Abstract
T lymphocytes and B lymphocytes are present in jawed vertebrates, including cartilaginous fishes, but not in jawless vertebrates or invertebrates. The origins of these lineages may be understood in terms of evolutionary changes in the structure and regulation of transcription factors that control lymphocyte development, such as PU.1. The identification and characterization of three members of the PU.1 family of transcription factors in a cartilaginous fish, Raja eglanteria, are described here. Two of these genes are orthologs of mammalian PU.1 and Spi-C, respectively, whereas the third gene, Spi-D, is a different family member. In addition, a PU.1-like gene has been identified in a jawless vertebrate, Petromyzon marinus (sea lamprey). Both DNA-binding and transactivation domains are highly conserved between mammalian and skate PU.1, in marked contrast to lamprey Spi, in which similarity is evident only in the DNA-binding domain. Phylogenetic analysis of sequence data suggests that the appearance of Spi-C may predate the divergence of the jawed and jawless vertebrates and that Spi-D arose before the divergence of the cartilaginous fish from the lineage leading to the mammals. The tissue-specific expression patterns of skate PU.1 and Spi-C suggest that these genes share regulatory as well as structural properties with their mammalian orthologs.
Lymphocytes, as defined by expression of the rearranging antigen receptor (Ig) or T cell receptor genes, are found in all of the major classes of jawed vertebrates including the cartilaginous fishes. Although cells with lymphocyte-like morphology exist in jawless vertebrates such as the lamprey (reviewed in refs. 1 and 2), numerous attempts to identify Ig and T cell receptor genes in these animals have been unsuccessful (3, 4). Until other molecular criteria can be applied, the question will remain open as to whether or not the lymphocyte-like cells in jawless vertebrates are homologous to lymphocytes in jawed vertebrates. The failure to identify cells with lymphocyte-like morphology in other deuterostomes such as ascidians and echinoderms restricts the timing of the emergence of lymphocytes to the base of the vertebrate lineage. This time frame also corresponds to the time at which two rounds of at least partial genome duplication are hypothesized to have occurred (5, 6). These duplication events presumably created a large pool of genes that were free to diverge and assume new functions and expression patterns. In support of this theory, vertebrate gene families often contain three to four members for each invertebrate member (5, 7, 8). It is possible that the duplication and divergence of certain key gene families in the vertebrate lineage facilitated the emergence of specialized cell types, such as lymphocytes. To test this theory in a gene family that may have been crucial for the emergence of lymphocytes, we have defined the phylogenetic distribution and expression patterns of the PU.1 transcription factor family.
The members of the PU.1 transcription factor family belong to a divergent subclass of the Ets transcription factor family. Whereas the Ets family is found throughout the metazoans, the PU.1 family members have been identified only in vertebrates (9–13). Three members of the PU.1 family have been described thus far, and their expression is largely restricted to hematopoietic cells and tissues. PU.1/Spi-1 itself is essential for normal development of T cells, B cells, macrophages, and granulocytes in the mouse (14, 15). Spi-B is expressed primarily in the B lineage (16) and is necessary for both B cell activation and germinal center formation (17). Spi-C/Prf, a very divergent member of the PU.1 family, is expressed in B cells but also at lower levels in macrophages (12, 18). The critical roles of these genes in lymphocyte development suggest that one or more of them should be present in all animals with lymphocytes. The role of PU.1 in myeloid development also suggests that its functions may predate the appearance of lymphocytes, as phagocytic cells are widespread throughout the metazoans. However, no PU.1 family member has been identified thus far using PCR (19) or sequence searches (data not shown) in representative nonvertebrate deuterostomes or protostomes, despite the large genomic databases that now exist for Drosophila and Caenorhabditis elegans.
Therefore, investigations into the evolutionary history of the PU.1 family, which may have arisen suddenly in the vertebrate lineage, may provide a better understanding of the origins of vertebrate hematopoiesis. Furthermore, the identification of PU.1 family homologs in jawless vertebrates and other deuterostomes will provide potential markers for identifying cells that share a common ancestor with hematopoietic stem cells in mammals. Toward these ends, we have initiated a search for members of the PU.1 family in a cartilaginous fish (skate) and a jawless vertebrate (lamprey). We report here the sequences and expression patterns of three PU.1 family members in the skate, one of which appears to be an additional family member not represented by a known ortholog in mammals.
Materials and Methods
Library Screening.
