Abstract
Elucidating the genetic basis of adaptation on a genomewide scale has evaded biologists, but complete genome sequences and DNA high-density array technology make genomewide surveys more tractable. Six lines of Escherichia coli adapted for 2,000 generations to a stressful high temperature of 41.5°C were examined on a genomewide scale for duplication/deletion events by using DNA high-density arrays. A total of five duplication and deletion events were detected. These five events occurred in three of the six lines, whereas the remaining three lines contained no detectable events. Three of the duplications were at 2.85 Mb of the E. coli chromosome, providing evidence for the replicability of the adaptation to high temperature. Four candidate genes previously shown to play roles in stress and starvation survival were identified in the region of common duplication. Expression of the two candidate genes examined is elevated over expression levels in the ancestral lines or the lines without the duplication. In the two cases where the duplication at 2.85 Mb has been further characterized, the timing of the genome reorganization is coincident with significant increases in relative fitness. In both of these cases, the model for the origin of the duplication is a complex recombination event involving insertion sequences and repeat sequences. These results provide additional evidence for the idea that gene duplication plays an integral role in adaptation, specifically as a means for gene amplification.
Both the short generation time and large population sizes of microorganisms allow them to respond quickly to selection imposed in the laboratory and in nature. Random mutations in conjunction with large population sizes and short generation times allow microorganisms to overcome limits to adaptation associated with a primarily asexual mode of reproduction (1). Numerous studies successfully have used microorganisms to test evolutionary theory and address the evolution of physiological systems; they are now beginning to address the genetic basis of adaptation (2–7). The genetic changes underlying the process of adaptation can be mutations, small insertion/deletion events, large insertion/deletion events, or chromosomal inversions. Recent studies examining the genetic basis of adaptation at the level of single nucleotide mutations have shown high levels of convergent evolution in viruses adapted to different hosts and environments (2, 3). Here, we examine the genetic basis of adaptation at the level of large insertion/deletion events by using a whole genome approach and determine whether comparable levels of convergent evolution occur in a more complex enteric prokaryote or whether greater developmental and physiological complexity result in inherently less replicable patterns of evolution.
Gene duplications are a common evolutionary response in bacteria exposed to different selection pressures of the laboratory and presumably in nature (8, 9). Generally, gene regulatory elements allow cells to adjust their physiology to maintain homeostasis during the exposure to stress; however, when conditions exist that cannot be compensated by an alteration in gene expression, selection may favor an increase in the copy number of a gene or group of genes. These duplications are dynamically similar to a regulatory response because they are reversible. When growth conditions change and the individuals containing the duplication no longer have a growth advantage, revertants may again have a selective advantage. In this way, gene duplication can be thought of as a primitive regulatory mechanism whose specificity is governed by natural selection (4). Aware of the prominent role gene duplications play in the adaptation of microorganisms, in this study we examine patterns of gene duplication and deletion in six lines of Escherichia coli adapted to stressful high temperatures (Fig. 1, ref. 6) and examine the role of identified duplications in the adaptation to this temperature. These lines have been previously characterized relative to their ancestors with respect to their growth rates, fitness, and a number of physiological measures of performance (4–7). These six lines showed a 33.5% (range 20–65%) increase in fitness on average relative to the ancestor when assayed at 41.5°C (6). The ability to compare ancestral and derived organisms directly over small time intervals, to detect a potentially small number of genetic changes associated with their evolution, and to examine the replicate nature of their evolution among lineages makes this system a powerful resource for investigating the genetic basis of adaptation.
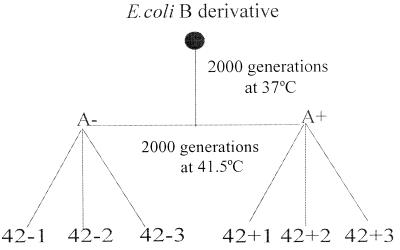
Figure 1.
Evolutionary history of thermally adapted lines of E. coli.
The size and complexity of the genomes of most organisms make studies of the genetic basis of adaptation on a genomewide scale a formidable task. However, the complete genome sequence (10) in combination with DNA high-density arrays allows all 4,290 ORFs of the E. coli genome to be examined simultaneously (11–13). This whole genome approach allows us to estimate the amount of evolutionary convergence that occurs during adaptation and determine the relative importance of gene duplications in this adaptation while analyzing the overall genetic basis of adaptation.
Materials and Methods
Derivation of E. coli Lines (Fig. 1).
