Abstract
Patterns of nucleotide sequence diversity in the predominantly self-fertilizing species Hordeum vulgare subspecies spontaneum (wild barley) are compared between the putative alcohol dehydrogenase 3 locus (denoted “adh3”) and alcohol dehydrogenase 1 (adh1), two related but unlinked loci. The data consist of a sequence sample of 1,873 bp of “adh3” drawn from 25 accessions that span the species range. There were 104 polymorphic sites in the sequenced region of “adh3.” The data reveal a strong geographic pattern of diversity at “adh3” despite geographic uniformity at adh1. Moreover, levels of nucleotide sequence diversity differ by nearly an order of magnitude between the two loci. Genealogical analysis resolved two distinct clusters of “adh3” alleles (dimorphic sequence types) that coalesce roughly 3 million years ago. One type consists of accessions from the Middle East, and the other consists of accessions predominantly from the Near East. The two “adh3” sequence types are characterized by a high level of differentiation between clusters (≈2.2%), which induces an overall excess of intermediate frequency variants in the pooled sample. Finally, there is evidence of intralocus recombination in the “adh3” data, despite the high level of self-fertilization characteristic of wild barley.
The development of coalescence theory in population genetics has provided a powerful theoretical construct for the analysis of the distribution of mutational variation at single genetic loci (1, 2). Coalescence theory combined with the technology to sample relatively large numbers of gene sequences drawn from single genetic loci, has allowed population geneticists to draw inferences about the historical forces that have shaped the distribution of variation at a locus. Moreover, a number of statistical tools are available to test for selection on gene sequences (3). A particularly informative approach is to compare sequence samples drawn from different loci within the same genome. As a first approximation, the statistical descriptors of variation are expected to be homogeneous over loci within a genome for neutral loci. Sources of heterogeneity among loci should arise from differences in patterns of selection at the target loci themselves or at nearby loci (3).
The goal of this article is to examine the correlation in nucleotide sequence diversity between functionally similar genes in the predominantly self-fertilizing species Hordeum vulgare subspecies spontaneum, or wild barley. Wild barley is an annual diploid species with seven pairs of chromosomes and is the progenitor of cultivated barley, H. vulgare subspecies vulgare. Its natural distribution ranges from southwestern Asia through the Middle East to the Mediterranean area (4). The average outcrossing rate of wild barley was estimated to be 1.6% based on isozyme surveys of samples throughout the native range of the species (5).
This study focuses on the genes coding for alcohol dehydrogenase (adh; EC 1.1.1.1) because many molecular population genetic studies in plants have concentrated on adh genes, providing a strong context of comparative data (6–11). In addition, alcohol dehydrogenase is an important enzyme in anaerobic metabolism and is usually encoded by a small multigene family in higher plants, so we can study the behavior of duplicate genes within the same genome. Two or three adh loci are commonly found in grass genomes where adh1 and adh2 duplicated shortly before the origin of the grass family, approximately 70 million years ago (12, 13). The barley genome has three functional adh loci (14) and possibly several pseudogenes or related alcohol dehydrogenases that no longer use ethanol as a substrate (15). Barley adh3 duplicated from adh2 approximately 16 million years ago within the barley lineage (16). The more ancient adh1 and adh2 genes are closely linked on chromosome 4 (17, 18), whereas the more recently derived adh3 is freely recombining with adh1 and adh2 (14, 19, 20).
Trick et al. (16) cloned three adh genes from a genomic library of barley. They established that one of these genes is homologous to maize adh1, thus confirming this gene as barley adh1. The identities of the two maize-adh2-like genes in barley are questionable. Our analysis of DNA sequences from wild barley lines with known allozyme phenotypes (14) revealed that a frameshift at the locus called adh2 by Trick et al. (16) corresponds to a null adh3 allozyme allele (unpublished data), suggesting that this gene actually represents the adh3 isozyme locus. Thus, the two maize-adh2-like genes in barley may have been mislabeled. The sequences of the adh2 gene of Trick et al. (16) will be studied in the text. We shall denote this gene “adh3” where the quotation marks indicate some uncertainty about the actual locus designation.
Cummings and Clegg (10) studied nucleotide sequence diversity at the adh1 locus for a sample of 45 accessions of wild barley drawn from throughout the species range. They found a substantial excess of singletons at nonsynonymous sites in a region of 1,362 bp at adh1, which appears to be consistent with the background selection model (21). They concluded that the distribution of amino acid replacement polymorphism at adh1 was consistent with a weak selection/mutation balance. There was no evidence for geographic substructuring or intragenic recombination at adh. In this article, we contrast the adh1 sample to data based on an “adh3” sequence sample of 1,873 bp drawn from 25 accessions that derive from the same lineages studied by Cummings and Clegg (10). The data reveal a remarkable pattern of heterogeneity between these two loci with respect to geographic distributions, levels of nucleotide sequence diversity, and associated test statistics. Unlike adh1, there is evidence of intralocus recombination within the “adh3” sample.
