all accessions were further aligned and visually inspected using the Se-Al program (Rambaut 1996). An alignment gap of a 67-bp G-rich region (coordinates 1061–1127; bracketed region in Figure 2) was excluded from analysis because of poor sequence quality in this simple sequence region. The program DnaSP 4.10 (Rozas and Rozas 1999; Rozas et al. 2003) was used to calculate Tajima's D statistic (Tajima 1989). Nucleotide and indel diversity θw was calculated following the method of Watterson (1975). For the DV (degenerate and variant telomere repeat) region of the centromere-proximal telomere, sequences were aligned manually and organized into repeat units starting with signature of TTH (H: A or T) or TYY (Y: T or C) (Richards et al. 1992, 1993). Gaps, grouped into consecutive repeat units, were generated to maximize the alignment of contiguous conserved repeats as well as to minimize the mutational steps within a repeat unit.
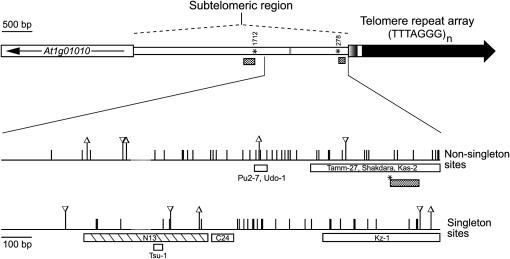
Figure 2.
Summary of polymorphic sites over the 1442-bp telomere-proximal region of chromosome 1N among 35 accessions. A map of chromosome 1N subtelomeric region from accession Columbia is shown on the top (as in Figure 1). At the bottom, an expanded 1442-bp region is diagrammed. Vertical bars represent single nucleotide substitutions; vertical bars with triangules indicate insertions (∇) and deletions (Δ) of ≤11 bp. Open boxes below the lines indicate larger-scale deletions (>30 bp) and the striped box denotes a deletion (409 bp), which is replaced by a copia-like LTR sequence (470 bp); accessions carrying these alterations are labeled. Asterisks mark the sites where a sequence inversion occurred (vertical numbers indicate the nucleotide coordinates of the breakpoints) along with deletions of the flanking sequences (stippled boxes); this complex rearrangement is observed in accessions Bur-0, Ct-1, Cvi-0, Oy-0, and Ws-2. Singleton sites are polymorphisms found in only one accession, whereas nonsingleton sites are found in more than one accession. The dashed regions along the chromosomal lines are sequence gaps at the poly G tract (the shaded region on the map at the top).
A haplotype tree was constructed for the subtelomeric region on the basis of single nucleotide substitutions and small indel polymorphisms, omitting the larger indels (>30 bp) and large-scale DNA rearrangements. The tree was generated with PAUP* (4.0b10) software using a maximum parsimony criterion. A separate phylogenetic analysis was performed for the DV region on the basis of numbers of telomeric repeat units in combination with nucleotide substitutions and indel variations within individual telomeric repeats. The timing of divergence among accessions since the mtDNA capture in the telomere was estimated on the basis of the mean pair-wise genetic distance over the distal 1.4-kb subtelomeric region (0.00102 by the Kimura two-parameter model) (Kimura 1980).
RESULTS
General organization of the subtelomeric region of chromosome 1N in A. thaliana:
In studies of organisms with complex genomic structures at the chromosome ends (e.g., humans, fungi, trypanosomes), the subtelomeric region is defined as the domain between the telomere and the most distal chromosome-specific sequence (Pryde et al. 1997; Mefford and Trask 2002). Because many chromosomal ends in A. thaliana carry unique sequences adjacent to the telomeric sequences, we define the subtelomeric region as the genomic sequences between the first chromosome-specific coding sequence and the telomeric sequence. In the current version of the A. thaliana genome sequence (TAIR 6.0) from accession Columbia (Col), the telomeric repetitive sequence on chromosome 1N spans coordinates 1–113, and the transcription start site of the first annotated expressed gene At1g01010 lies at coordinate 3631 (Figure 1). The intervening 3517-bp subtelomeric region comprises a single-copy sequence that shares no significant similarity with any sequence present in public nucleotide databases or other subtelomeric regions in A. thaliana accession Col (Heacock et al. 2004). The GC content of this region is 32%, comparable to the GC composition in noncoding regions of the genome (Arabidopsis Genome Initiative 2000). Among the few distinguishing features in the Col chromosome 1N subtelomeric region are two small islands of simple sequence, including a short stretch of BAAAA repeats (where B = C, T, or G) and a 32-bp tract composed almost entirely of G (Figure 1).
We examined the organization of chromosome 1N ends in 35 wild accessions of A. thaliana (Table 1) by PCR analysis using primers corresponding to various positions throughout At1g01010 and the subtelomeric region (Figure 1). Amplification using a combination of centromere-proximal primers (F3883 with R3270, R2789, or R2364) generated DNA products of predicted sizes from genomic DNA of all 35 accessions (data not shown). This finding indicates that the sequence corresponding to At1g01010 is adjacent to the chromosome 1N subtelomeric region for all accessions surveyed and that the accessions do not show detectable length variation in the centromere-proximal portion of the subtelomeric region. In contrast, genomic amplifications using the telomere-proximal primers F1640, F784, or F555 together with a primer corresponding to the canonical telomeric repeat [TeloRep(R): (CCCTAAA)3] resulted in predominant products of variable length among some accessions and no amplification in five accessions (Bur-0, Ct-1, Cvi-0, Oy-0, and Ws-2; data not shown). Subsequent nucleotide sequence analysis indicated that these five accessions contain a complex rearrangement of the subtelomeric region (see below). Thus, our tiling PCR analysis suggested that the general organization of the centromere-proximal portion of the chromosome 1N subtelomeric region is similar among the 35 accessions, while the telomere-proximal region has a higher degree of variation among accessions.
Sequence variation in the 1.4-kb telomere-proximal subtelomeric region:
The PCR fragment-length variation apparent in the telomere-proximal portion of the subtelomeric region led us to investigate this variation at the nucleotide sequence level. The polymorphisms observed in our sample of 35 accessions are depicted in Figure 2. We detected a total of 87 polymorphic sites, including 70 sites of single-nucleotide substitutions, 10 small indels (≤11 bp), and 7 larger-scale DNA rearrangements. The single nucleotide substitutions include 44 transitions and 28 transversions; 2 sites had three segregating nucleotides. Two of the 10 small indels occurred at mononucleotide sequences [(dA)7 and (dA)4], and another small indel resulted from a deletion/duplication of adjacent sequence.
Among the larger-scale DNA rearrangements, deletions ranging from 31 to 418 bp were detected at five different positions (Figure 2), which corresponded to the length variation of predominant products observed in the PCR analysis. A sixth rearrangement corresponded to a replacement of a 409-bp sequence with an insertion of a 470-bp copia-like LTR (described below). The final large-scale rearrangement detected was a 1432-bp sequence inversion flanked by two deletions of 191 and 97 bp at the centromeric and telomeric boundaries of the inversion, respectively. This complex rearrangement underlies the distinct variation found in the five accessions that showed no amplification in the tiling PCR analysis.
Nucleotide diversity in the 1.4-kb subtelomeric region measured θw = 0.012, on the basis of the number of segregating sites (Watterson 1975) (supplemental Table 1, http://www.genetics.org/supplemental/). As shown in Figure 2, the polymorphic sites are not evenly distributed throughout the 1.4-kb region. A sliding-window analysis showed that nucleotide diversity is lowest in an ∼500-bp region closest to the centromere and peaks at the center of the 1.4-kb region (within a 250-bp region; data not shown). The central region with higher diversity is characterized by a cluster of singleton single nucleotide substitutions that are not associated with any particular sequence structure. The pattern of polymorphism in this region is consistent with a null hypothesis of neutral evolution (Tajima's D = 1.02, P > 0.1) (Tajima 1989). We did not detect evidence for homologous recombination events in this region on the basis of the pattern of polymorphism and the haplotype relationships among the accessions (see next section).