An E. coli B derivative (strain Bc251; ref. 14) was grown under laboratory conditions (37°C in Davis minimal medium supplemented with 25 μg/ml−1 glucose) for 2,000 generations with daily transfer into fresh medium as described (6). Each day, cultures were diluted 100-fold into fresh medium requiring an estimated 6.6 (log2100 = 6.6) generations of binary fission to maintain a stationary phase cell density of ≈4 × 107 cells/ml. Adaptation to culture conditions was evident by a 30% increase in the fitness of A− relative to the original E. coli B derivative. After 2,000 generations, two genotypes (A− and A+) that differed only in a neutral genetic marker were isolated, and each became the ancestor of three lines (−1, −2, −3 and +1, +2, +3, respectively) that were propagated in the lab for another 2,000 generations at 41.5°C.
DNA High-Density Array Hybridization.
Genomic DNA (gDNA) was isolated by using standard procedures (15) from each E. coli line, grown overnight at 37°C in Luria broth, RNase-treated (Promega), and labeled. Each labeling reaction included 500 ng of gDNA, 25 μCi of 33P-labeled dCTP (New England Nuclear), 0.225 mM nucleotide mix (dGTP, dTTP, dATP, 0.075 mM each), 2.5 units of DNA polymerase (New England Biolabs), and 1× random hexamers (Boehringer Mannheim) and was incubated for 1 h at 37°C. Labeled gDNA was separated from unincorporated nucleotides by filtration over G-50 Sephadex columns (Amersham Pharmacia). E. coli DNA high-density array (Sigma Genosys) nylon filters were soaked in 2× standard saline phosphate/EDTA (SSPE, 0.18 M NaCl/10 mM phosphate, pH 7.4/1 mM EDTA) for 10 min and prehybridized in 10 ml of hybridization solution (5× SSPE/2% SDS/1× Denhardt's solution/0.1 mg/ml sheared salmon sperm DNA) for 1 h at 65°C. After separation from unincorporated nucleotides, the probe was boiled for 10 min and snap-cooled on ice. The probe was added to 7.5 ml of hybridization solution. Hybridization was performed overnight at 65°C. After hybridization, each filter was rinsed with 50 ml of 0.5× SSPE containing 0.2% SDS at room temperature for 5 min, followed by three washes in the same solution at 65°C for 20 min each. The filters were wrapped in plastic wrap and exposed to a phosphor screen for 24–48 h. Filters were stripped by microwaving at the low/defrost setting in 500 ml of a previously boiled solution of 10 mM Tris (pH 8.0), containing 1 mM EDTA and 1% SDS for 30 min. Each high-density array was probed eight times (twice with separate gDNA preparations from each ancestor and its three high temperature-selected derivatives). It is important to point out that the use of K-12 genomic sequence (10) in the construction of the high-density array excludes the examination of regions of E. coli B missing from K-12; therefore, any genes or gene regions not present in K-12 are impossible to analyze with this technique.
DNA High-Density Array Analysis.
For every comparison, two independent gDNA preparations were each hybridized to a double-spotted high-density array. Intensity values for each spot were obtained by using DNA Arrayvision (Research Imaging, St. Catharines, ON, Canada), converted to a percentage of total signal on the blot, and natural log transformed; two spots from any given experiment then were averaged. This method resulted in four observations per gene: A1, A2 (for ancestor) and E1, E2 (for evolved). The regression model (over genes) A1 = m A2 + b was fitted to the ancestral data and used to obtain a total blot intensity-corrected replicate-averaged intensity value for each gene as A = (A1 + m A2 + b)/2. A similar value, E, was obtained for the evolved lines. An intensity-corrected difference intensity value, Di (for the ith gene) was defined as the residual obtained by fitting a second regression model, E = m A + b. The Di values were sorted by gene order in E. coli K-12, and running average difference scores (RADS) as a function of window location midpoint were obtained. RADS were based on a window size of 20 differences, with the most extreme difference thrown out for each window. Confidence intervals on the RADS were obtained by using a permutation testing procedure based on a larger sliding confidence interval window of 500 differences. The variance in RADS for a window of size 20 differences centered within the larger window was calculated over 500 random permutations of the order of the 500 differences contained in the sliding confidence window. Confidence intervals are the observed difference plus or minus four standard deviations obtained from the above variance estimate.
Relative Fitness Assays.