Materials and Methods
Plant Materials.
Seeds of wild barley were obtained from the U.S. Department of Agriculture National Small Grains Collection (Aberdeen, ID). The sample includes 25 accessions (Table 1) selected at random from the 45 accessions in the adh1 sample (10). Seeds were germinated and grown in a growth chamber with 14 h of light at 18°C and 10 h of darkness at 10°C. Leaf material from each accession was ground in liquid nitrogen in a 1.5-ml microfuge tube, and DNA was isolated by using an SDS buffer (50 mM Tris⋅HCl, pH 8.0/20 mM EDTA, pH 8.0/0.3 M NaCl/2% sarcosyl/0.5% SDS) following a standard protocol (22).
Table 1.
Accessions of H. vulgare subspecies spontaneum used in this study
| Accession no.* | PI no.† | Country of origin |
|---|---|---|
| 2 | 212305 | Afghanistan |
| 3 | 212306 | Afghanistan |
| 4 | 219796 | Iraq |
| 6 | 220523 | Afghanistan |
| 9 | 236388 | Syria |
| 10 | 253933 | Iraq |
| 11 | 254894 | Iraq |
| 12 | 268242 | Iran |
| 13 | 293402 | Turkmenistan |
| 16 | 293409 | Turkmenistan |
| 17 | 293411 | Tajikistan |
| 21 | 296926 | Israel |
| 22 | 366446 | Afghanistan |
| 24 | 401370 | Iran |
| 25 | 401371 | Iran |
| 27 | 406276 | Israel |
| 28 | 420911 | Jordan |
| 30 | 420913 | Jordan |
| 32 | 420916 | Jordan |
| 35 | 466460 | Israel |
| 36 | 531851 | Israel |
| 38 | 531853 | Israel |
| 39 | 531857 | Israel |
| 43 | 559556 | Turkey |
| 44 | 560559 | Turkey |
Accession no. in the order of adh1 sample (10).
National Plant Germplasm System Plant Introduction Number.
PCR and Sequencing.
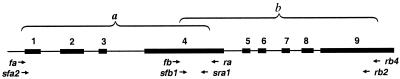
PCR primers were designed based on the published barley adh2 sequence of Trick et al. (16) that we now conclude to correspond actually to the adh3 isozyme locus. The gene was amplified as two segments, a and b, which overlap in exon 4 and begin in the 5′ flanking sequences and extend into exon 9 (Fig. 1). Templates for DNA sequencing were generated by using a two-step nested primer amplification procedure. Segment a was first amplified with primers fa (5′-GTGACCGGGAAAAAGAAGAA-3′) and ra (5′-GATACCACAGCTAAGGAGGCAGA-3′). The resulting PCR products were then used as templates to reamplify this region by using two nested primers, sfa2 (5′-AAGAAGAAACAGCA GGGGAGAT-3′) and sra1 (5′-CTTGGCGACGCACCCGACA-3′). Segment b was initially amplified with primers fb (5′-GTCGACCGTGGCGTGATGATT-3′) and rb4 (5′-GCAGGCTGTGGGTGATGAACTTGT-3′), then reamplified with primers sfb1 (5′-CAGTCCCGCTTCACCATC-3′) and rb2 (5′-ATGTCGACGACGCCGGGGAG-3′). PCR amplification was performed with an initial denaturing of 3 min at 94°C followed by 35 cycles of 45 s at 94°C, 1 min at 45–55°C (depending on the primers' Tm values), and 1 min at 72°C and ending with a 10-min extension at 72°C.
Figure 1.
Gene structure of “adh3” in barley. PCR primers are indicated by arrows.
The PCR products were purified by using a PCR purification kit (Qiagen, Chatsworth, CA). Nucleotide sequences were determined by direct sequencing of PCR products on a Li-Cor 4200L sequencer (Lincoln, NE) with end-labeled internal primers. All samples were sequenced at least twice on both strands, thus providing a minimum of 4-fold coverage for all sequenced regions.
Sequence Analysis.
The four replicates of each sequence were aligned to find the consensus by using sequencher (Gene Codes, Ann Arbor, MI), and any discrepancy was removed by resequencing. Polymorphism analysis and coalescent estimation of intragenic recombination were carried out on the consensus sequences of each of the 25 accessions by using the program sites (23). Tests of neutrality and determination of their associated significance levels were done by using the programs of Fu (24), and genealogical analysis was done by using paup* (25).