The complete coding sequence of mouse PU.1 was PCR-amplified from a full-length mouse cDNA. The gel-purified PCR product was radiolabeled and used to screen a clearnose skate (Raja eglanteria) spleen cDNA library constructed in the λZAPII vector (Stratagene) as described (20), under low stringency conditions. Positive phage cores were plaque-purified, and plasmids were in vivo-excised according to the manufacturer's instructions. Plasmid DNA was prepared by using Qiagen Spin Miniprep columns (Qiagen, Chatsworth, CA). EcoRI fragments containing the entire coding regions of skate PU.1 and Spi-C were obtained from plasmid DNA by restriction digest, gel-purified, and radiolabeled as above. Mouse PU.1 and skate PU.1 and Spi-C probes were used to screen a sea lamprey (Petromyzon marinus) protovertebral arch cDNA library constructed in λgt11 (Stratagene). cDNA inserts were subcloned from λDNA into the pBluescript vector (Stratagene).
Sequence Analysis.
DNA sequencing was performed by using the ABI Prism Dye Terminator Cycle Sequencing Ready Reaction Kit (Perkin–Elmer). Sequence analysis was performed by using the dnasis software package (Hitachi, San Francisco, CA). Alignments were generated by using the clustalw program (21). The neighbor-joining tree was generated by using the me tree program (22), using proportions of differences as a distance measure.
Real-Time PCR Expression Analysis.
One microgram of total RNA from each tissue was reverse-transcribed into cDNA using 5.5 mM MgCl2, 500 μM of each dNTP, 2.5 μM random hexamers, 0.4 unit/μl RNase inhibitor, and 1.25 units/μl MultiScribe reverse transcriptase (Perkin–Elmer). One microliter of each 100-μl reaction was used in each PCR with 10× SYBR green PCR buffer (25 mM MgCl2/2.5 mM dATP, dCTP, dGTP/5.0 mM dUTP/5 units per μl AmpliTaq Gold/300 nM of each primer). Reactions were run and analyzed by using the GeneAmp 5700 sequence detection system (Perkin–Elmer). Primer sequences were as follows (5′ to 3′): PU.1, GAAGCAAACGAGCACTCTGCT and GATTCCTACTGTGTCTGCGCC; Spi-C, CCCCCTCTCCAGTATCTCCG and GAGATTGTGCATGGAGTGCG; Spi-D, GGAGTCCGGAAGAAGGTTCG and CTTCAGCAGCTCCAGCAAGA. PCR products detected by SYBR green were compared with a standard curve for each primer combination generated using the appropriate plasmid DNA. To control for cDNA quality, each sample was amplified with 28S primers (23): AAGACCTCCTTTCTCCGACGA and TCGATCAGAAGGACTTGGGC. Cycle threshold values for positive samples ranged from 21 to 35 cycles as compared with no template control values of over 40 cycles. All PU.1 family member values were normalized to 28S levels.
Results
Three PU.1 Family Members Recovered from a Skate Spleen cDNA Library.
As a first step toward determining whether lymphocyte development in cartilaginous fish uses the same transcriptional networks that operate in mammalian lymphocytes, we screened a skate spleen cDNA library for homologs of PU.1 under low stringency conditions with a mouse PU.1 probe. Partial sequencing of 40 hybridizing clones indicated the presence of three PU.1-related classes of cDNA as defined by the DNA-binding domain. The longest representative cDNA of each of these classes was completely sequenced. As discussed in detail below, one of these classes most resembles PU.1, another most resembles Spi-C, and the third falls into a different class, which we refer to as Spi-D.
Phylogenetic Analysis of the Three PU.1 Subfamily Members in the Skate: PU.1, Spi-C, and Spi-D.
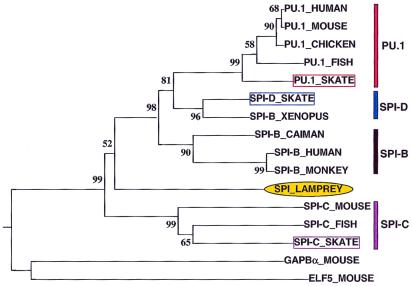
Phylogenetic analysis was performed to classify the three skate PU.1 family homologs in the context of the other known PU.1 family members (see Fig. 1 legend). An amino acid alignment of the Ets DNA-binding domains of representative vertebrate PU.1 family members and lamprey Spi was constructed. Mouse GABPα and Elf-5 sequences were used as an outgroup because their Ets domains are the most closely related to PU.1 outside the PU.1 family genes themselves. This alignment was used to build a neighbor-joining tree, shown in Fig. 1. The topology of the tree does not change when mouse Ets-1 is used as an outgroup or when the tree is constructed without an outgroup (data not shown). These trees confirm that one of the skate sequences is an ortholog of PU.1, another is an ortholog of Spi-C, and the third, Spi-D, represents a different member of the PU.1 family. The clustering of Xenopus Spi-B (13) with skate Spi-D indicates that this Xenopus gene is likely an ortholog of Spi-D. The placement of the Spi-D genes in the tree implies that Spi-D diverged from the other PU.1 family members before the divergence of the cartilaginous fish from the other vertebrate lineages. Furthermore, the presence of Spi-D in both a cartilaginous fish and an amphibian provides additional evidence that it may be present in other vertebrates, including mammals.
