Abstract
Adrenomedullin, a recently identified potent vasodilator, is expressed widely and has been suggested to have functions ranging from reproduction to blood pressure regulation. To elucidate these functions and define more precisely sites of Adm expression, we replaced the coding region of the Adm gene in mice with a sequence encoding enhanced green fluorescent protein while leaving the Adm promoter intact. We find that Adm−/− embryos die at midgestation with extreme hydrops fetalis and cardiovascular abnormalities, including overdeveloped ventricular trabeculae and underdeveloped arterial walls. These data suggest that genetically determined absence of Adm may be one cause of nonimmune hydrops fetalis in humans.
Adrenomedullin (Adm), a potent 52-aa peptide vasodilator, was isolated from a human pheochromocytoma on the basis of its ability to elevate platelet cAMP levels (1). Recently, Adm has gained much attention in clinical settings because of its elevated plasma levels in patients with various cardiovascular diseases (2). However, the variety of conditions associated with elevated Adm levels, which include essential hypertension, septic shock, and normal pregnancy, suggest that increases in Adm are secondary to other primary events rather than causative. For example, Adm may be elevated in certain hypertensive disease states as a compensatory mechanism to decrease blood pressure, but genetic experiments to distinguish between causation and compensation have not yet been performed.
On the basis of its nucleotide homologies with calcitonin gene-related peptide (CGRP), calcitonin, and amylin, Adm is considered to be the fourth CGRP family member. The Adm gene contains four exons spanning approximately 2 kb and codes for two biologically active peptides: proadrenomedullin N-terminal 20 peptide (PAMP) and Adm. Recent studies to identify the receptors for Adm and CGRP have led to the discovery of a mechanism of signal transduction whereby a series of single transmembrane spanning receptor activity-modifying proteins (RAMPs) interact with a common receptor to impart specificity of binding for either Adm or CGRP (3). The differential expression of the receptor activity-modifying proteins may dictate the sites of function of the two ligands.
To explore the physiological functions of Adm in an in vivo model, we used homologous recombination in embryonic stem cells to generate mice that are deficient for the coding region of the Adm gene. At the same time, to provide a biological marker for sites of Adm gene expression, we replaced the Adm coding region with a sequence coding for enhanced green fluorescent protein (EGFP) while leaving the Adm promoter and Adm 5′ untranslated region intact. Here we show that Adm is essential for survival because Adm−/− embryos die at midgestation with extreme hydrops fetalis. In addition, Adm−/− embryos have unusual cardiovascular defects that are best characterized as overdeveloped ventricular trabeculae and underdeveloped vascular smooth muscle in the large arteries. Because the combination of cardiovascular defects and the severity of hydrops fetalis are, to our knowledge, unlike any previously described pathology in the mouse, our data demonstrate a unique role for Adm in development and suggest that the absence of Adm may be one cause of nonimmune hydrops fetalis in humans.
Materials and Methods
Gene Disruption by Homologous Recombination.
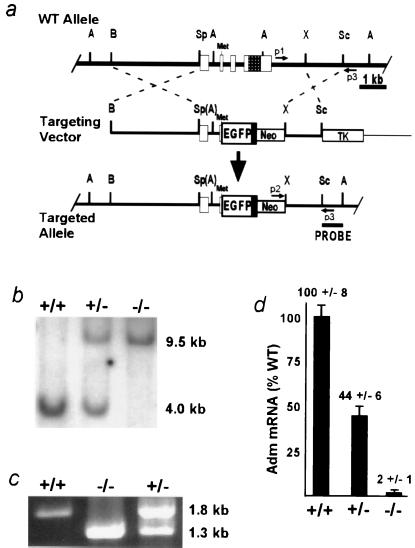
Mice lacking Adm were generated with standard gene targeting methods (4). The targeting construct was made (Fig. 1a) with (i) a 3.7-kb BamHI/SpeI genomic fragment that includes the Adm promoter; (ii) a 600-bp PCR-generated fragment that begins 5′ with the endogenous SpeI site, contains exon 1, intron 1, part of exon 2, and ends 3′ at the endogenous initiator methionine; (iii) a 700-bp cDNA encoding EGFP (CLONTECH); (iv) a 300-bp bovine growth hormone poly(A) addition region; and (v) a 1.3-kb XhoI/SacI genomic fragment from downstream of the Adm gene. 129-derived SvEv embryonic stem cells (TC-1, a gift from Phil Leder, Harvard University, Boston, MA) were electroporated; G418/ganciclovir-resistant colonies, initially identified by the PCR-based assay depicted in Fig. 1 a and c, were expanded; and homologous recombination was confirmed by Southern blot analysis with the use of an 800-bp SacI/Avr II probe external to the targeting construct sequence (Fig. 1 a and b). Male chimeras were mated to wild-type SvEv females to establish an isogenic Adm line, and all experiments were conducted on the resulting isogenic line.