Relative fitness (6) was assayed by performing competition experiments. The two competitors, of opposite genetic marker state, were taken from −80°C storage and grown separately in Luria broth for 1 day at 37°C. Cultures then were transferred to minimal media (Davis minimal media supplemented with 25 μg/ml glucose) and grown for 1 day at 37°C, transferred to fresh media and grown for 1 day at 41.5°C (the competition temperature). The two competitors then were mixed in a 1:1 volumetric ratio and diluted 100-fold into minimal media, where they grew and competed at 41.5°C during a standard daily growth cycle. Initial and final densities of each competitor were estimated by plating dilutions onto tetrazolium arabinose agar, on which the two competitors could be distinguished by colony color. Approximately 200 colonies were counted per fitness assay, and relative fitness was estimated from the average of 10–18 independent assays. Relative fitness was calculated from the ratio of the number of doublings achieved by each of the two competitors.
Expression of Candidate Genes.
Total RNA was isolated (Rnaqueous, Ambion, Austin, TX) from stationary phase cells after 12 h in minimal media culture (Davis minimal broth supplemented with 500 μg/ml−1 glucose) at 41.5°C. RNA was blotted (15), and Northern blot analysis was performed with PCR products for rpoA (control), rpoS (candidate), and surE (candidate) as probes. Procedures were generally the same as described previously in DNA high-density array hybridization, except that 25 μCi of 32P-labeled dCTP (New England Nuclear) was used for probe labeling, and prehybridization and hybridizations were performed at 57°C in 6× SSC, 2× Denhardt's solution, 0.1% SDS, and washing was done in 0.2× SSC, 0.1% SDS. RNA was isolated from four independent stationary phase cultures, and each RNA prep was separately spot-blotted onto four different nylon filters. Each blot was probed with all three probes. Blots were stripped with 0.1% SDS between probings. All RNA work was done with Rnase-free supplies and diethyl pyrocarbonate-treated liquids. Background-subtracted signal intensity was obtained by using DNA Arrayvision (Research Imaging), log transformed, and expressed as relative intensity. Intensities were compared by using ANOVA and post hoc tests (n = 6 for ancestors, 9 for selected lines with the 2.85-Mb duplication, 9 for the selected lines without duplication).
Standard PCR.
Reactions contained 50 mM KCl/10 mM Tris, pH 8.8/1.5 mM MgCl2/0.1% Triton X/0.5 μM each primer (E. coli ORFmers, Sigma Genosys)/0.05 mM each dNTP/2.5 units of Taq polymerase (Perkin–Elmer Biosystems)/20 ng of gDNA. gDNA was isolated from single colony isolates that were frozen at 100, 200, 400, 600, 800, 1,000, 1,200, 1,400, 1,600, 1,800, and 2,000 generations. PCR cycling conditions were 94°C for 2 min, 25 cycles of 94°C for 30 s, 64°C for 30 s, 72°C for 2 min, and a final extension step at 72°C for 10 min.
Long PCR.
Reactions contained 20 mM Tricine, pH 8.9/84.8 mM KOAc/2 mM Mg(OAc)2/0.5 μM each primer/0.2 mM each dNTP/5 units of Taq polymerase (Perkin–Elmer Biosystems)/2.5 units of Taq extender (Stratagene)/20 ng of gDNA. Amplification of the middle junction in the 42−1 line (see Fig. 3B, row 1) used the following primers: 5′-GCTCATTTCACCCGTAGACG-3′ and 5′-GGAAATAGCTGGCATGACG-3′ (One Trick Pony Oligos, Ransom Hill Bioscience, Ramoma, CA). The left breakpoint in the 42−1 line (see Fig. 3B, row 2) was amplified by using Sigma Genosys ORFmers for ascB. Amplification of the middle junction in the 42−2 line (see Fig. 3B, row 4) used the following primers: 5′-GGAAATAGCTGGCATGACG-3′ and 5′-GCACGTCCGTCTGAATGAT-3′. Amplification of the left breakpoint in the 42−2 line (see Fig. 3B, row 3) used the following primers: 5′-CGCAGATCGTTTCTGTCGT-3′ and 5′-GCACGTCCGTCTGAATGAT-3′. Cycling conditions for long PCR were 94°C for 2 min, 35 cycles of 94°C for 45 s, 55–57°C for 45 s, and 72°C for 2–3 min (1 min/2 kb) and a final extension step at 72°C for 10 min.