Figure 1.
Neighbor-joining tree of Ets DNA binding domain amino acid sequences of vertebrate PU.1 family transcription factors. This tree was constructed by using the me tree program, using proportions of differences as a distance measure and excluding gaps. This method finds a phylogenetic tree and statistically similar trees likely to represent a minimum total genetic difference (branch length) among the genes. The value at each node represents the probability that that branch length is not zero. GenBank accession nos.: mouse PU.1, L03215; human PU.1, X52056; chicken PU.1, Y12225; teleost fish PU.1, AF247366; Xenopus Spi-B, AF247365; caiman Spi-B, AF247364; human Spi-B, X66079; monkey Spi-B, AF025395; lamprey Spi, AF247362; mouse Spi-C, AF025395; fish Spi-C, AF247367; mouse GABPα, M74515; and mouse Elf-5, NM_010125.
All Known Functional Domains of Mouse PU.1 Are Conserved in Skate PU.1.
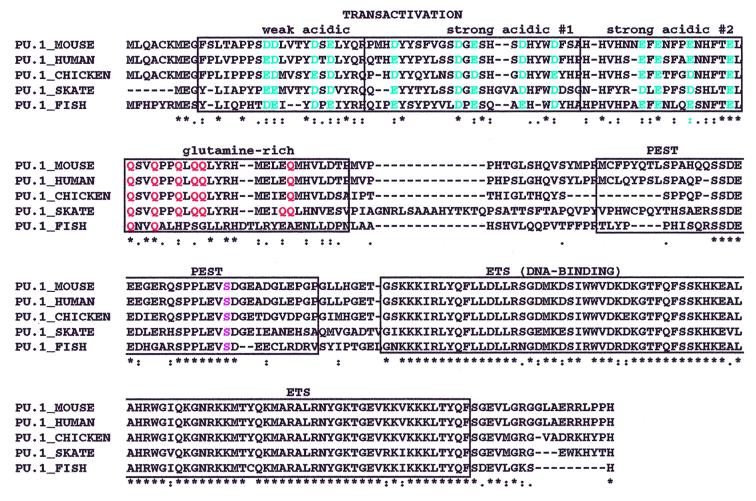
An alignment of vertebrate PU.1 genes is shown in Fig. 2. The entire coding sequence of skate PU.1 exhibits 61% overall amino acid identity with mouse PU.1. The Ets domains are exceptionally well conserved, with 92% identity at the amino acid level. Similarities also extend throughout the functional domains that have been rigorously mapped in murine PU.1 (reviewed in refs. 24 and 25). The N-terminal transactivation domain can be subdivided into a weakly acidic domain, two strongly acidic domains (SA1 and SA2), and a glutamine (Q)-rich region. The Q-rich region is especially critical for macrophage development (26, 27). All of the residues within these regions that are important in transactivation are conserved between mouse PU.1 and skate PU.1 (see Fig. 2). In the weakly acidic domain, four of four aspartic acids (D) and glutamic acids (E), which have been shown to be critical for in vitro transactivation function (28), are conserved as either D or E (blue in Fig. 2), although this region exhibits only 50% identity overall. The strongly acidic regions exhibit five of five and four of four conservation of D and E as either D or E. Likewise, all of the glutamines in the Q-rich region are conserved between mouse PU.1 and skate PU.1 (red in Fig. 2). This region is 75% identical overall with a 100% conserved core from residues 74–90.
Figure 2.
Amino acid alignments of vertebrate PU.1 genes. The predicted amino acid sequence of skate PU.1 is aligned with PU.1 sequences from mouse, human, chicken, and teleost fish by using clustalw (see Fig. 1 legend for GenBank accession nos.). Stars indicate identity in all sequences, and dots indicate conserved types of amino acids. The transactivation domain is divided into four functionally relevant subdomains indicated above the sequence. The colored amino acids have been shown to be necessary for mouse PU.1 transactivation activity; Ser-148 is pink.
The length of the linker region between the transactivation domain and the PEST domain differs between mouse PU.1 and skate PU.1 and exhibits very low identity (10%). However, a core portion of the PEST domain (residues 132–151) is highly conserved, exhibiting 85% amino acid identity. In PU.1, this domain is important for interaction of PU.1 with the IRF family member Pip (PU.1-interaction partner), which only binds to PU.1 if Ser-148 within the PEST domain is phosphorylated (29). This interaction appears to be important for regulation of B-cell genes (30, 31). Ser-148 (pink in Fig. 2) is conserved in the skate sequence as well as in all other jawed vertebrate PU.1 sequences.
Evolution of PU.1 Family Member Transactivation and Protein Interaction Domains.