Figure 1.
Generation of Adm−/− animals by homologous recombination. (a) Strategy to disrupt the Adm gene. (Top) Endogenous wild-type (WT) allele. (Middle) Linearized targeting vector. (Bottom) Targeted allele after homologous recombination. Open boxes represent exons 1–4; the checkered region in exon 4 codes for the mature Adm peptide; the initiator methionine of the propeptide is indicated (Met). The solid black box depicts a bovine growth hormone poly(A) addition sequence downstream of the EGFP cDNA. The locations of primer sequences for PCR (p1, p2, and p3) are shown by small arrows. The sequence used as a probe for the Southern-based detection strategy is indicated by a labeled line (PROBE). Restriction sites: A, AvrII; B, BamHI; Sc, SacI; Sp, SpeI; X, XhoI. Parentheses denote sites destroyed during cloning. (b) Detection of the targeted allele by Southern blot analysis. Genomic DNA from E9.5 embryos was digested with AvrII and probed with the SacI/AvrII fragment (PROBE) depicted in the bottom line of a. (c) Detection of the targeted allele by PCR. Genomic DNA from embryos was amplified with the primer sequences depicted in a. (d) Measurement of Adm RNA in total RNA extracts from E9.5 embryos by real-time quantitative reverse transcription–PCR. The relative quantity of Adm RNA in Adm+/− and Adm−/− embryos is expressed as a percentage of total Adm RNA in Adm+/+ embryos. Error bars represent SEM.
To generate Adm−/− embryos, timed heterozygous matings were set up, and the morning of vaginal plug detection was considered to be embryonic day 0.5 (E0.5). To genotype the embryos, DNA was isolated from yolk sac tissue (E8.5–E12.5) or from a small piece of tail tissue (E12.5–E14.5) and analyzed by the PCR-based strategy described above.
Expression Analysis by Using Real-Time Quantitative Reverse Transcription–PCR.
Expression of the Adm gene was characterized by real-time quantitative reverse transcription–PCR with the Sequence detection system (Perkin–Elmer). Primers for Adm amplification were 5′-GAGCGAAGCCCACATTCGT-3′ and 5′-GAAGCGGCATCCATTGCT-3′. The probe for Adm detection was 5′-FAM-CTACCGCCAGAGCATGAACCAGGG-Tamra-3′. All reactions included a β-actin internal standard. The primers used for β-actin amplification were 5′-CTGCCTGACGGCCAAGTC-3′ and 5′-CAAGAAGGAAGGCTGGAAAAGA-3′. The probe for β-actin detection was 5′-TET-CACTATTGGCAACGAGCGGTTCCG-Tamra-3′. RNA was isolated from E9.5 embryos with the use of the TRIzol Reagent (GIBCO/BRL). The reactions were performed with 5 μg total RNA with minor differences from ABI 7700 manufacturer's instructions. Relative levels of Adm expression were determined by the ΔΔCt method and expressed as a percentage of wild type (Fig. 1d). All assays were repeated twice, each with duplicates.
Expression Analysis by Using GFP Fluorescence.
Adult or embryonic heterozygote animals were euthanized, and organs or whole embryos were fixed in 4% paraformaldehyde. Tissues were embedded in Tissue-Tek OCT media (Sakura Finetek, Torrance, CA), and 8-μm frozen sections were analyzed under fluorescent microscopy. The slides were then stained with hematoxylin/eosin (H&E) to histologically confirm sites of expression. To visualize Adm expression during development, heterozygote embryos were fixed in 4% paraformaldehyde and analyzed by fluorescent confocal microscopy.