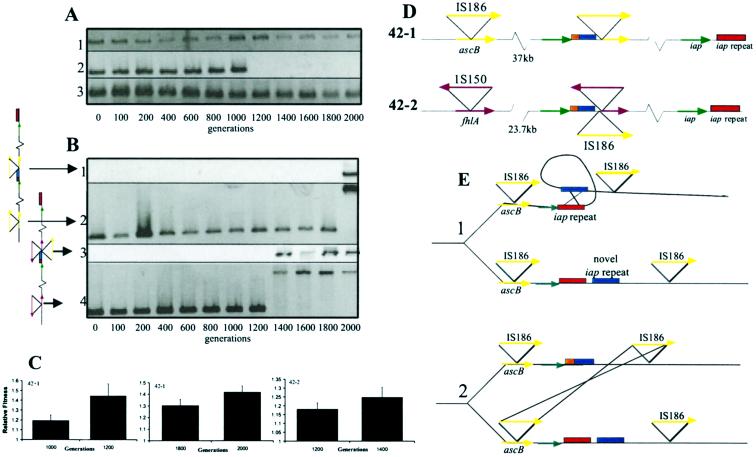
Figure 3.
Confirmation, timing, and molecular characterization of a subset of the duplications and deletions. (A) The 12-kb deletion in the 42+1 line. PCR products for amplifications of sbcB, upstream of the deletion (1.5 kb, row 1), galF, within the deletion (1.1 kb, row 2), and cspG, immediately downstream of the deletion (0.2 kb, row 3), are shown as a function of the number of generations at 41.5°C. (B) Identification of the breakpoints for the tandem duplications in the 42−1 and 42−2 lines. The 37-kb repeat in the 42−1 line occurred between generations 1,800 and 2,000 (middle junction, row 1, 2.5 kb; left breakpoint, row 2, 1.4–2.8 kb). The 23.7-kb repeat in the 42−2 line occurred between generations 1,200 and 1,400 (middle junction, row 3, 4.4 kb; left breakpoint, row 4, 1.1–2.5 kb). Primers used in the amplification of the products in rows 1 and 3 amplify the junction generated by the duplication. Primers used in the amplification of products in rows 2 and 4 amplify the gene at the left breakpoint. The appearance of IS at the left breakpoints is coincident with the origin of the duplications. (C). Relative fitness measurements before and after chromosome rearrangement events. Bars represent means ±95% confidence intervals. (D) Genetic structure of duplications. The unique junction generated by the duplication is represented by the juxtaposition of red and blue blocks. We depict a single duplication, but the number of tandem copies is not determined. (E) Proposed model for the origin of the duplication. The intrastrand recombination is presented in part 1 and the sister strand exchange in part 2. The figure depicts the model for the generation of the 23.7-kb tandem duplication; the model for the 37-kb tandem duplication is identical except that the second recombination event occurs between IS150 located adjacent to the iap repeat sequence and IS150 at the left breakpoint.
Results and Discussion
Comparing the intensity of hybridization signal for each ORF, we detect a total of five duplications and deletions in three of the evolved lines (Fig. 2). One of the lines contains three duplication and deletion events (42+1), two of the lines (42−1, 42−2) contain a single event, and three of the lines contain no detectable events. Of the five independent duplication and deletion events detected, three of the lines exhibit a duplication of at least 23.7 kb at 2.85 Mb on the E. coli chromosome. This duplication at 2.85 Mb is strong evidence for the replicability of specific genome reorganization events during thermal adaptation. Furthermore, observed duplication events occur only during adaptation to high temperature, as neither of the ancestral lines had similar duplications or deletions after culture for 2,000 generations at 37°C relative to their founding clone (A606) (see supplementary Fig. 5, which is published as supplemental data on the PNAS web site, www.pnas.org).
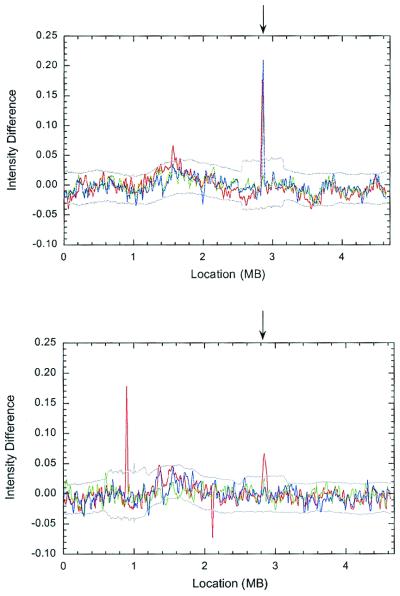
Figure 2.