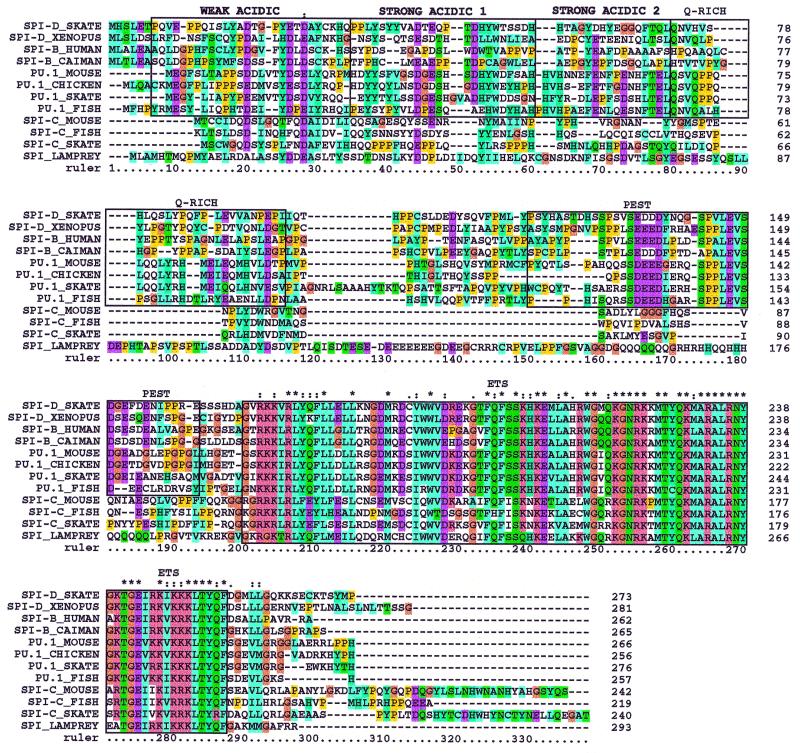
An alignment derived from the clustalx program (32) of skate Spi-D and the putative Xenopus Spi-D [originally designated Spi-B by Shintani et al. (13)] with the other PU.1 subfamily members (PU.1, Spi-B, and Spi-C) is shown in Fig. 3. PU.1, Spi-B, and Spi-D homologs all exhibit strong conservation of the Ets domain and the core of the PEST domain, whereas Spi-C does not. Ser-148 also is conserved in PU.1, Spi-B, and Spi-D but not in Spi-C. Pip has been shown to bind to Spi-B as well as PU.1 (33), confirming its functional relevance. However, both Spi-B and Spi-D have fewer acidic residues per acidic domain than PU.1. Additionally, Spi-D has fewer glutamines per Q-rich region than PU.1, whereas human Spi-B has only two glutamines, and caiman Spi-B has no glutamines in this region (Fig. 4). These differences indicate that the transactivation domains of PU.1, Spi-B, and Spi-D have diverged considerably, although they retain traces of a common ancestral gene.
Figure 3.
Amino acid alignment of jawed vertebrate PU.1 family members with lamprey Spi. This alignment was generated by using clustalw and includes the entire coding sequence of each gene. Boxes indicate known functional domains. Conserved types of amino acids are color-coded according to the clustalx program (32). See Fig. 1 legend for GenBank accession numbers.
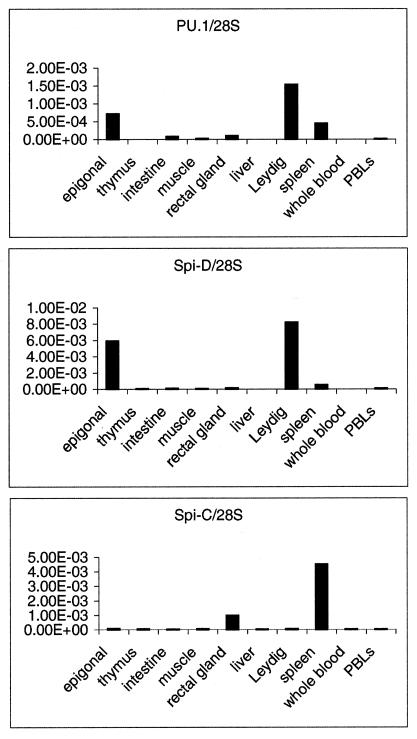
Figure 4.
Real-time reverse transcription–PCR analysis of PU.1 family member expression in adult skate tissues. Total RNA was random-primed to generate the cDNAs used as templates for SYBR green-based real-time PCRs. All results are normalized to levels of 28S rRNA.
Spi-C possesses not only the most divergent DNA-binding domain (see Fig. 1) but also a unique N-terminal region. The N-terminal regions differ significantly even between the Spi-C genes currently identified (mouse, fish, and skate) (Fig. 3), and thus are more difficult to use for unambiguous classification. However, conservation of residues is more readily apparent between two of three species in the N terminus when only the mouse, fish, and skate Spi-C genes are aligned (data not shown). The unusual nature of both the putative transactivation and DNA-binding domains supports the idea that Spi-C is subject to rapid divergence, as suggested previously (13).