Subtelomeric structure and phylogenetic relationships among haplotypes:
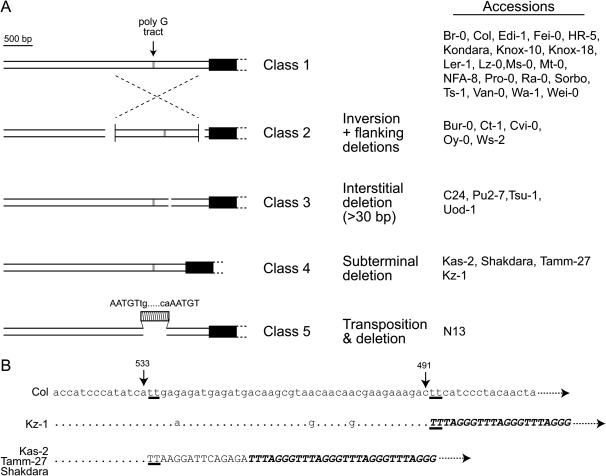
The large-scale DNA rearrangements over the 1.4-kb subtelomeric region were grouped into five distinct structural classes (Figure 3A). The most common class includes the standard lab strains Col and Ler-1 (class 1). The class 2 subtelomeric structure is characterized by the large sequence inversion event accompanied by flanking deletions. Class 3 subtelomeric structures are marked by three independent interstitial deletions (31, 43, and 76 bp) at various positions. Two of these deletions are flanked by short direct repeat sequences (4 and 5 bp, respectively). These interstitial deletions are distinguished from class 4 subtelomeric regions, which contain deletions immediately adjacent to the telomeric repeats (subterminal deletions; Figure 3). The class 4 subtelomeric structures include two subterminal deletions with endpoints that are 42 bp apart. One subterminal deletion resulted in a 376-bp deletion (distal to coordinate 491, Figure 3B); the breakpoint was followed immediately by canonical telomeric repeats (TTTAGGG). The other subterminal deletion removed 418 bp (distal to coordinate 533), resulting in a boundary containing a short degenerate telomeric sequence (TTAAGGATTCAGAGA) followed by canonical telomeric repeats. Finally, the insertion of a 470-bp copia-like LTR in replacement of a 409-bp sequence defines the class 5 subtelomeric structure, which was found in a single accession, N13. This solo LTR has inverted repeat termini (5′-tg … ca-3′) characteristic of retroviral and retrotransposon LTRs flanked by a 5-bp target site duplication (AATGT) (Figure 3).
Figure 3.
Five classes of subtelomeric structures at chromosome 1N among 35 accessions. (A) Diagrams depict the large-scale DNA rearrangements that define five classes of subtelomeric structures at chromosome 1N. Telomeric repeats are shown as solid boxes; the chromosome end is oriented toward the right. The accessions in each class are shown at the right. Intersecting dashed lines indicate the large 1432-bp inversion and deletions are shown as gaps. The striped box denotes a copia-class LTR insertion; putative 5-bp target site duplication (TSD) sequences flanking the LTR termini signature (5′-tg … ca-3′) are indicated. (B) Sequence alignment of Col and class 4 accessions showing two subterminal deletion breakpoints; nucleotide coordinates correspond to the Col genomic sequence (TAIR 6.0). Dots are conserved nucleotides; upper case letters denote discontinued sequence alignments, with canonical repeats (TTTAGGG) highlighted in boldface type and italics; dashed lines with arrowheads point to the chromosome end. TT dinucleotides at the novel subtelomeric–telomeric sequence junction are underlined.
The 1.4-kb distal subtelomeric sequences from 35 A. thaliana accessions characterize 19 haplotypes. A heuristic search based on the maximum parsimony criterion generated 16 equally parsimonious trees with a length of 89 steps [consistency index (CI) = 0.966, retention index (RI) = 0.958], and a consensus tree (>50% majority) is presented in Figure 4A. The major differences between the 16 trees are the branch positions of class 4 haplotypes that cannot be resolved unambiguously due to missing polymorphic data in the large subterminal sequence gaps. The very high CI indicates that there are no recombinant haplotypes in the sampled accessions that would generate homoplasy on the haplotype tree. Overall, there is no congruence between the geographic distribution and the phylogenetic relationship among these haplotypes (Table 1).
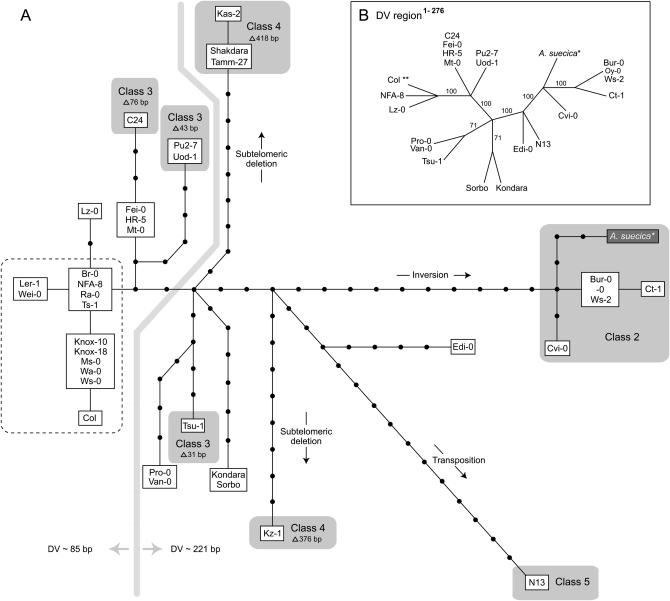
Figure 4.
Phylogenetic relationship among chromosome 1N subtelomeric and telomeric haplotypes. (A) A tree containing 19 A. thaliana haplotypes and 1 A. suecica haplotype was constructed on the basis of maximum parsimony criterion and a data set covering the 1442-bp distal subtelomeric region. Each mutation step (including single nucleotide substitutions and indels <11 bp) is represented by a line segment and nodes represent unknown/missing haplotypes. Shaded areas highlight accessions with class 2 and class 4 subtelomeric structures. The dashed lines enclose accessions/haplotypes with a mitochondrial DNA (mtDNA) sequence captured in the telomeric array. The thick shaded line distinguishes two clusters of subtelomeric haplotypes that represent two major length morphs of the DV regions DV ∼85 bp and DV ∼221 bp as indicated (not including Kas-2, Tamm-27, Shakdara, and Kz-1). (B) A consensus parsimony tree (>50% majority) based on the DV region (positions 1–276 in Figure 5). Four A. thaliana accessions (Kas-2, Tamm-27, Shakdara, and Kz-1) were not included because of large deletions over the aligned region. The tree is generated by a heuristic search, excluding the gap features at position 222 and 231 in Figure 5 due to ambiguous alignments. Col** haplotype includes 11 accessions: Br-0, Col, Knox-10, Knox-18, Ler-1, Ms-0, Ra-0, Wa-0, Wei-0, Ws-0, and Ts-1. The numbers along the branches indicate percentage of total trees showing the topology indicated by the flanking nodes.
The subtelomeric haplotype tree provides a framework for examining the evolutionary history of the large-scale DNA rearrangements detected in this region (illustrated by shaded haplotypes in Figure 4A). The class 2 subtelomeric structure was found in a cluster of related haplotypes at the tip of a lineage, indicating that these haplotypes originated from a single common inversion event. The three interstitial deletions grouped in the class 3 subtelomeric structure corresponded to three different lineages on the tree and represent three independent deletion events. The class 4 subtelomeric structure mapped to two separate lineages corresponding to different deletion breakpoints (Figure 3B), illustrating two independent subterminal deletion events in the evolutionary history represented by the sampled accessions.
Linkage disequilibrium between the subtelomeric region and the adjacent telomeric region:
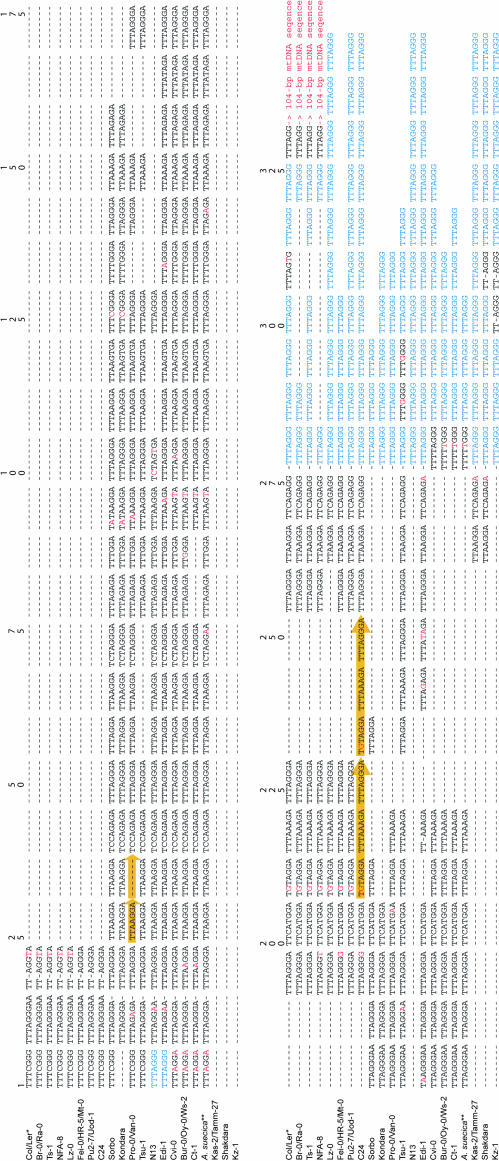
We extended our DNA sequence analysis into the telomeric region adjacent to the subtelomeric sequence, where motifs were found that conform to a greater or lesser degree to canonical telomeric repeats (TTTAGGG) (Richards et al. 1993). In most of the accessions (with the exception of Kz-1) this centromere-proximal telomeric region consisted of a mosaic of degenerate repeats and variant repeats (Figure 5). We refer to this heterogeneous zone as the DV region, which is followed by more distal canonical repeats. Despite the complexity of the DV region, we were able to align the different sequences in this region by referring to nucleotide substitutions characteristic of particular degenerate and variant repeat motifs (Figure 5). The length of the DV region is variable among the accessions, ranging from 14 to 280 bp. However, two major DV region length morphs, ∼85 and ∼221 bp, exist that distinguish two clusters of subtelomeric haplotypes on the haplotype tree (distinguished by the dividing shaded vertical line shown in Figure 4A). Accessions sharing a particular subtelomeric haplotype contain nearly identical sequences over the DV region (Figure 5), indicating that the subtelomeric region and the adjacent DV region are in linkage disequilibrium. To compare explicitly the haplotype structure of the two regions, we performed a phylogenetic analysis on the DV region, which yielded 24 equally parsimonious trees (tree length = 50 steps; CI = 1; RI = 1). As with the subtelomeric tree, ambiguities in the haplotype topology are attributable to gaps (in this case, those generated by indels of telomeric repeats). A DV consensus tree (Figure 4B) has a topology that is completely congruent with the subtelomeric region haplotype tree (Figure 4A), consistent with our observation that these two adjacent regions share the same underlying haplotype structure and are in strong linkage disequilibrium. The alignment shown in Figure 5 also highlights the duplication and deletion of variable numbers of repeats, and in some cases (marked by gold arrows) we could detect duplication/deletion of adjacent repeats by referring to their unique sequence signatures.