Detection of duplication and deletion events using DNA high-density array data. (Upper) The A− derived lines: 42−1 in red, 42−2 in blue, and 42−3 in green. (Lower) The A+ derived lines: 42+1 in red, 42+2 in blue, and 42+3 in green. The gray lines are confidence intervals calculated by using a permutation testing approach (Upper, confidence interval for 42−2; Lower, 42+1). Confidence intervals are similar for the other two lines in a panel. Regions in which signal intensity is above the gray line indicate a duplication in the selected line, and regions in which it is below indicate a deletion. An arrow indicates the common duplication at 2.85 Mb present in three of the six selected lines. Note the presence of a red peak beneath the blue peak (Upper).
Verification of these duplications or deletions by Southern blotting (not shown) and/or PCR (Fig. 3) provides strong evidence that DNA high-density arrays can be reliably used to detect regions in which gene copy number has changed through evolution. Confirmation by PCR also allowed us to develop simple assays for examining the timing of the genomic reorganizations. The 12-kb deletion in the 42+1 line occurred between generations 1,000 and 1,200 (Fig. 3A); a 25% increase (P < 0.005) in relative fitness (6) (Fig. 3C) at 41.5°C in the 42+1 line is associated with this time interval. Tandem duplications at 2.85 Mb, with repeat sizes of 37 and 23.7 kb, occurred in the 42−1 and 42−2 lines between generations 1,800–2,000 and 1,200–1,400, respectively (Fig. 3B). In these lines, the time intervals in which the duplication events have occurred also are associated with significant increments in relative fitness of 12% (P < 0.01, 42−1) and 7% (P < 0.05, 42−2) (Fig. 3C). Although these are not the only generational intervals associated with fitness increments, their coincidence with each chromosomal rearrangement is again supportive evidence of their adaptive implication.
PCR analysis, Southern blotting, and subsequent DNA sequencing suggests that the duplications at 2.85 Mb did not arise as a result of a simple ectopic exchange event between bacterial insertion sequences (ISs) associated with the endpoints of the duplication. DNA sequencing of the breakpoints associated with the duplication in the 42−1 and 42−2 lines shows that an IS appeared at the left breakpoint during the same 200 generation period as the duplication event and interrupts an ORF (Fig. 3B, IS186 at bp812 in ascB in the 42−1 line and IS150 at bp802 of fhlA in the 42−2 line). In both cases examined, the right breakpoint is immediately downstream of the iap gene in the iap repeat region, a region that contains a number of nearly identical 29-bp repeats, each separated by 32 bp of unique sequence speculated to play a role in chromosomal rearrangement (16, 17). There is no evidence for bacterial insertion sequences at the right breakpoint in either the 42−1 line or the 42−2 line (Fig. 3D), making exchange between IS elements an unlikely explanation for the origin of these duplications.
To infer the nature of the molecular mechanisms that gave rise to the duplication in the 42−1 and 42−2 lines, we sequenced the unique middle junction created by the duplication event (Fig. 3B, rows 1 and 3). The middle junction in the 42−1 line contains two iap repeats present in E. coli K-12, four iap repeats absent in E. coli K-12, and IS186. The middle junction in the 42−2 line contains two iap repeats present in E. coli K-12, four iap repeats absent in E. coli K-12, IS186, 226 bp of sequence from the cysJIH region (11 kb downstream of iap, ref. 18), and IS150. The novelty of the iap repeat unit at the junction can be inferred from a complete lack of homology of the unique 32-bp spacers to any sequence in the K-12 genome. A proposed model for the origin of the duplications involves two homologous exchange events (Fig. 3E). First, an intrastrand exchange occurs between the iap repeats immediately downstream of iap and a hypothesized second set of iap repeats unique to E. coli B located elsewhere in the genome. Second, a sister chromosome exchange event occurs between an IS element located in the second set of iap repeats and the homologous IS element located in the gene at the left breakpoint of the duplication. Additional support for this model comes from the observation that the sequences of the iap repeats, although not found anywhere else in E. coli, are identical up to and including IS186 in the 42−1 and 42−2 lines. Based on this model, the molecular events leading to the origin of the duplications in the 42−1 and 42−2 lines are likely to be rare occurrences. That such a rare molecular event occurred and was fixed in two different populations makes selection a more parsimonious explanation for its fixation than random mutation and genetic drift.