Lamprey Spi Gene Compared with Skate Genes.
The diversified PU.1 family defined by the jawed vertebrate genes contrasts with the single, highly divergent Spi gene in the lamprey isolated from larval gut, a tissue rich in lymphocyte-like cells (13). To identify potential additional family members in a jawless vertebrate, a lamprey protovertebral arch (supraneural body) cDNA library was screened with a pool of mouse PU.1 and skate PU.1 and Spi-C probes under low stringency conditions. Two cDNA clones of different lengths, which represent putative splice isoforms of the same PU.1 family member reported by Shintani et al. (13), were identified. Additional PU.1 subfamily members were not found; however, it is possible that additional related genes are expressed in other tissues.
The Ets domain of the lamprey gene falls outside the PU.1/Spi-D/Spi-B cluster (Fig. 1). The most striking feature of the lamprey Spi gene, however, is the complete divergence of its N-terminal sequence from the N-terminal regions of any of the other genes (Fig. 3). The lamprey Spi gene does include acidic and glutamine-rich domains in the N-terminal regions (Fig. 3), but there is no sequence evidence for shared ancestry with the corresponding functional domains of the jawed vertebrate PU.1 family members. In contrast, the N-terminal domains of skate PU.1 show a high degree of similarity with the corresponding regions in all of the other vertebrate PU.1 orthologs (see Fig. 2). Furthermore, the N-terminal domains of jawed vertebrate Spi-B and Spi-D also show strong evidence for shared ancestry with the N-terminal domains of the PU.1 orthologs. Thus, a substantial change in the “effector” domain structure of PU.1 family members appears to have occurred between the time of divergence of the ancestors of jawless and jawed vertebrates and the time of divergence of the ancestors of cartilaginous and bony vertebrates.
Regulation of Expression of Skate PU.1 Family Members.
In mammals, PU.1 family genes are expressed only in the hematopoietic lineages, in contrast to other members of the Ets family, which are more widely used. To characterize the cell types that express the skate PU.1 family members, reverse transcriptase-dependent PCR was used to analyze a series of RNA samples from skate tissues that have been characterized in terms of their histology (2) and their expression of B-cell and T-cell receptor genes (42). Fig. 4 shows that the skate PU.1 family members are expressed specifically in a number of hematopoietic tissues, whereas little to no expression is detected in the nonhematopoietic muscle. PU.1 is most abundant in the epigonal and Leydig organs, which are enriched in B cells and myeloid cells and depleted of T cells (2, 42). PU.1 also is expressed at lower levels in intestine and spleen but is not detectable in the thymus, which is the site of T cell development as in mammals. By contrast, Spi-C is expressed abundantly only in the spleen, a tissue enriched for T cells and a particular subclass of B lineage cells distinct from those in Leydig and epigonal organs (42). The expression of Spi-D closely resembles that of PU.1, with abundant expression in the Leydig and epigonal organs and only low expression in spleen. These results indicate strong resemblances between the skate and mammalian PU.1 family members in terms of the tissues where they are expressed.
Discussion
The sudden appearance of the adaptive immune response in the jawed vertebrates has been referred to as the “Big Bang” (34). Polymorphic MHC, all four major classes of T cell receptors (α, β, γ, and δ), multiple Ig isotypes, and an additional type of rearranging antigen receptor (NAR) are found in the cartilaginous fish, whereas no rearranging genes have been detected in jawless vertebrates or any invertebrates (3, 4). Furthermore, Rag-1, which is essential for rearrangement, also is restricted to the jawed vertebrates and may have been acquired in a jawed vertebrate ancestor by horizontal transfer (35, 36). A plausible scenario for the origin of lymphocytes involves recruitment of recently duplicated transcription factors for the developmental control of the new rearranging genes in cell types that previously served a related purpose but lacked clonal antigen-specific responses. The existence of at least three PU.1 family members in a cartilaginous fish, the skate, indicates that by the time T cells and B cells had evolved, one of the key transcription factors involved in their development had duplicated several times. Furthermore, the identification thus far of only one PU.1 family member far in the sea lamprey (ref. 13 and this report) indicates that the duplication of at least some of the family members occurred during the time that Ig, T cell receptor, and MHC arose.