Figure 5.
Alignment of the centromere-proximal telomeric region of chromosome 1N from 35 A. thaliana accessions and 9 accessions of A. suecica. Accessions are indicated on the left of the corresponding nucleotide sequences, which are shown in two tiers, oriented 5′ to 3′ toward the end of the chromosome. Vertical numbers on the top indicate the nucleotide positions within the telomeric repeat sequences distal to the subtelomeric sequence. Canonical telomeric sequence (TTTAGGG) is highlighted in blue; nucleotide polymorphisms are shown in red, as is the position of the 104-bp mtDNA sequence. Dashes denote alignment gaps. Arrowheads in gold indicate examples of indels of adjacent direct repeats. The haplotype represented by Col/Ler-1* also includes accessions: Knox-10, Knox-18, Ms-0, Wa-1, Wei-0, and Ws-0. The A. suecica** haplotype includes all 9 accessions of this species sampled in this study.
Capture of mitochondrial DNA in the proximal telomere:
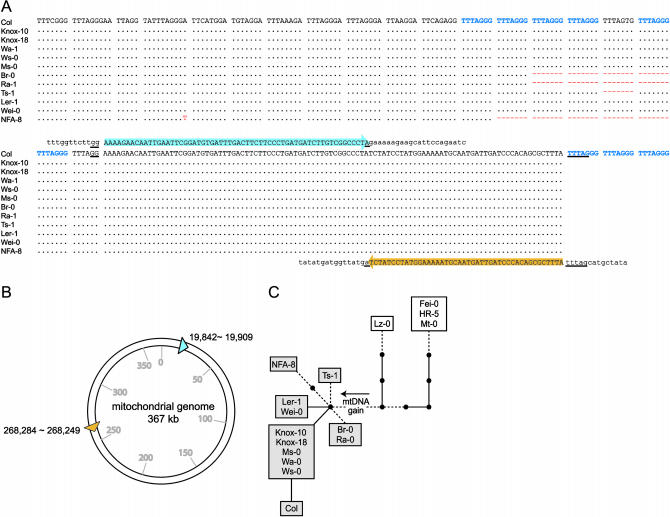
We previously isolated and partially characterized a set of A. thaliana telomeric genomic clones that corresponded to junctions between the terminal telomeric repeat arrays and the adjacent subtelomeric sequence (Richards et al. 1992). One of these clones derived from accession Ler had a small island of nontelomeric sequence embedded in the telomeric repeat array (E. J. Richards, unpublished data). A comparison with the genomic sequence from the accession Col indicated that this Ler telomere clone corresponds to the termini of chromosome 1N; however, the current version of the sequence database does not cover the position of the nontelomeric sequence. We confirmed the presence of this unique sequence distal to the subtelomeric region of chromosome 1N by PCR analysis with the genomic DNA isolated from Ler (data not shown). This nontelomeric sequence is 104 bp in length and is located in the homogeneous telomeric repeat array distal to the DV region (sequence marked by colored arrows in Figure 6A). A BLAST search of the database showed that this nontelomeric sequence consists of two fragments (a proximal 68-bp segment and a distal 36-bp segment) identical to two noncontiguous sequences in the mitochondrial genome of A. thaliana (Figure 6B) (Unseld et al. 1997). In addition, the distal mtDNA fragment also matched a sequence in the pericentromeric region of chromosome 2 (coordinates 3262720–3262755) where a 620-kb duplicated and rearranged mitochondrial genomic sequence was transferred (Lin et al. 1999; Stupar et al. 2001). The proximal 68-bp mtDNA segment is identical to an intergenic region between orf315 and NAD5 of the mitochondrial genome and overlaps with an intronic region between exon b and exon c of NAD2. The distal 36-bp fragment is identical to an intergenic region between orf106e and orf107f.
Figure 6.
Sequence alignment of the DV region and 104-bp nontelomeric sequence in the Chr1N telomere among 12 accessions. (A) The mitochondrial genomic sequences that are identical to the 104-bp sequence in the telomeric array are highlighted in blue and gold, and the flanking genomic sequences are in lowercase letters. Nucleotides with identity at the junction of the breakpoints are underlined. The canonical telomeric repeats are shown in blue, dots denote nucleotides that match the reference sequence corresponding to accession Col, and alignment gaps are indicated by red dashes. (B) A map of the A. thaliana mitochondrial genome (based on sequence from the C24 accession) showing the positions and orientations of the two mtDNA segments that compose the 104-bp sequence captured in the telomeric repeat array. (C) An expansion of a tip of the phylogenetic tree shown in Figure 4A, based on incorporation of polymorphisms within the centromere-proximal domain of the telomeric repeat array shown in A. Layout is the same as that described for Figure 4A, except that polymorphisms within the telomeric region (indels of telomeric repeats and one single nucleotide substitution) are marked by dashed lines to distinguish them from subtelomeric polymorphisms (solid lines).
We further investigated the distribution of this unique arrangement of mtDNA in other accessions using PCR analysis and found that 11 accessions in addition to Ler had this insertion within the chromosome 1N telomere. These 12 accessions represent subtelomeric haplotypes that are closely related (enclosed by dashed lines in Figure 4A) and the adjacent DV regions are also highly conserved among them (with only one nucleotide polymorphism; see Figure 6A). Moreover, the 104-bp mtDNA sequence is itself invariant among the 12 accessions. These patterns suggest that the mtDNA captured in the telomeric arrays of these accessions arose recently through a single evolutionary event. On the basis of the assumption of a nuclear nucleotide substitution rate of 1.5 × 10−8/site/year (Koch et al. 2000), we estimate that the mtDNA capture occurred approximately 30,000 years ago (YA).
Among the 12 accessions containing the mtDNA insertion event, we observed variation corresponding to apparent deletions of one to five canonical telomeric repeats (alignment gaps indicated by red dashes in Figure 6A). This variation was found in a group of accessions sharing a common subtelomeric haplotype (Br-0, NFA-8, Ra-1, and Ts-1) (Figures 4A and 6A), implying that the canonical telomeric repeat deletion and amplification is proceeding at a comparatively fast rate. We incorporated the polymorphisms within the proximal telomeric region (DV plus the region extending to the mtDNA) to resolve the phylogenetic relationship among a number of closely related class 1 subtelomeric haplotypes (Figure 6C). The refined tree suggests that the mtDNA insertion is a unique event that occurred after the divergence of accession Lz-0 from the ancestral haplotype of the 12 accessions (indicated in the shaded boxes in Figure 6C).