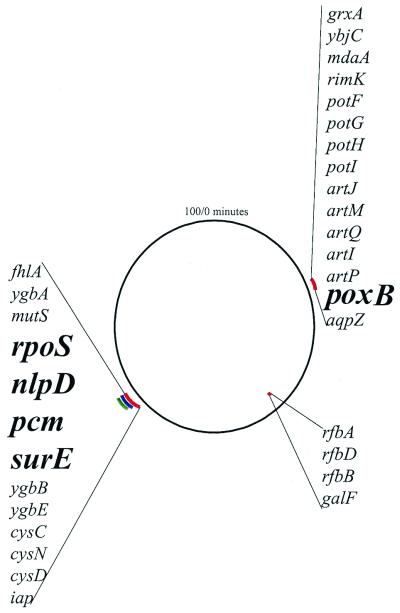
We wanted to determine whether there are candidate genes in the observed duplications and deletion with functions consistent with an adaptive response to high temperature. The five duplications and deletions contain 55 named and 30 unnamed ORFs (Fig. 4), or approximately only 2% of all E. coli genes. The ability to focus our efforts on specific genes or gene regions based on high-density array data makes the search for the physiological basis of adaptation considerably more efficient. The 12-kb deletion in the 42+1 line contains a suite of genes (Fig. 4) primarily involved in lipopolysaccharide synthesis, specifically the conversion of D-glucose 1-phosphate to dTDP-L-rhamnose and O-antigen assembly (19). None of the deleted genes have functions that make them obvious candidate genes for high-temperature adaptation. The tandem duplication with a repeat size of 30.7 kb in the 42+1 line contains poxB (pyruvate oxidase; EC 1.2.2.2), which converts pyruvate to acetate and carbon dioxide. Under certain growth conditions, this process can provide a source of energy from pyruvate catabolism. poxB expression is maximal in early stationary phase and strictly depends on the rpoS gene (20), which is located in the duplication region at 2.85 Mb. The region of the common duplication at 2.85 Mb (in lines 42+1, 42−1, 42−2) contains 13 named ORFs and 10 unnamed ORFs. Four ORFs (rpoS, nlpD, pcm, and surE) are candidate genes that harbor functions that may be involved in adaptation to high temperature. rpoS encodes a sigma factor (σs) involved in stationary phase gene expression (21), and nlpD encodes a lipoprotein (22). rpoS and its protein product σs are controlled at the level of transcription, translation, and proteolysis. Exposure to high temperature and expression of DnaK, a prokaryotic heat shock protein, has been shown to inhibit σs degradation and allow for the expression of stress and stationary phase genes controlled by σs (23). pcm (protein-L-isoaspartate(D-aspartate) O-methyltransferase; EC 2.1.1.77) is likely to play a role in protein repair or degradation pathways that metabolize proteins containing damaged aspartyl residues (24). pcm acts to maintain protein structure by converting L-isoaspartyl residues that can place kinks in peptide backbones preventing refolding, back to normal L-aspartyl residues. Without pcm, L-isoaspartyl residues accumulate during stressful conditions, increasing the unfolding of proteins and preventing their refolding (25). surE is less characterized than pcm, although mutant studies confirm that surE also plays a role in stress and starvation survival (26). Single gene mutants for all four genes grow normally during exponential phase, but all exhibit decreased stationary phase survival (21, 22, 24, 26), and some do not survive well when exposed to high temperature (21, 24, 26, 27). Additionally, lines exhibiting a growth advantage in stationary phase phenotype have been shown to have allelic variations in rpoS, resulting in reduced function of the allele while conferring an increased growth rate on mixtures of amino acids available after stationary phase cell death (28).
Figure 4.
Genes present in the regions of genome reorganization. The circle represents the E. coli chromosome. Lines outside of the circle represent regions of duplication, and lines inside the circle represent regions of deletion. Red lines are reorganization events in the 42+1 line, the blue line represents the duplication in the 42−1 line, and the green line represents the duplication in the 42−2 line. ORFs of known function are listed for each duplication or deletion. (For a complete list of ORFs for each duplication and deletion, see supplemental text.)
Did the three lines (42+2, 42+3, 42−3) that did not duplicate the genomic region at 2.85 Mb adapt to high temperature through pathways other than those encoded by these candidate genes or did they up-regulate the expression of these genes through cis- or trans-regulatory mutations? Northern blot analysis was carried out to quantify expression of two candidate genes, rpoS and surE. The lines containing the duplication at 2.85 Mb (42+1, 42−1, 42−2) have higher surE and rpoS expression than the ancestral lines (surE P < 0.05; rpoS P < 0.05). Lines without the duplication have expression levels indistinguishable from the ancestor (surE P > 0.90; rpoS P > 0.65) and lower than the lines with the duplication (surE P < 0.02; rpoS P < 0.06). We conclude that lines without the duplication at 2.85 Mb have adapted to high temperature via pathways other than those encoded by the candidate genes located in the duplication at 2.85 Mb.