Results
Nucleotide Sequence Polymorphism.
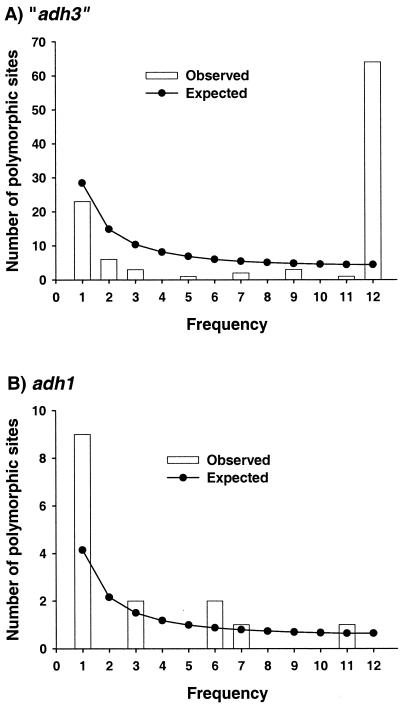
The “adh3” region sequenced was 1,873 bp, including 929 bp of noncoding sequence and 944 bp of coding sequence (≈82% of the total coding sequences). The distributions of polymorphic sites in the entire sequenced region and in various partitions (intron, exon, replacement, and synonymous sites) are summarized in Table 2. For the whole sample, mutations at 65 of the 104 polymorphic sites in the entire sequenced region occurred 12 times in the sample. This frequency is significantly higher than that expected under the neutral mutation model (ref. 26; Fig. 2A). The excess of intermediate frequency alleles at “adh3” was also observed when the data were partitioned into exon and intron sites.
Table 2.
Number of polymorphic sites at various frequencies in regions of “adh3” and adh1 in wild barley
| Gene | Region | Total* | Number of
polymorphic sites at frequency of
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |||
| “adh3” whole sample | ||||||||||||||
| (n = 25 accessions) | ||||||||||||||
| All | 104 | 23 | 6 | 3 | 0 | 1 | 0 | 2 | 0 | 3 | 0 | 1 | 65 | |
| Intron | 56 | 7 | 4 | 2 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 41 | |
| Exon | 48 | 16 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 3 | 0 | 1 | 24 | |
| Replacement | 22 | 9 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 7 | |
| Synonymous | 26 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 17 | |
| “adh3” cluster 1 | ||||||||||||||
| (n = 13 accessions) | ||||||||||||||
| All | 28 | 22 | 4 | 0 | 0 | 1 | 1 | |||||||
| Intron | 11 | 7 | 2 | 0 | 0 | 1 | 1 | |||||||
| Exon | 17 | 15 | 2 | |||||||||||
| Replacement | 11 | 9 | 2 | |||||||||||
| Synonymous | 6 | 6 | ||||||||||||
| “adh3” cluster 2 | ||||||||||||||
| (n = 12 accessions) | ||||||||||||||
| All | 13 | 3 | 3 | 6 | 0 | 0 | 1 | |||||||
| Intron | 5 | 1 | 2 | 2 | ||||||||||
| Exon | 8 | 2 | 1 | 4 | 0 | 0 | 1 | |||||||
| Replacement | 5 | 1 | 1 | 2 | 0 | 0 | 1 | |||||||
| Synonymous | 3 | 1 | 0 | 2 | ||||||||||
| adh1 | ||||||||||||||
| (n = 25 accessions)† | ||||||||||||||
| All | 15 | 9 | 0 | 2 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 1 | ||
| Intron | 7 | 3 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | ||
| Exon | 8 | 6 | 0 | 0 | 0 | 0 | 1 | 1 | ||||||
| Replacement | 5 | 5 | ||||||||||||
| Synonymous | 3 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | ||||||
Total number of polymorphic sites.
adh1 data from ref. 10.
Figure 2.
Expected number of polymorphic sites under a neutral model calculated by using equation 51 of Tajima (26) and observed number of polymorphic sites at various frequencies in the whole sample and in all regions of “adh3” (A) and adh1 (B) in wild barley.
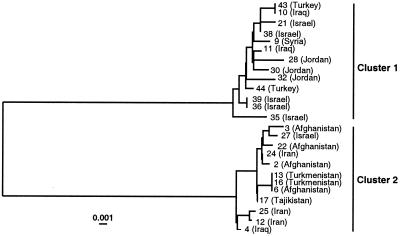
Genealogical analysis resolved two distinct clusters of sequences (Fig. 3). The two clusters are of nearly equal frequency with 13 accessions in cluster 1 and 12 accessions in cluster 2. Cluster 1 consists of accessions from the Near East (Israel, Jordan, Turkey, Syria, and Iraq), whereas cluster 2 consists of accessions mostly from the Middle East (Iran, Afghanistan, Turkmenistan, and Tajikistan), except for accession 27 from Israel and accession 4 from Iraq. Three accessions (accession 4 from Iraq and accessions 12 and 25 from Iran) within cluster 2 have been shown to be recombinants between the two sequence types (see below). Thus, there appears to be an extreme pattern of geographic differentiation, with one sequence type distributed in the Near East in the Mediterranean habitats and the other distributed in the Middle East in the high desert habitat, and Iraq and Iran appear to be a contact zone.