DISCUSSION
Our study of nucleotide sequence variation of the unique subtelomeric region of chromosome 1N among 35 accessions of A. thaliana demonstrates the genomic dynamics of chromosome termini. A recent study highlighted the variation in telomere length regulation among natural Arabidopsis accessions (Shakirov and Shippen 2004); here, we focused on natural variation of the genomic region adjacent to the telomere. As hypothesized for a noncoding genomic region (Graur and Li 2000), the investigated chromosome 1N subtelomeric region is evolving neutrally (Taijima's D = −1.02, P > 0.1) and exhibits diversified haplotypes. The negative Tajima's D value for the chromosome 1N subtelomeric region indicates an excess of low-frequency polymorphisms, similar to the trend observed at many other loci in A. thaliana (Nordborg et al. 2005; Schmid et al. 2005). This observation, as well as the lack of association between the geographical distribution of accessions and haplotypes, is consistent with the hypothesis that this species experienced a recent population expansion (Price et al. 1994; Innan et al. 1996 Bergelson et al. 1998). Overall, the level of nucleotide polymorphism in this region (θw = 0.012) is comparable to the average genomewide silent nucleotide diversity [θw = 0.00896 from Schmid et al. (2005); θw = ∼0.007–0.01 from Nordborg et al. (2005)]. The indel diversity in this subtelomeric region is also comparable to that present in other noncoding regions along chromosome 1 (Nordborg et al. 2005; (see supplemental Table 1, http://www.genetics.org/supplemental/). Therefore, the subtelomeric region of chromosome 1N appears to be evolving in a manner similar to that seen in other noncoding regions of the genome. We note that the subtelomeric region of chromosome 1N displays an elevated frequency of larger-scale rearrangements in the distal region adjacent to the telomeric repeats. As discussed below, these larger-scale rearrangements were caused by diverse molecular mechanisms. This observation is reminiscent of a recent report that the repertoire of DNA damage-induced rearrangements in budding yeast increases in subtelomeric regions (Ricchetti et al. 2003).
A wealth of cytological observations indicates that initiation of homologous chromosome synapsis and recombination occurs toward the chromosome ends (Scherthan 2001; Schwarzacher 2003; Harper et al. 2004). These observations suggest that homologous recombination in subtelomeric regions may be frequent. Consistent with this expectation, the ratio of genetic to physical distance increases toward the ends of linkage maps in many organisms (NIH/CEPH Collaborative Mapping Group 1992; Lukaszewski and Curtis 1993; Schwarzacher 1996; Lin et al. 1999; Mayer et al. 1999). It is not clear, however, whether the elevated recombination extends to the extreme terminal region of the chromosome. The presence of strong linkage disequilibrium between the distal subtelomeric region and the adjacent proximal telomere, which is observed in humans (Baird et al. 2000) and Arabidopsis (this study), argues that recombination is infrequent at the junctions between the subtelomeric regions and the telomeres.
The evolutionary neutrality and well-defined haplotype structure of the Arabidopsis chromosome 1N subtelomeric region suggest that the region could be useful for addressing evolutionary questions in this genus. We applied this marker to examine the evolutionary origins of the allotetraploid A. suecica (2n = 4X = 26), a putative hybrid of A. thaliana (2n = 10) and A. arenosa (2n = 16) (Hylander 1957; O'Kane et al. 1996). Several studies indicate A. thaliana was the maternal parent of A. suecica (Mummenhof and Hurka 1994; Säll et al. 2003); on the basis of chloroplast DNA sequences, Säll et al. (2003) suggested that A. suecica (endemic populations in Sweden and Finland) arose through a single hybridization event. To test this hypothesis, we analyzed a set of 10 A. suecica accessions collected from endemic geographic locations and 1 of unknown origin (Table 1). We found that all accessions shared identical sequences over the 2-kb telomere-proximal region and contained sequence rearrangement characteristic of class 2 subtelomeric structures (Figure 3A). Phylogenetic analysis indicated that these A. suecica accessions originated from a common ancestral A. thaliana haplotype closely related to the extant class 2 haplotypes (Figure 4A). Thus, our data are consistent with the single-origin hypothesis for A. suecica.
Molecular mechanisms underlying the dynamics of the proximal telomeric region:
Mutations in telomeric repeats have been shown to alter the interaction between the telomere and its associated proteins, resulting in alterations in telomere structure and developmental anomalies (Yu et al. 1990; McEachern and Blackburn 1995; Prescott and Blackburn 1997). The accumulation of mutations in the centromere-proximal portion of the telomere suggests that this region may be under less functional constraint. In addition to the ongoing nucleotide substitutions and small indels within the repeats, comparison of the proximal telomeric region among natural accessions revealed that deletion and expansion of blocks of repeats also contribute to polymorphisms. This mutational pattern is reminiscent of the instability of microsatellites (Schlotterer 2000; Ellegren 2004). The molecular mechanisms underlying the instability of microsatellites and other tandem repetitive sequences include unequal exchange and replication slippage (Bzymek and Lovett 2001; Schlotterer and Tautz 1992). The expansion of human telomeric repeat sequences (TTAGGGn) via replication slippage has been demonstrated in an in vitro DNA synthesis assay, and instability of telomeric repeat sequences (expansions and deletions) carried out on plasmid DNA was also observed during propagation in bacterial cells (Nozawa et al. 2000). In humans, the proximal telomeric region is also characterized by accumulation of variant telomeric repeats, which are postulated to arise from intra-allelic mutational processes, such as replication slippage or unequal sister chromatid exchange, in light of the evidence that inter-allelic homologous recombination is suppressed in this region (Baird et al. 1995, 2000). As discussed above, we have found no evidence for reciprocal homologous recombination in the Arabidopsis chromosome 1N subtelomeric-proximal telomere region that would accompany unequal exchange. However, it is important to note that this plant's predominant selfing habit (and associated high homozygosity) would make detection of these homologous recombination events less likely. Regardless of the precise molecular mechanisms at work, the pattern of genetic variation suggests that similar evolutionary processes are acting on the proximal telomeric regions in humans and Arabidopsis.
An unusual type of polymorphism identified within the Arabidopsis chromosome 1N proximal telomere repeat array is the insertion of mitochondrial DNA. The composite structure of the 104-bp fragment is similar to structures generated by filler DNA captured in double-strand break (DSB) repaired sites through NHEJ. Insertion of filler sequences (e.g., nuclear genomic sequences, reverse transcribed products of retrotransposons, mitochondrial DNA) at repaired DSB sites is found in many eukaryotes (Nassif et al. 1994; Moore and Haber 1996; Gorbunova and Levy 1997; Salomon and Puchta 1998; Ricchetti et al. 1999; Yu and Gabriel 1999; Lin and Waldman 2001). These filler sequences integrate either as one contiguous segment or as an assemblage of scrambled segments. The synthesis-dependent strand annealing model has been proposed to explain the insertion of filler sequences at DSB sites. In this model, a 3′-protruding strand of a broken end primes DNA synthesis and copies a stretch of DNA using an ectopic template before rejoining the other broken terminus (Formosa and Alberts 1986; Nassif et al. 1994; Gorbunova and Levy 1997). This model could explain the two noncontiguous mtDNA sequences captured in a reverse orientation in the telomeric array found here: repair synthesis had switched between two ectopic mitochondrial genomic fragments, using short stretches of complementarity at the junctions between the telomeric sequence and the flanking genomic sequences of the captured fragments (Figure 6B, underlined nucleotides).
We estimated that this mtDNA insertion is a recent evolutionary event (30,000 YA) supporting the view that mtDNA integration into the nuclear genome is an ongoing process (Yu and Gabriel 1999; Adams et al. 2000; Ricchetti et al. 2004). Furthermore, our results demonstrate that the telomeres can be a port of entry for organellar DNA into the nuclear genome. Interestingly, another example of a transfer of a short, scrambled mitochondrial sequence to the chromosome termini has been reported in yeast, but in this case the mtDNA was inserted into the subtelomeric X–Y′ element boundary rather than the terminal telomeric repeat array (Louis and Haber 1991). The inclusion of filler sequences into the telomere repeat array has been reported in a special circumstance: NHEJ-mediated end-to-end fusions of critically shortened telomeres (∼100–400 bp) in an Arabidopsis telomerase mutant (Heacock et al. 2004). This result suggests that when the telomere length falls below a minimum threshold it will be processed by DSB-initiated DNA repair in the absence of telomerase. The captured mtDNA described in the present study occurred ∼140 bp from the subtelomeric–telomeric boundary, suggesting that DSB repair machinery can compete with telomerase-mediated telomere addition for access to the proximal region of telomere to repair the broken telomere in wild-type backgrounds where telomerase is expected to be active.
Molecular mechanisms underlying the dynamics of the subtelomeric region:
The larger-scale DNA rearrangements found over the distal subtelomeric region in this study are associated with deletions of >30 bp. All of these deletions, with the exception of the inversion characterizing class 2 subtelomeric regions, occur without additional rearrangements, including the transposition event seen in class 5 (see below). Deletions at nonrepetitive random sequences can be derived from replication slippage between short direct repeats or NHEJ after exonucleolytic processing of DSB sites. Although little is known about the requirements for replication slippage in plants, deletions caused by replication slippage are associated with 3- to 9-bp perfect or imperfect direct repeats in the budding yeast (Tran et al. 1995). Among the seven characterized DNA rearrangements in the distal subtelomeric region, the two smallest deletions, the 31- and 43-bp interstitial deletions, are flanked by 4-bp perfect direct repeats and could have resulted from either replication slippage or an NHEJ event. In contrast, the 76-bp interstitial deletion shows no recognizable sequence similarity flanking the breakpoints and is likely to have resulted from NHEJ. The remaining four rearrangements show 0- to 3-bp identity flanking the breakpoints and are likely to be caused by an NHEJ process rather than replication slippage, a conclusion further supported by additional characteristics of these rearrangements. For example, in the class 2 subtelomeric inversion event, we observed limited regions of identity at the sequences flanking the rejoined sites (1 and 3 bp for the proximal and the distal deletion breakpoints, respectively). This rearrangement appears to involve two DSB events, followed by exonucleolytic processing and NHEJ rejoining of three chromosome fragments, with the central fragment inverted. This rearrangement resembles the irradiation-induced DNA rearrangements found in the A. thaliana transparent testa 3 (tt3) allele (Shirley et al. 1992).