Three of six high-temperature adapted lines of E. coli exhibited duplications spanning the same region of the genome, providing evidence for the replicability of adaptation at the genome level. This amount of evolutionary convergence as an evolutionary response to the same environmental stress suggests that a limited number of adaptive pathways are available to E. coli or that one adaptive solution is highly preferred. The work of Wichman and colleagues (2) also showed high levels of evolutionary convergence during the adaptation of two lines of bacteriophage to high temperature. In two replicate lines of bacteriophage, about 50% of the nucleotide changes (15 in one replicate and 14 in the other) that occurred during the adaptation to high temperature did so in parallel.
Our results also provide additional evidence for the idea that gene duplication may play an integral role in the adaptation of microorganisms (8, 9, 29, 30) as a method for gene amplification. Models suggest that cells carrying multiple copies of certain genes or gene regions of the chromosome are favored under certain environmental conditions and make their possessors more fit than the part of the population lacking the additional copies of the region. If the environmental conditions change, the population would not be committed to this new genotype because homologous recombination could return cells to their original haploid state (29).
Four of the five chromosomal changes detected in this study were previously localized to large genomic regions in a study that used rare cutting enzymes and pulse field electrophoresis (31). Only high-density arrays allowed us to infer that three of the duplication events included the same small genomic region. Once we had obtained the high-density array data, we were able to use other molecular techniques to pinpoint the duplication breakpoints and determine the timing and fitness effects of the duplication. Another study has used DNA microarrays to detect regions deleted in Bacille Calmette–Guérin vaccines against tuberculosis and provides evidence for the evolution of Bacille Calmette–Guérin strains since their original isolation, a finding integral to the optimization of future tuberculosis vaccines (32).
This experimental evolutionary system in which duplications occur in a relatively short time is useful not only for examining the genetic basis of adaptation and the link between physiology and genetics, it may allow experimental testing of models concerning the evolution of new gene function after gene duplication. High-density arrays have allowed us to identify regions of genomic deletion and duplication and assess the role of candidate genes in the adaptation to high temperature. This application of DNA high-density arrays to a longstanding problem in evolution is complementary to the approach of expression profiling and provides insight into the evolutionary process of adaptation at a larger scale.
Supplementary Material
Acknowledgments
We thank A. S. Peek for helpful discussions at the beginning and throughout this project. We thank G. W. Hatfield and members of his laboratory for advice and equipment and members of the Bennett, Gaut, and Long labs for helpful discussions and assistance. This work was funded by National Science Foundation Grants IBN 9507416 and IBN 9905980 (to A.F.B.), National Institutes of Health Grant GM58564 (to A.D.L.), and a National Science Foundation Predoctoral Fellowship (to M.M.R.).
Abbreviations
- IS
insertion sequence
- RADS
running average difference score
- gDNA
genomic DNA
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.021448998.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.021448998
References
- 1.Muller H J. Am Nat. 1932;68:118–138. [Google Scholar]
- 2.Wichman H A, Badgett M R, Scott L A, Boulianne C M, Bull J J. Science. 1999;285:422–424. doi: 10.1126/science.285.5426.422. [DOI] [PubMed] [Google Scholar]
- 3.Bull J J, Badgett M R, Wichmann H A, Huelsenbeck J P, Hillis D M, Gulati A, Ho C, Molineux I J. Genetics. 1997;147:1497–1507. doi: 10.1093/genetics/147.4.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett A F, Lenski R E. Evolution (Lawrence, Kans) 1993;47:1–12. doi: 10.1111/j.1558-5646.1993.tb01194.x. [DOI] [PubMed] [Google Scholar]