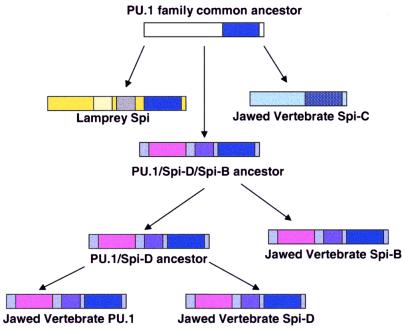
Our identification of an additional member of the PU.1 family, Spi-D, brings this family to four members. The neighbor-joining tree of PU.1 family members indicates that the four paralogs described thus far (PU.1, Spi-B, Spi-C, Spi-D) did not arise by a simple series of binary duplications. A summary of the potential series of duplications leading to the current vertebrate complement of PU.1 family members, assuming relatively similar divergence rates, is shown in Fig. 5. This scheme predicts that a PU.1 family member that resembles Spi-C more than PU.1 might be present in the lamprey or in a nonvertebrate deuterostome. The recent discovery of Spi-C in the mouse (12, 18), fish (13), and skate (this report) should facilitate efforts to isolate PU.1 family members from nonvertebrates.
Figure 5.
Proposed scheme for evolution of vertebrate PU.1 family members. A common ancestor of the PU.1 family members was duplicated at least once before the emergence of the jawed vertebrates. In the lamprey, an N-terminal domain (yellow) containing a stretch of acidic residues (light yellow) and a stretch of glutamines (gray) was incorporated. In the jawed vertebrates, two different types of N-terminal regions evolved, neither of which is detectably related to the lamprey N-terminal region. The PU.1/Spi-D/Spi-B precursor (pink and purple N terminus) duplicated once, resulting in a PU.1/Spi-D precursor and a Spi-B type gene, followed by duplication of the PU.1/Spi-D precursor into two genes: PU.1 and Spi-D. These two duplications may have occurred between the emergence of the jawless and the jawed vertebrates. The other type of N-terminal domain (light blue) is associated with a significantly more divergent Ets domain (hatched dark blue), represented by Spi-C.
The tissue specificities of the skate PU.1 family members are similar to those of their mammalian orthologs. In mice, PU.1 is expressed broadly in hematopoietic precursors and specifically in B cells and myeloid cells (24), whereas Spi-B is largely lymphoid-specific with a strong bias toward expression in B cells (16). Immature subsets of developing T cells express these genes transiently (37), but they are barely detectable in the whole thymus population (9, 16). Murine Spi-C appears to be expressed most restrictively in specific stages of B cell development, in the spleen much more than in bone marrow or lymph node. Spi-C is not expressed in the thymus (12). Although the ontogeny of the skate hematopoietic system is not understood in detail, and myeloid cells have not yet been tracked specifically, the tissues in which these genes are expressed do provide some indication as to cell-type specificity. The Leydig and epigonal organs, in which skate PU.1 and Spi-D are strongly expressed, are far more enriched for B cells than for T cells (42), in agreement with the bias of mammalian PU.1 (and Spi-B) toward expression in the B lineage. However, the abundance of myeloid cells identified at the histological level in these tissues (2) is consistent with a role for PU.1 and/or Spi-D in myeloid cells as well. As in mammals, all of the skate PU.1 family genes are largely silent in the thymus, in contrast to other hematopoietic sites. In addition, the spleen-specific expression pattern of skate Spi-C is strikingly reminiscent of the murine pattern. Analysis of the cell-type specificity of expression in the skate is at an early stage. However, the data presented here indicate that both the structure and the cell-type specific regulation of the PU.1 type genes are conserved.
The extent of structural conservation between the skate and mouse orthologs strongly implies functional conservation. This is especially compelling for PU.1. The mouse PU.1 gene has been mapped extensively in terms of its functional domains and their roles in myeloid development (26–28), enabling a comparison of critical residues between the mouse and skate PU.1 orthologs. The conservation not only of the DNA-binding domain but also of the domains involved in transactivation and other key protein–protein interactions (30, 38–40) in all PU.1 orthologs isolated thus far indicate that the cooperative interactions between PU.1 and other DNA-binding proteins probably have been conserved as well. The N-terminal regions of PU.1 mediate delicate balancing functions in mammalian hematopoietic development, for example, deciding whether a cell will develop as a B cell or as a macrophage (24, 27, 41), or as an erythroid or myeloid precursor (40). These central regulatory activities appear to result from finely titrated competitions among different regulatory partner proteins that interact with particular subregions of PU.1. The conservation seen in the skate PU.1 N-terminal domains emphasizes the strong selective pressure to maintain the sequences involved in these interactions even over long evolutionary spans. This implies that the basic ground rules for hematopoiesis are roughly similar in all jawed vertebrates. If the transcriptional networks in which PU.1 is involved are similar, then an ancient ancestor of jawed vertebrates already may have used many of the same regulatory components for hematopoietic development that are found in modern vertebrates.