Histological Analysis.
Embryos, dissected from the uterus at the desired gestational times, were fixed in 4% paraformaldehyde, dehydrated, and embedded in paraffin. Five-micrometer-thick sections were mounted on slides and stained with H&E.
Results
Targeted disruption of the Adm gene replaced the protein-coding region of the gene with a cDNA coding for EGFP but left the 5′ regions of the gene intact, so that EGFP expression is Adm specific (Fig. 1a). The disrupted allele was detected by Southern blot analysis (Fig. 1b) and by PCR (Fig. 1c). To confirm that the targeting inactivated the Adm gene, quantitative reverse transcription–PCR was performed on embryonic RNA, with the use of an Adm TaqMan probe and the ABI7700 Sequence Detection system. Adm−/− embryos express Adm mRNA at levels indistinguishable from zero, thus confirming complete loss of Adm production; heterozygotes have half-normal expression (Fig. 1d).
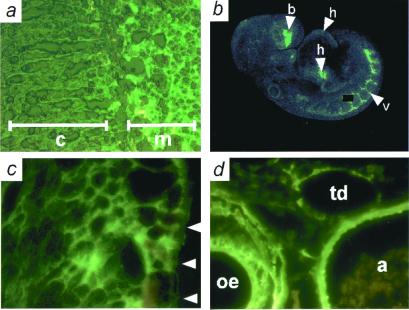
Mice heterozygous for Adm showed no overt differences from wild-type mice and were used to determine the pattern of Adm expression by observing fluorescence from its surrogate, EGFP. As expected from previous studies (5, 6), the adult adrenal medulla showed strong fluorescence, but no fluorescence was detected in the zona fasciculata/reticularis of the adrenal cortex (Fig. 2a). Confocal microscopy of E9.5 Adm+/− embryos showed moderate expression in the heart and strong expression in the developing vasculature (Fig. 2b). E13.5 Adm+/− embryos express Adm in the cardiac myocytes of the developing ventricle (Fig. 2c). The vascular smooth muscle cells of the aorta and esophagus and the endothelial cells of the aorta and thoracic duct strongly express Adm (Fig. 2d).
Figure 2.
Sites of Adm expression determined by EGFP fluorescence. (a) Frozen section of adrenal gland from an adult Adm+/−. Note intense fluorescence in adrenal medulla (m) and the absence of fluorescence in adrenal cortex (c). (b) Confocal microscopy of an E9.5 Adm+/− embryo. Moderate expression is seen in the heart (arrowhead h), and strong expression in the developing vasculature (arrowhead v) and forebrain (arrowhead b); a speck of nonspecific fluorescence has been covered with a black box. (c) Frozen section of an E13.5 Adm+/− embryo, showing expression in the developing left ventricle. Arrowheads indicate the pericardial surface of the left ventricle. (d) Frozen section of an E16.5 Adm+/− embryo, showing expression in the aorta (a), esophagus (oe), and thoracic duct (td).
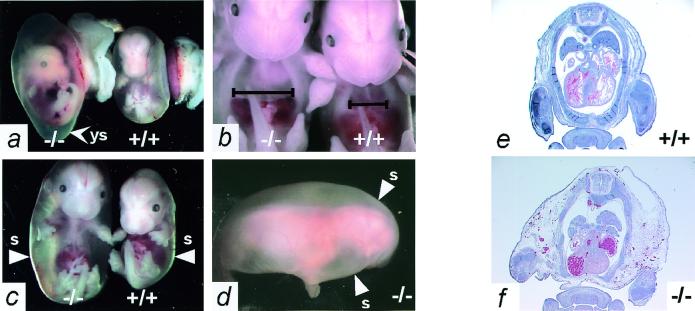
Mice homozygous for the targeted allele die in utero, as shown by our finding that among the progeny of Adm+/− × Adm+/− matings 32% were Adm+/+, 68% were Adm+/−, but none were Adm−/−. Timed matings between Adm+/− mice showed that at E12.5 the Adm−/− embryos are indistinguishable from their Adm+/− and Adm+/+ littermates. However, the Adm−/− embryos die between E13.5 and E14.5, as evidenced by increased numbers of resorbing Adm−/− embryos (Table 1). Early in this period, the yolk sac cavity becomes distended (Fig. 3a), and Adm−/− embryos exhibit modest general edema with a notable increase in the size of their thoracic cavities relative to control littermates (Fig. 3b). By E14.5, Adm−/− embryos suffer from extreme hydrops fetalis (Fig. 3c) that encompasses the entire body and neural structures (Fig. 3d). Note that the yolk sacs have been removed from the embryos illustrated in Fig. 3 b–d. Histological examination of the Adm−/− embryos (Fig. 3f) confirms the enlargement of their thoracic cavities and their extreme and generalized hydrops compared with control littermates (Fig. 3e).