Telomere addition at a broken chromosome end (chromosome healing) has been observed in a number of organisms (Haber and Thorburn 1984; Pologe and Ravetch 1988; Wilkie et al. 1990; Yu and Blackburn 1991; Werner et al. 1992; Sprung et al. 1999). Chromosome healing can occur through various processes, including de novo telomere synthesis by telomerase or NHEJ with a preexisting telomere (telomere capturing). The class 4 subtelomeric structures (subterminal deletions) resemble healed chromosome ends resulting from DSBs and deletions of the distal subtelomeric region. Intriguingly, such deletions occurred twice independently at nearby positions in the evolutionary history represented by the accessions examined (Figures 3B and 4). The different signatures of telomeric sequences adjacent to the subterminal breakpoints suggest that different processes underlie the two independent events.
In one accession, Kz-1, the deletion breakpoint was followed by an array of homogeneous repeats, the majority of which match the canonical telomere repeat motif. This structure is most easily explained by de novo telomere synthesis by telomerase. Under such a model, the telomere addition could have initiated using 5′-TT-3′ as a priming site (underlined TT dinucleotide in Figure 3B), which is in phase with the first added telomere repeat sequence (TTTAGGG). Alternatively, telomerase can also add telomeric repeats to a DNA 3′ terminus lacking any complementarities to the telomerase RNA template using a so-called default mechanism (Melek and Shippen 1996). Under such a scenario, the 5′-AC-3′ dinucleotide directly proximal to the first complete canonical telomere repeat could have served as a priming site for telomerase-mediated telomere addition. Although G-rich sequences in the proximity of a noncomplementary 3′ terminus are postulated to recruit telomerase by the default mechanism (Harrington and Greider 1991; Muller et al. 1991; Bottius et al. 1998), we did not observe such a feature in the flanking sequence of the Kz-1 subterminal deletion breakpoint. It is noteworthy that an in vitro study demonstrated that Arabidopsis telomerase has a relaxed specificity for DNA recognition (Fitzgerald et al. 2001). While 5′-G1–3-3′ is a preferred priming site for Arabidopsis telomerase in vitro, elongation of nontelomeric 3′ ends can occur by positioning the DNA at a default site in the RNA template, leading to the addition of 5′-GGTTTAG-3′ as the first telomeric sequence. However, the novel subtelomeric–telomeric junction created in Kz-1 does not show the sequence signature predicted by the in vitro result. This discrepancy may be due to the different biochemical activity of the telomerase in vivo or a nontelomerase-mediated mechanism (e.g., NHEJ-mediated telomere capture) could underlie this healing event.
In contrast to the situation in Kz-1, the junction of the subterminal deletion shared by accessions Kas-2, Shakdara, and Tamm-27 contains two degenerate telomeric repeats before transitioning into a more homogeneous array of repeats conforming to the canonical telomere sequence (Figure 3B). Moreover, these two degenerate telomeric repeats are nearly identical to repeats that are also located adjacent to more uniform canonical telomere repeat arrays in the DV region of other accessions with different subtelomeric structures (Figure 5). These observations make NHEJ-mediated telomere capture a more plausible mechanism to explain the origin of this subterminal deletion. If a telomerase-mediated mechanism is invoked, it is also necessary to postulate that an accumulation of mutations at the subtelomeric–telomeric region boundary occurred subsequent to de novo telomere synthesis.
The other complex rearrangement found in the subtelomeric regions of chromosome 1N is a deletion apparently coupled with an insertion of a solo LTR of a copia class retrotransposon in accession N13. Retrotransposons make up a significant portion of plant genomes and are important players in plant genome evolution (Kumar and Bennetzen 1999). In A. thaliana, retrotransposons predominantly cluster in the heterochromatic centromeres and pericentromeric regions but not in the subtelomeric regions, although subtelomeric regions have been regarded as heterochromatic and hot spots for retroelement insertion in some organisms (Pearce et al. 1996; Zou et al. 1996; Arabidopsis Genome Initiative 2000; Peterson-Burch et al. 2004). A genomewide study of retrotransposon distribution in A. thaliana showed that solo LTRs are abundant (composing 1.57% of the genome) and localize predominantly in pericentromeric regions (Peterson-Burch et al. 2004). These elements can be derived from various processes, such as recombination between the 5′ and 3′ LTRs of a single element (intra-element) to generate a recombinant LTR flanked by identical target site duplications (TSDs). Alternatively, an unequal reciprocal recombination event between two elements (either intrachromatid or interchromosomal) will result in a recombinant solo LTR flanked by different TSDs (Devos et al. 2002). The copia class solo LTR found in N13 is flanked by identical 5-bp TSDs and is also associated with a 409-bp deletion of the subtelomeric sequence at the insertion site. A simple intra-element recombination cannot explain the associated deletion at the same location. The presence of the same TSD sequence also makes inter-element recombination (either intra- or interchromatid) unlikely, because this scenario requires two independent transposition events targeting the same 5-bp sequence motif at a nearby location. Capture of LTR sequences (derived from incomplete reverse transcribed products) at induced DSB sites by illegitimate recombination has been observed in yeast (Moore and Haber 1996; Teng et al. 1996; Yu and Gabriel 1999), but the retention of an intact solo LTR together with identical flanking TSDs argues against such an explanation. These considerations suggest that the N13 subtelomeric haplotype arose from a sequence of events, starting with a 409-bp deletion, followed by an insertion of a copia element, and subsequent deletion of the internal region of the retrotransposon by intra-element recombination.
Implications for genome evolution:
The organization of the subtelomeric region of chromosome 1N is characteristic of the apparent simplicity of subtelomeric regions in Arabidopsis. Although there is sequence similarity shared between some subtelomeric regions (Kotani et al. 1999; Heacock et al. 2004), most Arabidopsis subtelomeric regions are characterized by their small size and paucity of repetitive sequences. In contrast, subtelomeric regions found in many other organisms, such as yeast and human, are mosaics of repetitive sequences, many of which share extensive similarity among subtelomeric regions of nonhomologous chromosome (Flint et al. 1997). Complex mechanisms operate to generate these patchwork structures, including ectopic sequence translocations, NHEJ, and homology-mediated recombination between nonhomologous chromosome ends (see Introduction). The simple organization of the chromosome 1N subtelomeric region in Arabidopsis is not due to the lack of fluidity of this genomic region, as we observed diverse large-scale rearrangements over the distal end of this subtelomeric region. The structures of the naturally occurring DNA rearrangements that we observed indicate that the predominant mechanism operating on this region is simple deletion-associated NHEJ repair (at least five of seven rearrangements are rejoined at sites with limited identity).
In plants, NHEJ is a major mechanism underlying DSB repair (Gorbunova and Levy 1999; Puchta 2005). Characterization of experimentally induced DSBs has demonstrated that complex processes are employed during NHEJ in plants. In tobacco, two independent experimental systems demonstrate that many DSB repair events involve deletions and approximately one-third of the events result in addition of filler DNA (Gorbunova and Levy 1997; Salomon and Puchta 1998). In a direct comparison of NHEJ processing at DSB in Arabidopsis and tobacco (with a genome size ∼20-fold greater that of Arabidopsis), the average length of deletions recovered in tobacco were relatively smaller and often accompanied by sequence insertions, whereas in A. thaliana deletions were frequently larger without insertions (Kirik et al. 2000). This observation led to a model that the species-specific difference in DSB repair is a cause for genome size evolution in plants (Kirik et al. 2000). This hypothesis fits the inverse relationship between deletion and genome size previously demonstrated in insects (Petrov et al. 2000). The observations of Kirik et al. (2000) are consistent with spontaneous deletions found in other plants with large genomes, such as that described in the maize Waxy locus that was associated with insertion of filler DNA (Wessler et al. 1990). The hypothesis also gains further support from the study of turnover of retrotransposons, which have contributed significantly to the genome expansion in higher plants (Devos et al. 2002; Kumar and Bennetzen 1999). Devos et al. (2002) estimated that deletion-associated illegitimate recombination is > fivefold more frequent than homologous recombination-mediated elimination of retroelements in A. thaliana and suggested that such a mechanism may counteract the genome expansion in A. thaliana. The predominant simple deletion-associated NHEJ process evident in our study of natural variation in the subtelomeric region of Arabidopsis chromosome 1N parallels the process that appears to be constraining the growth of the genome of this species through the diminution of retrotransposon sequences. It is likely that this bias in processing NHEJ events dictates not only the small size but also the simple organization of the subtelomeric regions in Arabidopsis, in light of the studies that show how translocation-associated NHEJ leads to an accumulation of a complex patchwork of subtelomeric sequences shared between nonhomologous chromosome ends (Linardopoulou et al. 2005). Further investigation of natural variation in A. thaliana and other organisms with small and tightly managed genomes will determine how well simple deletion-associated NHEJ repair explains the dynamics and maintenance of simple subtelomeric structures.