It is this structural evidence for conserved interactions that is most conspicuously missing from the lamprey Spi gene. Although the Ets domain is conserved, there is no evidence that the remaining regions of the molecule have a common ancestry with the PU.1 family genes of jawed vertebrates. Therefore, it is unlikely that the lamprey gene encodes a full functional homolog of mammalian PU.1. Interestingly, the lamprey sequence contains both highly acidic and highly glutamine-rich domains, which may serve transactivation functions similar to those of the PU.1 domains of jawed vertebrates. However, these sequences differ completely from those of jawed-vertebrate PU.1 or Spi-B, in sharp contrast to the skate PU.1 in which blocks of highly conserved residues are found throughout the Q-rich and PEST core domains. In the lamprey Spi gene, the intron/exon border flanking the Ets domain coding sequence corresponds to the beginning of the discontinuity between conserved and unrelated sequences. The most straightforward scenario is that the Ets domain exon became associated with new 5′ flanking exons at some time between the divergence of the jawless vertebrates and the divergence of the cartilaginous fish, and that it is these new exons that conferred upon it the distinctive regulatory functions of the jawed vertebrate PU.1.
Acknowledgments
We thank Carl Luer and Cathy Walsh of the Mote Marine Laboratory in Sarasota, FL for their valuable assistance in identification and retrieval of hematopoietic skate tissues. We also thank Rashmi Pant for technical assistance. This work was supported by National Aeronautics and Space Administration Grant NAG2-1370 (to M.K.A. and E.V.R.) and National Institutes of Health Grant R37 AI2338 (to G.W.L.). M.K.A. is supported by the Stowers Institute for Medical Research. A.L.M. has been supported in part by the Institute for Biomolecular Science, University of South Florida.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF320627, AF320628, and AF320629).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.021478998.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.021478998
A.L.M., M.K.A., R. T. Litman, C. J. Walsh, C. A. Luer, E.V.R., and G.W.L., unpublished work.
References
- 1.Zapata A, Amemiya C T. Curr Top Microbiol Immunol. 2000;248:67–107. doi: 10.1007/978-3-642-59674-2_5. [DOI] [PubMed] [Google Scholar]
- 2.Zapata A, Cooper E. The Immune System: Comparative Histophysiology. New York: Wiley; 1990. [Google Scholar]
- 3.Litman G W, Anderson M K, Rast J P. Annu Rev Immunol. 1999;17:109–147. doi: 10.1146/annurev.immunol.17.1.109. [DOI] [PubMed] [Google Scholar]
- 4.Du Pasquier L, Flajnik M. In: Fundamental Immunology. Paul W, editor. Philadelphia: Lippincott; 1999. pp. 605–650. [Google Scholar]
- 5.Holland P W, Garcia-Fernandez J, Williams N A, Sidow A. Development (Cambridge, U.K.) 1994;,Suppl. S:125–133. [PubMed] [Google Scholar]
- 6.Pebusque M J, Coulier F, Birnbaum D, Pontarotti P. Mol Biol Evol. 1998;15:1145–1159. doi: 10.1093/oxfordjournals.molbev.a026022. [DOI] [PubMed] [Google Scholar]
- 7.Anderson M K, Rothenberg E V. Curr Top Microbiol Immunol. 2000;248:137–155. doi: 10.1007/978-3-642-59674-2_7. [DOI] [PubMed] [Google Scholar]
- 8.Kasahara M. Curr Top Microbiol Immunol. 2000;248:53–66. doi: 10.1007/978-3-642-59674-2_4. [DOI] [PubMed] [Google Scholar]
- 9.Klemsz M J, McKercher S R, Celada A, Van Beveren C, Maki R A. Cell. 1990;61:113–124. doi: 10.1016/0092-8674(90)90219-5. [DOI] [PubMed] [Google Scholar]
- 10.Ray D, Bosselut R, Ghysdael J, Mattei M G, Tavitian A, Moreau-Gachelin F. Mol Cell Biol. 1992;12:4297–4304. doi: 10.1128/mcb.12.10.4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kherrouche Z, Beuscart A, Huguet C, Flourens A, Moreau-Gachelin F, Stehelin D, Coll J. Oncogene. 1998;16:1357–1367. doi: 10.1038/sj.onc.1201650. [DOI] [PubMed] [Google Scholar]
- 12.Bemark M, Martensson A, Liberg D, Leanderson T. J Biol Chem. 1999;274:10259–10267. doi: 10.1074/jbc.274.15.10259. [DOI] [PubMed] [Google Scholar]