Table 1.
Genotype of embryos from Adm+/− × Adm+/− matings
| Age | Genotypes of embryos
|
Total | ||
|---|---|---|---|---|
| +/+ | +/− | −/− | ||
| E9.5 | 37 | 53 | 23 | 113 |
| E10.5 | 11 | 15 | 6 | 32 |
| E12.5 | 6 | 9 | 5 | 20 |
| E13.5 | 11 | 14 | 7 | 32 |
| E14.5 | 12 | 30 | 18* | 60 |
| E17.5 | 4 | 9 | 1 | 14 |
| P10 | 47 | 101 | 0 | 148 |
Dying or resorbed embryos.
Figure 3.
Adm−/− embryos have massive generalized edema. (a) Appearance of E14.5 Adm−/− (Left) and Adm+/+ (Right) embryos in their yolk sacs shortly after dissection from the uterus. Note that the yolk sac (arrowhead ys) of the Adm−/− embryo is distended with fluid, and its blood vessels are thinner than those of its wild-type littermate. (b) The thoracic cavity of E13.5 Adm−/− embryos is considerably enlarged (Left) compared with a wild-type littermate (Right). The black lines indicate the width of the thoracic cavity just above the diaphragm. (c) Severe hydrops fetalis is apparent in the E14.5 Adm−/− embryos (Left), readily visible after their yolk sacs are removed. The arrowheads (s) indicate the outer skin layer of the embryos. (d) The extreme hydrops of the Adm−/− embryos, dissected away from their yolk sacs, is completely general, as indicated by the uniformly swollen back and head. The arrowheads (s) again indicate the outer skin layer. (e and f) H&E stain of transverse sections through E14.5 Adm+/+ (e) and Adm−/− (f) embryos. The Adm−/− embryo has a fluid-filled thoracic cavity, and the tissues external to the rib cage are markedly swollen (e and f, ×1).
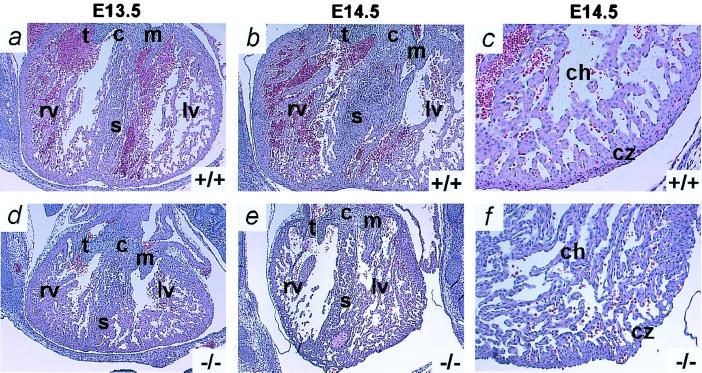
Edema in mice embryos has previously been described in association with cardiovascular defects, although we find no reports of edema of this severity (7, 8). We therefore examined the condition of the heart in transverse sections of E13.5 and E14.5 Adm−/− embryos. Fig. 4 illustrates that the E13.5 Adm−/− heart (Fig. 4d) is typically only two-thirds the size of the wild-type heart (Fig. 4a) but shows signs of increased left ventricular trabecular development leading to smaller chamber size. By E14.5, this pathological overdevelopment becomes more marked (Fig. 4 b and e): the compact zone appears thin and convoluted, the number of trabeculae is increased, and the myocardium has a generally disorganized appearance (Fig. 4 c and f). The mitral and tricuspid valves, intraventricular septum, and atria of Adm−/− embryos show no obvious abnormalities at any time.
Figure 4.