Acknowledgments
We thank James Beck, Kuo-Fang Chung, Yu-Chung Chiang, Taylor Maxwell, and William Martin for helpful discussions. Seeds were kindly provided by Craig Pikaard and James Beck, as well as the Arabidopsis Biological Resource Center at The Ohio State University. This work was supported by a grant from the National Science Foundation to E.J.R. (MCB-0321990).
References
- Adams, K. L., D. O. Daley, Y. L. Qiu, J. Whelan and J. D. Palmer, 2000. Repeated, recent and diverse transfers of a mitochondrial gene to the nucleus in flowering plants. Nature 408: 354–357. [DOI] [PubMed] [Google Scholar]
- Alkhimova, O. G., N. A. Mazurok, T. A. Potapova, S. M. Zakian, J. S. Heslop-Harrison et al., 2004. Diverse patterns of the tandem repeats organization in rye chromosomes. Chromosoma 113: 42–52. [DOI] [PubMed] [Google Scholar]
- Allshire, R. C., M. Dempster and N. D. Hastie, 1989. Human telomeres contain at least three types of G-rich repeat distributed non-randomly. Nucleic Acids Res. 17: 4611–4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarger, V., D. Gauguier, M. Yerle, F. Apiou, P. Pinton et al., 1998. Analysis of distribution in the human, pig, and rat genomes points toward a general subtelomeric origin of minisatellite structures. Genomics 52: 62–71. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative, 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815. [DOI] [PubMed] [Google Scholar]
- Baird, D. M., and N. J. Royle, 1997. Sequences from higher primates orthologous to the human Xp/Yp telomere junction region reveal gross rearrangements and high levels of divergence. Hum. Mol. Genet. 6: 2291–2299. [DOI] [PubMed] [Google Scholar]
- Baird, D. M., A. J. Jeffreys and N. J. Royle, 1995. Mechanisms underlying telomere repeat turnover, revealed by hypervariable variant repeat distribution patterns in the human Xp/Yp telomere. EMBO J. 14: 5433–5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird, D. M., J. Coleman, Z. H. Rosser and N. J. Royle, 2000. High levels of sequence polymorphism and linkage disequilibrium at the telomere of 12q: implications for telomere biology and human evolution. Am. J. Hum.Genet. 66: 235–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry, J. D., M. L. Ginger, P. Burton and R. McCulloch, 2003. Why are parasite contingency genes often associated with telomeres? Int. J. Parasitol. 33: 29–45. [DOI] [PubMed] [Google Scholar]
- Baur, J. A., Y. Zou, J. W. Shay and W. E. Wright, 2001. Telomere position effect in human cells. Science 292: 2075–2077. [DOI] [PubMed] [Google Scholar]
- Bergelson, J., E. Stahl, S. Dudek and M. Kreitman, 1998. Genetic variation within and among populations of Arabidopsis thaliana. Genetics 148: 1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn, E. H., 2000. Telomere states and cell fates. Nature 408: 53–56. [DOI] [PubMed] [Google Scholar]
- Blackburn, E. H., 2001. Switching and signaling at the telomere. Cell 106: 661–673. [DOI] [PubMed] [Google Scholar]
- Borst, P., G. Rudenko, M. C. Taylor, P. A. Blundell, F. Van Leeuwen et al., 1996. Antigenic variation in trypanosomes. Arch. Med. Res. 27: 379–388. [PubMed] [Google Scholar]
- Bottius, E., N. Bakhsis and A. Scherf, 1998. Plasmodium falciparum telomerase: de novo telomere addition to telomeric and nontelomeric sequences and role in chromosome healing. Mol. Cell. Biol. 18: 919–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broun, P., M. W. Ganal and S. D. Tanksley, 1992. Telomeric arrays display high levels of heritable polymorphism among closely related plant varieties. Proc. Natl. Acad. Sci. USA 89: 1354–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucholc, M., Y. Park and A. J. Lustig, 2001. Intrachromatid excision of telomeric DNA as a mechanism for telomere size control in Saccharomyces cerevisiae. Mol. Cell. Biol. 21: 6559–6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bzymek, M., and S. T. Lovett, 2001. Instability of repetitive DNA sequences: The role of replication in multiple mechanisms. Proc. Natl. Acad. Sci. USA 98: 8319–8325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, M., J. L. Celenza and F. J. Eng, 1985. Evolution of the dispersed SUC gene family of Saccharomyces by rearrangements of chromosome telomeres. Mol. Cell. Biol. 5: 2894–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai, L., A. P. Tyagi, K. Winter, R. Holmes-Davis, S. H. Reynolds et al., 2000. Phenotypic instability and rapid gene silencing in newly formed Arabidopsis allotetraploids. Plant Cell 12: 1551–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copenhaver, G. P., and C. S. Pikaard, 1996. RFLP and physical mapping with an rDNA-specific endonuclease reveals that nucleolus organizer regions of Arabidopsis thaliana adjoin the telomeres on chromosomes 2 and 4. Plant J. 9: 259–272. [DOI] [PubMed] [Google Scholar]
- Devos, K. M., J. K. Brown and J. L. Bennetzen, 2002. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 12: 1075–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duraisingh, M. T., T. S. Voss, A. J. Marty, M. F. Duffy, R. T. Good et al., 2005. Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum. Cell 121: 13–24. [DOI] [PubMed] [Google Scholar]
- Eichler, E. E., and D. Sankoff, 2003. Structural dynamics of eukaryotic chromosome evolution. Science 301: 793–797. [DOI] [PubMed] [Google Scholar]
- Ellegren, H., 2004. Microsatellites: simple sequences with complex evolution. Nat. Rev. Genet. 5: 435–445. [DOI] [PubMed] [Google Scholar]
- Epel, E. S., E. H. Blackburn, J. Lin, F. S. Dhabhar, N. E. Adler et al., 2004. Accelerated telomere shortening in response to life stress. Proc. Natl. Acad. Sci. USA 101: 17312–17315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald, M. S., E. V. Shakirov, E. E. Hood, T. D. McKnight and D. E. Shippen, 2001. Different modes of de novo telomere formation by plant telomerases. Plant J. 26: 77–87. [DOI] [PubMed] [Google Scholar]
- Flint, J., G. P. Bates, K. Clark, A. Dorman, D. Willingham et al., 1997. Sequence comparison of human and yeast telomeres identifies structurally distinct subtelomeric domains. Hum. Mol. Genet. 6: 1305–1313. [DOI] [PubMed] [Google Scholar]
- Formosa, T., and B. M. Alberts, 1986. DNA synthesis dependent on genetic recombination: characterization of a reaction catalyzed by purified bacteriophage T4 proteins. Cell 47: 793–806. [DOI] [PubMed] [Google Scholar]
- Freitas-Junior, L. H., E. Bottius, L. A. Pirrit, K. W. Deitsch, C. Scheidig et al., 2000. Frequent ectopic recombination of virulence factor genes in telomeric chromosome clusters of P. falciparum. Nature 407: 1018–1022. [DOI] [PubMed] [Google Scholar]
- Freitas-Junior, L. H., R. Hernandez-Rivas, S. A. Ralph, D. Montiel-Condado, O. K. Ruvalcaba-Salazar et al., 2005. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell 121: 25–36. [DOI] [PubMed] [Google Scholar]
- Garcia-Cao, M., R. O'Sullivan, A. H. Peters, T. Jenuwein and M. A. Blasco, 2004. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat. Genet. 36: 94–99. [DOI] [PubMed] [Google Scholar]
- Gorbunova, V., and A. A. Levy, 1997. Non-homologous DNA end joining in plant cells is associated with deletions and filler DNA insertions. Nucleic Acids Res. 25: 4650–4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbunova, V. V., and A. A. Levy, 1999. How plants make ends meet: DNA double-strand break repair. Trends Plant Sci. 4: 263–269. [DOI] [PubMed] [Google Scholar]
- Gottschling, D. E., O. M. Aparicio, B. L. Billington and V. A. Zakian, 1990. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63: 751–762. [DOI] [PubMed] [Google Scholar]
- Graur, D., and W.-H. Li, 2000. Fundamentals of Molecular Evolution. Sinauer Associates, Sunderland, MA.