- 13.Shintani S, Terzic J, Sato A, Saraga-Babic M, O'Huigin C, Tichy H, Klein J. Proc Natl Acad Sci USA. 2000;97:7417–7422. doi: 10.1073/pnas.110505597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scott E W, Simon M C, Anastasi J, Singh H. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 15.McKercher S R, Torbett B E, Anderson K L, Henkel G W, Vestal D J, Baribault H, Klemsz M, Feeney A J, Wu G E, Paige C J, Maki R A. EMBO J. 1996;15:5647–5658. [PMC free article] [PubMed] [Google Scholar]
- 16.Su G H, Ip H S, Cobb B S, Lu M M, Chen H M, Simon M C. J Exp Med. 1996;184:203–214. doi: 10.1084/jem.184.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su G H, Chen H M, Muthusamy N, Garrett-Sinha L A, Baunoch D, Tenen D G, Simon M C. EMBO J. 1997;16:7118–7129. doi: 10.1093/emboj/16.23.7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hashimoto S, Nishizumi H, Hayashi R, Tsuboi A, Nagawa F, Takemori T, Sakano H. Int Immunol. 1999;11:1423–1429. doi: 10.1093/intimm/11.9.1423. [DOI] [PubMed] [Google Scholar]
- 19.Degnan B M, Degnan S M, Naganuma T, Morse D E. Nucleic Acids Res. 1993;21:3479–3484. doi: 10.1093/nar/21.15.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rast J P, Anderson M K, Strong S J, Luer C, Litman R T, Litman G W. Immunity. 1997;6:1–11. doi: 10.1016/s1074-7613(00)80237-x. [DOI] [PubMed] [Google Scholar]
- 21.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rzhetsky A, Nei M. J Mol Evol. 1992;35:367–375. doi: 10.1007/BF00161174. [DOI] [PubMed] [Google Scholar]
- 23.Le H L, Lecointre G, Perasso R. Mol Phylogenet Evol. 1993;2:31–51. doi: 10.1006/mpev.1993.1005. [DOI] [PubMed] [Google Scholar]
- 24.Fisher R C, Scott E W. Stem Cells (Dayton) 1998;16:25–37. doi: 10.1002/stem.160025. [DOI] [PubMed] [Google Scholar]
- 25.Tenen D G, Hromas R, Licht J D, Zhang D E. Blood. 1997;90:489–519. [PubMed] [Google Scholar]
- 26.Fisher R C, Olson M C, Pongubala J M, Perkel J M, Atchison M L, Scott E W, Simon M C. Mol Cell Biol. 1998;18:4347–4357. doi: 10.1128/mcb.18.7.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeKoter R P, Singh H. Science. 2000;288:1439–1441. doi: 10.1126/science.288.5470.1439. [DOI] [PubMed] [Google Scholar]
- 28.Klemsz M J, Maki R A. Mol Cell Biol. 1996;16:390–397. doi: 10.1128/mcb.16.1.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pongubala J M, Van Beveren C, Nagulapalli S, Klemsz M J, McKercher S R, Maki R A, Atchison M L. Science. 1993;259:1622–1625. doi: 10.1126/science.8456286. [DOI] [PubMed] [Google Scholar]
- 30.Eisenbeis C F, Singh H, Storb U. Genes Dev. 1995;9:1377–1387. doi: 10.1101/gad.9.11.1377. [DOI] [PubMed] [Google Scholar]
- 31.Brass A L, Zhu A Q, Singh H. EMBO J. 1999;18:977–991. doi: 10.1093/emboj/18.4.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rao S, Matsumura A, Yoon J, Simon M C. J Biol Chem. 1999;274:11115–11124. doi: 10.1074/jbc.274.16.11115. [DOI] [PubMed] [Google Scholar]
- 34.Schluter S F, Bernstein R M, Bernstein H, Marchalonis J J. Dev Comp Immunol. 1999;23:107–111. doi: 10.1016/s0145-305x(99)00002-6. [DOI] [PubMed] [Google Scholar]
- 35.Bernstein R M, Schluter S F, Bernstein H, Marchalonis J J. Proc Natl Acad Sci USA. 1996;93:9454–9459. doi: 10.1073/pnas.93.18.9454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Agrawal A, Eastman Q M, Schatz D G. Nature (London) 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- 37.Anderson M K, Hernandez-Hoyos G, Diamond R A, Rothenberg E V. Development (Cambridge, UK) 1999;126:3131–3148. doi: 10.1242/dev.126.14.3131. [DOI] [PubMed] [Google Scholar]
- 38.Behre G, Whitmarsh A J, Coghlan M P, Hoang T, Carpenter C L, Zhang D E, Davis R J, Tenen D G. J Biol Chem. 1999;274:4939–4946. doi: 10.1074/jbc.274.8.4939. [DOI] [PubMed] [Google Scholar]
- 39.Petrovick M S, Hiebert S W, Friedman A D, Hetherington C J, Tenen D G, Zhang D E. Mol Cell Biol. 1998;18:3915–3925. doi: 10.1128/mcb.18.7.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rekhtman N, Radparvar F, Evans T, Skoultchi A I. Genes Dev. 1999;13:1398–1411. doi: 10.1101/gad.13.11.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maitra S, Atchison M. Mol Cell Biol. 2000;20:1911–1922. doi: 10.1128/mcb.20.6.1911-1922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miracle, A. L., Anderson, M. K., Litman, R. T., Walsh, C. J., Luer, C. A., Rothenberg, E. V. & Litman, G. W. (2001) Int. Immunol., in press. [DOI] [PubMed]