- 5.Bennett A F, Lenski R E. Evolution (Lawrence, Kans) 1996;50:493–503. doi: 10.1111/j.1558-5646.1996.tb03862.x. [DOI] [PubMed] [Google Scholar]
- 6.Bennett A F, Lenski R E, Mittler J E. Evolution (Lawrence, Kans) 1992;46:16–30. doi: 10.1111/j.1558-5646.1992.tb01981.x. [DOI] [PubMed] [Google Scholar]
- 7.Bennett A F, Dao K M, Lenski R E. Nature (London) 1994;346:79–81. doi: 10.1038/346079a0. [DOI] [PubMed] [Google Scholar]
- 8.Roth J R, Benson N, Galitski T, Haack K, Lawrence J G, Miesel L. In: Escherichia coli and Salmonella Cellular and Molecular Biology. Neidhardt F C, editor. Vol. 2. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 2256–2276. [Google Scholar]
- 9.Sonti R V, Roth J R. Genetics. 1989;123:19–28. doi: 10.1093/genetics/123.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blattner F R, Plunkett G, 3rd, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Mayhew G F, et al. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 11.Arfin S M, Long A D, Ito E T, Tolleri L, Riehle M M, Paegle E S, Hatfield G W. J Biol Chem. 2000;275:29672–29684. doi: 10.1074/jbc.M002247200. [DOI] [PubMed] [Google Scholar]
- 12.Richmond C S, Glasner J D, Mau R, Jin H, Blattner F R. Nucleic Acids Res. 1999;27:3821–3835. doi: 10.1093/nar/27.19.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tao H, Bausch C, Richmond C, Blattner F R, Conway T. J Bacteriol. 1999;181:6425–6440. doi: 10.1128/jb.181.20.6425-6440.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lederberg S. J Bacteriol. 1966;91:1029–1036. doi: 10.1128/jb.91.3.1029-1036.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 16.Nakata A, Amemura M, Makino K. J Bacteriol. 1989;171:3553–3556. doi: 10.1128/jb.171.6.3553-3556.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bachellier S, Gilson E, Hofnung M, Hill C. In: Escherichia coli and Salmonella Cellular and Molecular Biology. Neidhardt F C, editor. Vol. 2. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 2012–2040. [Google Scholar]
- 18.Ostrowski J, Wu J, Rueger D, Miller B, Siegel L, Kredich N. J Biol Chem. 1989;264:15726–15732. [PubMed] [Google Scholar]
- 19.Raetz C R H. In: Escherichia coli and Salmonella Cellular and Molecular Biology. Neidhardt F, editor. Vol. 2. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1035–1063. [Google Scholar]
- 20.Gennis R, Stewart V. In: Escherichia coli and Salmonella Cellular and Molecular Biology. Neidhardt F C, editor. Vol. 2. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 217–261. [Google Scholar]
- 21.Hengee-Aronis R. Cell. 1993;72:165–168. doi: 10.1016/0092-8674(93)90655-a. [DOI] [PubMed] [Google Scholar]
- 22.Ichikawa J K, Li C, Fu J, Clarke S. J Bacteriol. 1994;176:1630–1638. doi: 10.1128/jb.176.6.1630-1638.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hengge-Aronis R. In: Bacterial Stress Responses. Storz G, Hengee-Aronis R, editors. Washington, DC: Am. Soc. Microbiol.; 2000. pp. 161–178. [Google Scholar]
- 24.Li C, Clarke S. Proc Natl Acad Sci USA. 1992;89:9885–9889. doi: 10.1073/pnas.89.20.9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Visick J E, Clarke S. Mol Microbiol. 1995;16:835–845. doi: 10.1111/j.1365-2958.1995.tb02311.x. [DOI] [PubMed] [Google Scholar]
- 26.Li C, Ichikawa J K, Ravetto J J, Kuo H-C, Fu J C, Clarke S. J Bacteriol. 1994;176:6015–6022. doi: 10.1128/jb.176.19.6015-6022.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Visick J E, Cai H, Clarke S. J Bacteriol. 1998;180:2623–2629. doi: 10.1128/jb.180.10.2623-2629.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zambrano M M, Siegele D A, Almiron M, Tormo A, Kolter R. Science. 1993;259:1757–1760. doi: 10.1126/science.7681219. [DOI] [PubMed] [Google Scholar]
- 29.Anderson R P, Roth J R. Annu Rev Microbiol. 1977;31:473–505. doi: 10.1146/annurev.mi.31.100177.002353. [DOI] [PubMed] [Google Scholar]
- 30.Brown C J, Todd K M, Rosenzweig R F. Mol Biol Evol. 1998;15:931–942. doi: 10.1093/oxfordjournals.molbev.a026009. [DOI] [PubMed] [Google Scholar]
- 31.Bergthorsson U, Ochman H. J Bacteriol. 1999;181:1360–1363. doi: 10.1128/jb.181.4.1360-1363.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Behr M A, Wilson M A, Gill W P, Salamon H, Schoolnik G K, Rane S, Small P M. Science. 1999;284:1520–1523. doi: 10.1126/science.284.5419.1520. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.