- Haber, J. E., and P. C. Thorburn, 1984. Healing of broken linear dicentric chromosomes in yeast. Genetics 106: 207–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper, L., I. Golubovskaya and W. Z. Cande, 2004. A bouquet of chromosomes. J. Cell Sci. 117: 4025–4032. [DOI] [PubMed] [Google Scholar]
- Harrington, L. A., and C. W. Greider, 1991. Telomerase primer specificity and chromosome healing. Nature 353: 451–454. [DOI] [PubMed] [Google Scholar]
- Heacock, M., E. Spangler, K. Riha, J. Puizina and D. E. Shippen, 2004. Molecular analysis of telomere fusions in Arabidopsis: multiple pathways for chromosome end-joining. EMBO J. 23: 2304–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hylander, N., 1957. Cardaminopsis suecica (Fr.) Hiit., a northern amphidiploid species. Bull. Jardin Botanique Bruxelles 27: 591–604. [Google Scholar]
- Innan, H., F. Tajima, R. Terauchi and N. T. Miyashita, 1996. Intragenic recombination in the Adh locus of the wild plant Arabidopsis thaliana. Genetics 143: 1761–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, M., 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16: 111–120. [DOI] [PubMed] [Google Scholar]
- Kirik, A., S. Salomon and H. Puchta, 2000. Species-specific double-strand break repair and genome evolution in plants. EMBO J. 19: 5562–5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk, K. E., and E. H. Blackburn, 1995. An unusual sequence arrangement in the telomeres of the germ-line micronucleus in Tetrahymena thermophila. Genes Dev. 9: 59–71. [DOI] [PubMed] [Google Scholar]
- Koch, M. A., B. Haubold and T. Mitchell-Olds, 2000. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol. Biol. Evol. 17: 1483–1498. [DOI] [PubMed] [Google Scholar]
- Kotani, H., T. Hosouchi and H. Tsuruoka, 1999. Structural analysis and complete physical map of Arabidopsis thaliana chromosome 5 including centromeric and telomeric regions. DNA Res. 6: 381–386. [DOI] [PubMed] [Google Scholar]
- Kumar, A., and J. L. Bennetzen, 1999. Plant retrotransposons. Annu. Rev. Genet. 33: 479–532. [DOI] [PubMed] [Google Scholar]
- Levis, R. W., R. Ganesan, K. Houtchens, L. A. Tolar and F. M. Sheen, 1993. Transposons in place of telomeric repeats at a Drosophila telomere. Cell 75: 1083–1093. [DOI] [PubMed] [Google Scholar]
- Lin, X., S. Kaul, S. Rounsley, T. P. Shea, M. I. Benito et al., 1999. Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature 402: 761–768. [DOI] [PubMed] [Google Scholar]
- Lin, Y., and A. S. Waldman, 2001. Capture of DNA sequences at double-strand breaks in mammalian chromosomes. Genetics 158: 1665–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linardopoulou, E. V., E. M. Williams, Y. Fan, C. Friedman, J. M. Young et al., 2005. Human subtelomeres are hot spots of interchromosomal recombination and segmental duplication. Nature 437: 94–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis, E. J., 1995. The chromosome ends of Saccharomyces cerevisiae. Yeast 11: 1553–1573. [DOI] [PubMed] [Google Scholar]
- Louis, E. J., and J. E. Haber, 1990. Mitotic recombination among subtelomeric Y′ repeats in Saccharomyces cerevisiae. Genetics 124: 547–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis, E. J., and J. E. Haber, 1991. Evolutionarily recent transfer of a group I mitochondrial intron to telomere regions in Saccharomyces cerevisiae. Curr. Genet. 20: 411–415. [DOI] [PubMed] [Google Scholar]
- Lukaszewski, A. J., and C. A. Curtis, 1993. Physical distribution of recombination in B-genome chromosomes of tetraploid wheat. Theor. Appl.Genet. 86: 121–127. [DOI] [PubMed] [Google Scholar]
- Lundblad, V., and E. H. Blackburn, 1993. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell 73: 347–360. [DOI] [PubMed] [Google Scholar]
- Mayer, K., C. Schuller, R. Wambutt, G. Murphy, G. Volckaert et al., 1999. Sequence and analysis of chromosome 4 of the plant Arabidopsis thaliana. Nature 402: 769–777. [DOI] [PubMed] [Google Scholar]
- McClintock, B., 1941. The stability of broken ends of chromosomes in Zea mays. Genetics 26: 234–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern, M. J., and E. H. Blackburn, 1995. Runaway telomere elongation caused by telomerase RNA gene mutations. Nature 376: 403–409. [DOI] [PubMed] [Google Scholar]
- Mefford, H. C., and B. J. Trask, 2002. The complex structure and dynamic evolution of human subtelomeres. Nat. Rev. Genet. 3: 91–102. [DOI] [PubMed] [Google Scholar]
- Melek, M., and D. E. Shippen, 1996. Chromosome healing: spontaneous and programmed de novo telomere formation by telomerase. BioEssays 18: 301–308. [DOI] [PubMed] [Google Scholar]
- Moore, J. K., and J. E. Haber, 1996. Capture of retrotransposon DNA at the sites of chromosomal double-strand breaks. Nature 383: 644–646. [DOI] [PubMed] [Google Scholar]
- Muller, F., C. Wicky, A. Spicher and H. Tobler, 1991. New telomere formation after developmentally regulated chromosomal breakage during the process of chromatin diminution in Ascaris lumbricoides. Cell 67: 815–822. [DOI] [PubMed] [Google Scholar]
- Muller, H. J., 1938. The re-making of chromosomes. Collecting Net, Woods Hole 13: 181–198. [Google Scholar]
- Mummenhof, K., and H. Hurka, 1994. Subunit polypeptide composition of Rubisco and the origin of allopolyploid Arabidobsis suecica (Brassicaceae). Biochem. Syst. Ecol. 22: 807–812. [Google Scholar]
- Murashige, T., and F. Skoog, 1962. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant 15: 473–497. [Google Scholar]
- Nassif, N., J. Penney, S. Pal, W. R. Engels and G. B. Gloor, 1994. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol. Cell. Biol. 14: 1613–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIH/CEPH Collaborative Mapping Group, 1992. A comprehensive genetic linkage map of the human genome. Science 258: 67–86. [PubMed] [Google Scholar]
- Nordborg, M., T. T. Hu, Y. Ishino, J. Jhaveri, C. Toomajian et al., 2005. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 3: e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa, K., M. Suzuki, M. Takemura and S. Yoshida, 2000. In vitro expansion of mammalian telomere repeats by DNA polymerase alpha-primase. Nucleic Acids Res. 28: 3117–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmido, N., K. Kijima, I. Ashikawa, J. H. de Jong and K. Fukui, 2001. Visualization of the terminal structure of rice chromosomes 6 and 12 with multicolor FISH to chromosomes and extended DNA fibers. Plant Mol. Biol. 47: 413–421. [DOI] [PubMed] [Google Scholar]
- O'Kane, S. L., B. A. Schaal and I. A. Al-Shebaz, 1996. The origins of Arabidopsis suecica (Brassicaceae) as indicated by nuclear rDNA sequences. Syst. Bot. 21: 559–566. [Google Scholar]
- Pearce, S. R., U. Pich, G. Harrison, A. J. Flavell, J. S. Heslop-Harrison et al., 1996. The Ty1-copia group retrotransposons of Allium cepa are distributed throughout the chromosomes but are enriched in the terminal heterochromatin. Chromosome Res. 4: 357–364. [DOI] [PubMed] [Google Scholar]
- Peterson-Burch, B. D., D. Nettleton and D. F. Voytas, 2004. Genomic neighborhoods for Arabidopsis retrotransposons: a role for targeted integration in the distribution of the Metaviridae. Genome Biol. 5: R78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov, D. A., T. A. Sangster, J. S. Johnston, D. L. Hartl and K. L. Shaw, 2000. Evidence for DNA loss as a determinant of genome size. Science 287: 1060–1062. [DOI] [PubMed] [Google Scholar]
- Pologe, L. G., and J. V. Ravetch, 1988. Large deletions result from breakage and healing of P. falciparum chromosomes. Cell 55: 869–874. [DOI] [PubMed] [Google Scholar]
- Pontes, O., R. J. Lawrence, N. Neves, M. Silva, J. H. Lee et al., 2003. Natural variation in nucleolar dominance reveals the relationship between nucleolus organizer chromatin topology and rRNA gene transcription in Arabidopsis. Proc. Natl. Acad. Sci. USA 100: 11418–11423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott, J., and E. H. Blackburn, 1997. Telomerase RNA mutations in Saccharomyces cerevisiae alter telomerase action and reveal nonprocessivity in vivo and in vitro. Genes Dev. 11: 528–540. [DOI] [PubMed] [Google Scholar]
- Price, R. A., J. D. Palmer and I. A. Al-Shehbaz, 1994. Systematic relationships of Arabidopsis: a molecular and morphological perspective, pp. 7–19 in Arabidopsis, edited by E. M. Meyerowitz and C. R. Somerville. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Pryde, F. E., H. C. Gorham and E. J. Louis, 1997. Chromosome ends: all the same under their caps. Curr. Opin. Genet. Dev. 7: 822–828. [DOI] [PubMed] [Google Scholar]
- Puchta, H., 2005. The repair of double-strand breaks in plants: mechanisms and consequences for genome evolution. J. Exp. Bot. 56: 1–14. [DOI] [PubMed] [Google Scholar]
- Rambaut, A., 1996. Se-Al: Sequence Alignment Editor (http://evolve.zoo.ox.ac.uk/).