Figure 3.
Neighbor-joining tree depicting the genealogical relationships of alleles of “adh3” in wild barley. Country of origin is shown in parentheses. Scale indicates percentage of variation.
Numbers of polymorphic sites within clusters are considerably smaller than those observed between the two major clusters (Table 2). The number of unique polymorphic sites within cluster 1 was substantially greater than expected under a neutral model, whereas no excess of unique polymorphisms was observed among the 13 polymorphic sites in cluster 2.
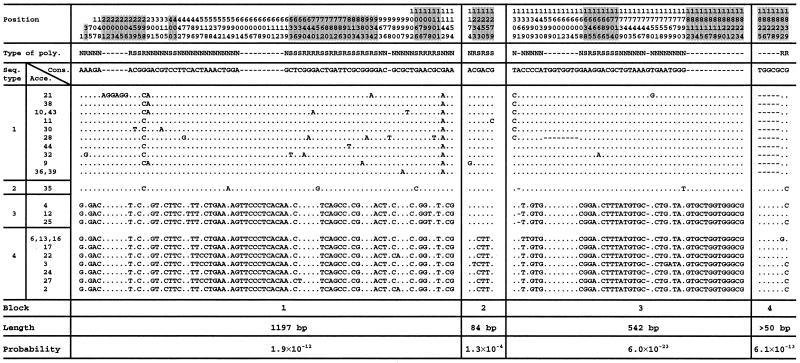
There were eight insertion/deletion events in the sequenced region of “adh3” (Fig. 4). A 10-bp deletion in intron 3 and an 18-bp deletion in exon 9 occurred only in cluster 1 sequences except for accession 35 for which a 5-bp insertion in exon 9 appears to be a recombination product from cluster 2 sequences (Fig. 4). As a result, cluster 1 sequences are shorter than cluster 2 sequences. Translation of the accession 35 sequence indicated that the 5-bp frameshift insertion has resulted in an early termination of the gene (data not shown). Thus, accession 35 could be a null allele.
Figure 4.
Polymorphic sites in the “adh3” region among 25 accessions of wild barley. Sequences are divided into four types with four blocks of sequences of different origins. Boundary point between two blocks was determined as the mid-point between two adjacent polymorphisms belonging to the two blocks. Sites in exons are highlighted. Type of polymorphism: N, noncoding; S, synonymous; R, replacement. Dash denotes deletion, and dot denotes consensus sequence.
The distribution of polymorphic sites at adh1 is in stark contrast to that at “adh3.” Only 15 polymorphic sites were detected at adh1 for the same accessions, and there was a substantial excess of rare alleles, particularly at nonsynonymous sites (Table 2 and Fig. 2B). Moreover, there was no evident geographic substructuring for adh1 alleles.
Estimates of Nucleotide Diversity and Statistical Tests for Natural Selection.
Nucleotide diversity for the whole sequenced “adh3” region was estimated by Tajima's π (26) and Watterson's θ (27) statistics to be 0.0219/bp and 0.0149/bp, respectively, for the whole sample (Table 3). There was a higher diversity in the intron region (π = 0.0257/bp) than in the exon region (π = 0.0179/bp). The several test statistics (T, D*, F, and F*) for deviation from a neutral distribution were not significant, except for Tajima's test, where a marginally significant T in the whole region was accounted for by a highly significant T in the intron region and a marginally significant T at the synonymous sites.
Table 3.