- Ricchetti, M., C. Fairhead and B. Dujon, 1999. Mitochondrial DNA repairs double-strand breaks in yeast chromosomes. Nature 402: 96–100. [DOI] [PubMed] [Google Scholar]
- Ricchetti, M., B. Dujon and C. Fairhead, 2003. Distance from the chromosome end determines the efficiency of double strand break repair in subtelomeres of haploid yeast. J. Mol. Biol. 328: 847–862. [DOI] [PubMed] [Google Scholar]
- Ricchetti, M., F. Tekaia and B. Dujon, 2004. Continued colonization of the human genome by mitochondrial DNA. PLoS Biol. 2: E273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, E. J., S. Chao, A. Vongs and J. Yang, 1992. Characterization of Arabidopsis thaliana telomeres isolated in yeast. Nucleic Acids Res. 20: 4039–4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, E. J., A. Vongs, M. Walsh, J. Yang and S. Chao, 1993. Substructure of telomere repeat arrays, pp. 103–114 in The Chromosome, edited by J. S. Heslop-Harrison and R. Flavell, BIOS Scientific Publishers, Oxford.
- Riha, K., and D. E. Shippen, 2003. Telomere structure, function and maintenance in Arabidopsis. Chromosome Res. 11: 263–275. [DOI] [PubMed] [Google Scholar]
- Roder, M. S., N. L. Lapitan, M. E. Sorrells and S. D. Tanksley, 1993. Genetic and physical mapping of barley telomeres. Mol. Gen. Genet. 238: 294–303. [DOI] [PubMed] [Google Scholar]
- Rogers, S. O., and A. J. Bendich, 1985. Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol. Biol. 5: 69–76. [DOI] [PubMed] [Google Scholar]
- Royle, N. J., D. M. Baird and A. J. Jeffreys, 1994. A subterminal satellite located adjacent to telomeres in chimpanzees is absent from the human genome. Nat. Genet. 6: 52–56. [DOI] [PubMed] [Google Scholar]
- Rozas, J., and R. Rozas, 1999. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15: 174–175. [DOI] [PubMed] [Google Scholar]
- Rozas, J., J. C. Sanchez-DelBarrio, X. Messeguer and R. Rozas, 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19: 2496–2497. [DOI] [PubMed] [Google Scholar]
- Säll, T., M. Jakobsson, C. Lind-Hallden and C. Hallden, 2003. Chloroplast DNA indicates a single origin of the allotetraploid Arabidopsis suecica. J. Evol. Biol. 16: 1019–1029. [DOI] [PubMed] [Google Scholar]
- Salomon, S., and H. Puchta, 1998. Capture of genomic and T-DNA sequences during double-strand break repair in somatic plant cells. EMBO J. 17: 6086–6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan, H., 2001. A bouquet makes ends meet. Nat. Rev. Mol. Cell Biol. 2: 621–627. [DOI] [PubMed] [Google Scholar]
- Schlotterer, C., 2000. Evolutionary dynamics of microsatellite DNA. Chromosoma 109: 365–371. [DOI] [PubMed] [Google Scholar]
- Schlotterer, C., and D. Tautz, 1992. Slippage synthesis of simple sequence DNA. Nucleic Acids Res. 20: 211–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, K. J., S. Ramos-Onsins, H. Ringys-Beckstein, B. Weisshaar and T. Mitchell-Olds, 2005. A multilocus sequence survey in Arabidopsis thaliana reveals a genome-wide departure from a neutral model of DNA sequence polymorphism. Genetics 169: 1601–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzacher, T., 1996. The physical organization of Triticeae chromosomes. Symp. Soc. Exp. Biol. 50: 71–75. [PubMed] [Google Scholar]
- Schwarzacher, T., 2003. Meiosis, recombination and chromosomes: a review of gene isolation and fluorescent in situ hybridization data in plants. J. Exp. Bot. 54: 11–23. [DOI] [PubMed] [Google Scholar]
- Shakirov, E. V., and D. E. Shippen, 2004. Length regulation and dynamics of individual telomere tracts in wild-type Arabidopsis. Plant Cell 16: 1959–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley, B. W., S. Hanley and H. M. Goodman, 1992. Effects of ionizing radiation on a plant genome: analysis of two Arabidopsis transparent testa mutations. Plant Cell 4: 333–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprung, C. N., G. E. Reynolds, M. Jasin and J. P. Murnane, 1999. Chromosome healing in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA 96: 6781–6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupar, R. M., J. W. Lilly, C. D. Town, Z. Cheng, S. Kaul et al., 2001. Complex mtDNA constitutes an approximate 620-kb insertion on Arabidopsis thaliana chromosome 2: implication of potential sequencing errors caused by large-unit repeats. Proc. Natl. Acad. Sci. USA 98: 5099–5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima, F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng, S. C., B. Kim and A. Gabriel, 1996. Retrotransposon reverse-transcriptase-mediated repair of chromosomal breaks. Nature 383: 641–644. [DOI] [PubMed] [Google Scholar]
- Thompson, J. K., J. P. Rubio, S. Caruana, A. Brockman, M. E. Wickham et al., 1997. The chromosomal organization of the Plasmodium falciparum var gene family is conserved. Mol. Biochem. Parasitol. 87: 49–60. [DOI] [PubMed] [Google Scholar]
- Tran, H. T., N. P. Degtyareva, N. N. Koloteva, A. Sugino, H. Masumoto et al., 1995. Replication slippage between distant short repeats in Saccharomyces cerevisiae depends on the direction of replication and the RAD50 and RAD52 genes. Mol. Cell. Biol. 15: 5607–5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unseld, M., J. R. Marienfeld, P. Brandt and A. Brennicke, 1997. The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat. Genet. 15: 57–61. [DOI] [PubMed] [Google Scholar]
- Vershinin, A. V., T. Schwarzacher and J. S. Heslop-Harrison, 1995. The large-scale genomic organization of repetitive DNA families at the telomeres of rye chromosomes. Plant Cell 7: 1823–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S. S., and V. A. Zakian, 1990. Telomere-telomere recombination provides an express pathway for telomere acquisition. Nature 345: 456–458. [DOI] [PubMed] [Google Scholar]
- Watterson, G. A., 1975. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7: 256–276. [DOI] [PubMed] [Google Scholar]
- Werner, J., R. S. Kota, B. S. Gill and T. R. Endo, 1992. Distribution of telomeric repeats and their role in the healing of broken chromosome ends in wheat. Genome 35: 844–848. [Google Scholar]
- Wessler, S., A. Tarpley, M. Purugganan, M. Spell and R. Okagaki, 1990. Filler DNA is associated with spontaneous deletions in maize. Proc. Natl. Acad. Sci. USA 87: 8731–8735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie, A. O., J. Lamb, P. C. Harris, R. D. Finney and D. R. Higgs, 1990. A truncated human chromosome 16 associated with alpha thalassaemia is stabilized by addition of telomeric repeat (TTAGGG)n. Nature 346: 868–871. [DOI] [PubMed] [Google Scholar]
- Wu, K. S., and S. D. Tanksley, 1993. Genetic and physical mapping of telomeres and macrosatellites of rice. Plant Mol. Biol. 22: 861–872. [DOI] [PubMed] [Google Scholar]
- Yu, G. L., and E. H. Blackburn, 1991. Developmentally programmed healing of chromosomes by telomerase in Tetrahymena. Cell 67: 823–832. [DOI] [PubMed] [Google Scholar]
- Yu, G. L., J. D. Bradley, L. D. Attardi and E. H. Blackburn, 1990. In vivo alteration of telomere sequences and senescence caused by mutated Tetrahymena telomerase RNAs. Nature 344: 126–132. [DOI] [PubMed] [Google Scholar]
- Yu, X., and A. Gabriel, 1999. Patching broken chromosomes with extranuclear cellular DNA. Mol. Cell 4: 873–881. [DOI] [PubMed] [Google Scholar]
- Zou, S., N. Ke, J. M. Kim and D. F. Voytas, 1996. The Saccharomyces retrotransposon Ty5 integrates preferentially into regions of silent chromatin at the telomeres and mating loci. Genes Dev. 10: 634–645. [DOI] [PubMed] [Google Scholar]