Estimates of nucleotide diversity and test statistics for selection at “adh3” and adh1 in wild barley (length of the regions is the average number of bases excluding alignment gaps for all lines)
| Gene | Region | Length, bp | θ/bp | π/bp | T | D* | F | F* |
|---|---|---|---|---|---|---|---|---|
| “adh3” whole sample | ||||||||
| (n = 25 accessions) | ||||||||
| All | 1,842.8 | 0.0149 | 0.0219 | 1.734† | 0.348 | 0.931 | 0.858 | |
| Intron | 921.9 | 0.0161 | 0.0257 | 2.183‡ | 0.929 | 1.599 | 1.437 | |
| Exon | 929.1 | 0.0137 | 0.0179 | 1.127 | −0.350 | 0.073 | 0.127 | |
| Rep. | 715.0 | 0.0082 | 0.0088 | 0.279 | −0.746 | −0.588 | −0.462 | |
| Syn. | 212.1 | 0.0325 | 0.0488 | 1.759† | 0.038 | 0.636 | 0.621 | |
| “adh3” cluster 1 | ||||||||
| (n = 13 accessions) | ||||||||
| All | 1,836.6 | 0.0049 | 0.0030 | −1.544† | −1.843† | −2.367‡ | −1.835† | |
| Intron | 917.4 | 0.0039 | 0.0029 | −0.890 | −1.096 | −1.378 | −1.083 | |
| Exon | 920.8 | 0.0059 | 0.0031 | −1.847‡ | −2.153† | −2.706‡ | −2.154† | |
| Rep. | 703.8 | 0.0050 | 0.0028 | −1.666† | −1.790† | −2.256‡ | −1.826† | |
| Syn. | 212.4 | 0.0091 | 0.0043 | −1.755‡ | −2.221‡ | −2.585‡ | −2.178‡ | |
| “adh3” cluster 2 | ||||||||
| (n = 12 accessions) | ||||||||
| All | 1,864.7 | 0.0023 | 0.0024 | 0.094 | 0.492 | 0.385 | 0.402 | |
| Intron | 926.8 | 0.0018 | 0.0017 | −0.130 | 0.513 | 0.338 | 0.359 | |
| Exon | 938.0 | 0.0028 | 0.0030 | 0.232 | 0.390 | 0.330 | 0.361 | |
| Rep. | 721.8 | 0.0023 | 0.0025 | 0.357 | 0.513 | 0.476 | 0.489 | |
| Syn. | 217.2 | 0.0046 | 0.0045 | −0.025 | 0.078 | −0.012 | 0.053 | |
| adh1 | ||||||||
| (n = 25 accessions)§ | ||||||||
| All | 1,359.9 | 0.0029 | 0.0021 | −0.926 | −1.705 | −1.828† | −1.582 | |
| Intron | 574.2 | 0.0032 | 0.0027 | −0.436 | −0.674 | −0.747 | −0.648 | |
| Exon | 786.0 | 0.0027 | 0.0016 | −1.174 | −2.169† | −2.249† | −2.013† | |
| Rep. | 606.4 | 0.0022 | 0.0007 | −1.858‡ | −2.890‡ | −3.022‡ | −2.776‡ | |
| Syn. | 179.6 | 0.0044 | 0.0049 | 0.247 | −0.192 | −0.109 | −0.071 |
θ, Watterson's estimate; π, Tajima's estimate; T, Tajima's D test; D*, Fu and Li's D test without outgroup information; F, Fu and Li's F test; F*, Fu and Li's F* test.
0.01 < P < 0.05.
P < 0.01.
adh1 data from ref. 10.
Nucleotide diversity within clusters was substantially lower than that in the whole sample. Diversity in cluster 1 was similar in introns, exons, and overall (π = 0.0029/bp, 0.0031/bp, and 0.0030/bp, respectively). Statistical tests for deviation from a neutral distribution were marginally to highly significant for the whole regions in cluster 1, and when the data were partitioned, the tests were significant for exons but not for introns. In addition, the test statistics were negative, indicating an excess of rare variants.
The patterns of nucleotide diversity in cluster 2 differ greatly from cluster 1 in that there was a higher diversity in exons (π = 0.0030/bp) than in introns (π = 0.0017/bp) and that the test statistics were largely positive but nonsignificant in cluster 2.
The patterns of diversity at adh1 are strikingly different from “adh3” (Table 3 and ref. 10). Nucleotide diversity was only about one-tenth (π = 0.0021/bp) the overall “adh3” diversity. Within the exon region, diversity based on synonymous sites was substantially higher than that based on nonsynonymous sites. Statistical tests for selection were highly significant for the replacement sites but were not significant for either the introns or synonymous sites (see ref. 10 for details).
Tests for Intragenic Recombination and Linkage Disequilibrium.
Based on the distribution of polymorphic sites, the “adh3” region was partitioned into four blocks as described (9, 28): block 1 between nucleotide positions 1 and 1197; block 2 between 1198 and 1282; block 3 between 1283 and 1824; and block 4 between 1824 and 1873 (Fig. 4). Based on these partitions, the sequences can be divided into four types. Type 1 sequence consists mostly of the consensus nucleotides, whereas type 4 consists of the alternative bases. Type 2 sequence is of the same type as type 1 sequence in blocks 1, 2, and 3 but is of type 4 in block 4. Type 3 sequence is of type 4 in blocks 1, 3, and 4, but of type 1 in block 2 (Fig. 4). Statistical tests for clustering (28) in these blocks were highly significant, suggesting that sequences in blocks 4 and 3 in sequence types 2 and 3, respectively, resulted from recombination. Within each block, most polymorphic sites are segregating for two nucleotides in intermediate frequency (Fig. 4).
Most of the polymorphic sites within the “adh3” region exhibit highly significant linkage disequilibrium (test results not shown) owing to the extreme differentiation between clusters. In contrast, there was no significant linkage disequilibrium between polymorphic sites of “adh3” and adh1.
The “adh3” Sample Data Map to a Single Genetic Locus.
An important issue to resolve is whether sample data map to a single genetic locus. As noted above, several putative adh genes have been reported in the barley genome (15), and it is possible that the sample includes sequences from more than one locus. Moreover, the contrasting patterns of variation between clusters are consistent with the hypothesis that there are two “adh3”-like genes.
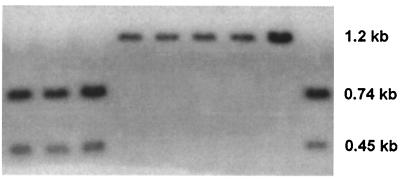
The hypothesis of two “adh3”-like genes can be tested by asking whether unique restriction sites that are associated with each haplotype are simultaneously present in Southern blots of single genomes. Accordingly, about 5 μg of genomic DNA was digested with the restriction enzyme BclI, for which a restriction site at nucleotide position 769 is present in cluster 2 sequences but absent in cluster 1 sequences. This restriction site can thus be used to determine whether both sequences are present in the genome. Probing the genomic blot with a 32P-labeled fa-ra PCR product, we would expect one band of ≈1.2 kb in cluster 1 sequences and two bands of ≈0.74 kb and ≈0.45 kb, respectively, in cluster 2 sequences. If both sequence types were present in the genome (i.e., if there are two loci), we would expect all three bands to be present in all accessions. The Southern blot results showed either one or two bands, but not three bands, present in any of the accessions tested (Fig. 5), suggesting that the sequences in this sample were from one locus. In addition, the geographic distribution of alleles further supports the one locus interpretation.
Figure 5.
Southern blot of genomic DNA from nine wild barley accessions digested with BclI and probed with a 32P-labeled fa-ra PCR product. Approximate sizes of the fragments are shown.
Discussion
The salient facts of this investigation are (i) the “adh3” sample is characterized by high levels of nucleotide polymorphism and an excess of intermediate frequency variants; (ii) a natural partition of the “adh3” sample into two clusters (clusters 1 and 2) with high linkage disequilibrium between clusters accounts for the excess of intermediate frequency variants; (iii) the two clusters are geographically distinct; (iv) test statistics for deviation from a drift/mutation model within cluster 1 are significantly negative, indicating an excess of rare variants within this cluster; (v) test statistics are nonsignificant and positive within cluster 2, suggesting a weak excess of intermediate frequency variants; (vi) there is clear evidence for interallelic recombination between clusters; and (vii) the distribution and level of polymorphism at “adh3” contrasts sharply with that observed at the adh1 locus.
It is important to note that the genealogical pattern observed at “adh3” is not unprecedented; two other studies in the highly self-fertilizing Arabidopsis thaliana (9, 29) have revealed patterns of variation similar to those observed in this study. Both studies were based on a sample of 17 ecotypes and two randomly selected loci, adh and acidic chitinase (chiA). A large number of polymorphic sites were identified between two major haplotypes, each in intermediate frequency at both loci. Intragenic recombination also appeared to have been important in generating allelic diversity within loci. Levels of polymorphism within each parental haplotype were considerably lower than the level of variation between haplotypes. This pattern was hypothesized to be a result of introgression of divergent lineages (9, 29).
An excess of intermediate frequency polymorphism and a deep coalescent are often viewed as evidence for balancing selection. However, simple heterozygous advantage is unlikely to be important in a highly self-fertilizing species like wild barley unless selection against homozygotes is symmetrical (30, 31). Even with symmetric overdominant selection, the gene tree will be similar to the topology of a neutral locus, although the time of the coalescent could be quite deep (32). In the present case, the ratio between coalescent time for the entire sample and the appearance of the next bifurcation is greater than seven, which is much larger than expected under neutral theory (see below and Fig. 3). This high ratio appears inconsistent with the predictions of a balancing selection model. Moreover, the distinct geographic distribution of the two clusters is not consistent with simple overdominant or frequency-dependent models of selection. A distinct geographic distribution is consistent with diversifying selection due to ecotypic adaptation to two different geographical regions/habitats. However, it is hard to reconcile this hypothesis with the facts that the adh3 gene is expressed at low levels and appears to have polymorphic null alleles.
Introgression at “adh3” Best Accounts for the Observations.
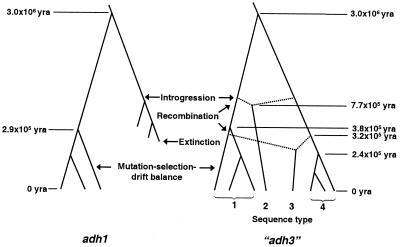
The scenario that appears to be most consistent with the data is the historical mixing of two long separated populations followed by a gradual increase in the frequency of one haplotype. It is possible to quantify this scenario by using some simple evolutionary calculations. Based on a mutation rate of 3.5 × 10−9 site/year for “adh3,” which is the average of mutation rates for synonymous and nonsynonymous sites for adh genes in grasses (12), the coalescent time of the two parental sequence types 1 and 4 can be calculated to be roughly 3.0 × 106 years and within types 1 and 4 to be 3.8 × 105 years and 2.4 × 105 years, respectively. The time of the recombination events can be estimated by using the average number of pairwise differences between the recombinant and parental types within each block (9). In the case of type 2 sequences, there are 111 differences between types 1 and 2 in blocks 1, 2, and 3 of 12 possible comparisons, and there are 8 differences between types 2 and 4 in block 4 of 9 possible comparisons (Fig. 4). The average number of pairwise differences per site between recombinant sequence type 2 and its parental sequence types 1 and 4 is d2 = (111/12 + 8/9)/1,873 bp = 0.0054/bp. Thus, the time of the recombination event leading to sequence type 2 is T2 ≈ 0.0054/(2 × 3.5 × 10−9) ≈ 7.7 × 105 years. Similarly, in the case of type 3 sequence, there are 54, 30, and 23 differences between types 3 and 4 in blocks 1, 3, and 4, respectively, of 27 possible comparisons, and there are 6 differences between types 3 and 1 in block 2 of 36 possible comparisons. Thus d3 = (54/27 + 6/36 + 30/27 + 23/27)/1,873 bp = 0.0022/bp, and T3 ≈ 0.0022/(2 × 3.5 × 10−9) ≈ 3.2 × 105 years.
Under the introgression hypothesis, the putative evolutionary history of wild barley could be as follows (Fig. 6): two divergent lineages of wild barley that had been separated for ≈3 million years came into contact, and introgression began ≈800 thousand years ago, probably in the Iraq–Iran region. A weak mutation-selection-drift balance has subsequently maintained nucleotide sequence variation at adh1 and “adh3” loci. A selection/mutation balance appears to be consistent with the cluster 1 test statistics, and it is consistent with the adh1 pattern. Cluster 2 of “adh3” is intriguing. The weak positive test statistics lead us to speculate that cluster 2 may be experiencing a demographic contraction and that the cluster 1 type may ultimately replace it. These data and interpretations are consistent with recent studies of barley domestication using AFLP loci and sequence data from a homeobox gene, BKn-3 (33). The geographic pattern of variation at the BKn-3 locus parallels the pattern reported here.
Figure 6.
Hypothetical evolutionary history of wild barley based on patterns of sequence variation at adh1 and “adh3.” Estimates of coalescent time are indicated. yra, years ago.
Under the introgression hypothesis, most loci in the genome are expected to show a similar pattern of variation, so how do we account for adh1? A plausible hypothesis is that one adh1 haplotype has replaced the other introgressant within the past 800 thousand years because it is better adapted. Adh1 makes up the majority of the adh protein (14, 34) and is likely to be a more important locus than “adh3” for the plant. Indeed, it is possible, as argued above, that “adh3” is also experiencing a selective replacement but on a much slower time scale. Thus, the ultimate patterns at these loci could become quite similar.
Acknowledgments
We thank B. Gaut, M. Cummings, N. Ellstrand, M. Durbin, P. Butcher, M. Ellis, and J. Miller for comments on the manuscript. We also thank M. Durbin, M. Kobayashi, B. McCaig, V. Oberholzer, G. Waines, and A. Denton for discussion and technical assistance. This project was supported in part by funds from the Alfred P. Sloan Foundation.
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF326691–AF326715).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.011537898.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.011537898
References
- 1.Kingman J F C. Stochastic Processes: Formalism. Appl Proc Winter Sch. 1982;13:235–248. [Google Scholar]
- 2.Hudson R. Oxford Surv Evol Biol. 1990;7:1–44. [Google Scholar]
- 3.Clegg M T. Heredity. 1997;88:1–7. doi: 10.1093/oxfordjournals.jhered.a023048. [DOI] [PubMed] [Google Scholar]
- 4.von Bothmer R, Jacobsen N, Baden C, Jorgensen R B, Linde-Laursen I. An Ecogeographical Study of the Genus Hordeum. 2nd Ed. Rome: International Plant Genetic Resources Institute; 1995. [Google Scholar]
- 5.Brown A H D, Zohary D, Nevo E. Heredity. 1978;41:49–62. [Google Scholar]
- 6.Gaut B S, Clegg M T. Proc Natl Acad Sci USA. 1993;90:5095–5099. doi: 10.1073/pnas.90.11.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaut B S, Clegg M T. Genetics. 1993;135:1091–1097. doi: 10.1093/genetics/135.4.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanfstingl U, Berry A, Kellog E A, Costa J T, III, Rüdiger W, Ausubel F M. Genetics. 1994;138:811–828. doi: 10.1093/genetics/138.3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Innan H, Tajima F, Terauchi R, Miyashita N T. Genetics. 1996;143:1761–1770. doi: 10.1093/genetics/143.4.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cummings M P, Clegg M T. Proc Natl Acad Sci USA. 1998;95:5637–5642. doi: 10.1073/pnas.95.10.5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charlesworth D, Liu F, Zhang L. Mol Biol Evol. 1998;15:552–559. doi: 10.1093/oxfordjournals.molbev.a025955. [DOI] [PubMed] [Google Scholar]
- 12.Gaut B S, Morton B R, McCaig B C, Clegg M T. Proc Natl Acad Sci USA. 1996;93:10274–10279. doi: 10.1073/pnas.93.19.10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaut B S, Peek A S, Morton B R, Clegg M T. Mol Biol Evol. 1999;16:1086–1097. doi: 10.1093/oxfordjournals.molbev.a026198. [DOI] [PubMed] [Google Scholar]
- 14.Hanson A D, Brown A H D. Biochem Genet. 1984;22:495–515. doi: 10.1007/BF00484519. [DOI] [PubMed] [Google Scholar]
- 15.Kleinhofs A, Kilian A, Maroof M A S, Biyashev R M, Hayes P, Chen F Q, Lapitan N, Fenwick A, Blake T K, Kanazin V, et al. Theor Appl Genet. 1993;86:705–712. doi: 10.1007/BF00222660. [DOI] [PubMed] [Google Scholar]
- 16.Trick M, Dennis E S, Edwards K J R, Peacock W J. Plant Mol Biol. 1988;11:147–160. doi: 10.1007/BF00015667. [DOI] [PubMed] [Google Scholar]
- 17.Brown A H D. J Hered. 1980;70:127–128. [Google Scholar]
- 18.Hart G E, Islam A K M R, Shepherd K W. Genet Res. 1980;36:311–325. [Google Scholar]
- 19.Harberd N P, Edwards K J R. Genet Res. 1983;41:109–116. [Google Scholar]
- 20.Brown A H D, Lawrence G J, Jenkin M, Douglass J, Gregory E. J Hered. 1989;80:234–239. [Google Scholar]
- 21.Charlesworth B, Morgan M T, Charlesworth D. Genetics. 1993;134:1289–1303. doi: 10.1093/genetics/134.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murray M G, Thompson W F. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hey J, Wakeley J. Genetics. 1997;145:833–846. doi: 10.1093/genetics/145.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu Y-X. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swofford D L. paup*, Phylogenetic Analysis Using Parsimony. Sunderland, MA: Sinauer Associates; 1999. , Version 4.0. [Google Scholar]
- 26.Tajima F. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watterson G A. Theor Popul Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- 28.Stephens J C. Mol Biol Evol. 1985;2:539–556. doi: 10.1093/oxfordjournals.molbev.a040371. [DOI] [PubMed] [Google Scholar]
- 29.Kawabe A, Innan H, Terauchi R, Miyashita N T. Mol Biol Evol. 1997;14:1303–1315. doi: 10.1093/oxfordjournals.molbev.a025740. [DOI] [PubMed] [Google Scholar]
- 30.Hayman B I. Heredity. 1953;7:185–192. [Google Scholar]
- 31.Kimura M, Ohta T. Theoretical Topics in Population Genetics. Princeton: Princeton Univ. Press; 1971. [Google Scholar]
- 32.Clark A G. Proc Natl Acad Sci USA. 1997;94:7730–7734. doi: 10.1073/pnas.94.15.7730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Badr A, Muller K, Schafer-Pregl R, El Rabey H, Efgen S, Ibrahim H H, Pozzi C, Rohde W, Salamini F. Mol Biol Evol. 2000;17:499–510. doi: 10.1093/oxfordjournals.molbev.a026330. [DOI] [PubMed] [Google Scholar]
- 34.Hanson A D, Jacobsen J V, Zwar J A. Plant Physiol. 1984;75:573–581. doi: 10.1104/pp.75.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]