Adm−/− embryos have developmental heart defects. Transverse sections through the hearts of Adm+/+ (Upper) and Adm−/− (Lower) embryos were stained with H&E. (a) E13.5 Adm+/+ littermate. (d) E13.5 Adm−/− embryo. The heart of the Adm−/− embryo is smaller than the heart of its wild-type littermate. (b) E14.5 Adm+/+ littermate. (e) E14.5 Adm−/− embryo. The heart of the Adm−/− embryo is still smaller, and the left ventricle is significantly occluded with increased trabeculae compared with its Adm+/+ littermate. (c) E14.5 Adm+/+ littermate. (f) E14.5 Adm−/− embryo. Higher-power magnification of the left ventricle at E14.5 shows an increase in the number of trabeculae, a thinner, convoluted compact zone (cz), and a generally delaminated appearance of the Adm−/− heart compared with wild type. c, endocardial cushion; s, septum; t, tricuspid valve; m, mitral valve; rv, right ventricle; lv, left ventricle; ch, chamber; cz, compact zone (a, b, d, and e, ×2; c and f, ×10).
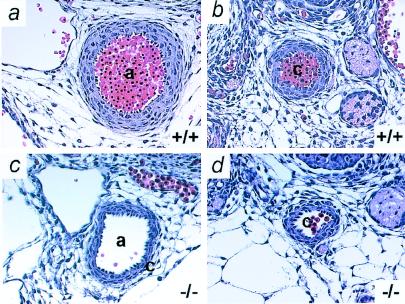
The aorta and carotid artery of the Adm−/− embryos are also abnormal (Fig. 5 c and d); they have markedly thinner walls and are irregular in shape when compared with wild type (Fig. 5 a and b). However, we observed no significant differences in the external appearance of the smaller vessels of the paws and tail (Fig. 2 b and d). The thinner major arteries and the cardiac pathology of the Adm−/− embryos demonstrate that cardiovascular development is abnormal in the absence of Adm.
Figure 5.
Adm−/− embryos have thin arterial walls. Transverse sections through vessels of Adm+/+ (Upper) and Adm−/− (Lower) embryos at E13.5 were stained with H&E. (a and c) aorta; (b and d) carotid artery. Note the thin vascular walls (approximately three cells thick) of the Adm−/− vessels compared with wild-type vessels (approximately five cells thick). a, aorta; c, carotid artery (×10).
Discussion
Because our Adm−/− animals express EGFP in place of Adm, we comment on a recent report that extremely high levels of a GFP transgene expressed in the heart of adult mice causes dilated cardiomyopathy (9). For several reasons, it is highly unlikely that the abnormal trabecular development in the ventricles of our Adm−/− embryos is caused by EGFP toxicity. First, the level of EGFP reported to be toxic appears much greater than in our mice, as judged by fluorescence. Second, our phenotype manifests itself over 24–48 h during embryonic development, as opposed to a several-month progression in the adult transgenic mice. Third, other investigators have previously shown that GFP is nontoxic in mice (10–12), and that moderately expressed cardiac-specific GFP transgenes do not cause adverse effects during embryogenesis or adulthood (13, 14).
Nonimmune hydrops fetalis has an incidence of 1:3,000 births and a high fetal mortality rate. One-quarter of these cases are associated with cardiovascular defects such as arrhythmias and malformations. Fifteen percent are of unknown etiology. Here we describe a type of nonimmune hydrops fetalis that is associated with cardiovascular defects and is caused by a single gene defect. Cardiovascular defects have been reported in mice having a wide variety of disrupted genes [RXR α (8, 15), α 4 integrin (16), PDGF α (17), BARK 1 (18), Hand-1 (19, 20), PPAR γ (21), calreticulin (22), jumonji (23), etc.] likely reflecting the many ways in which cardiac anomalies can be induced. However, the consequences of these gene deficiencies differ in two fundamental ways from the Adm−/− phenotype. First, any observed edema tends to be mild, localized, and subcutaneous. Second, the heart defects are generally characterized as involving hypoplasia indicative of impaired myocardial growth and subsequent ventricular dilation. In contrast, Adm−/− hearts show evidence of trabecular hyperplasia that can progress sufficiently to cause ventricular occlusion. Adm, secreted by neonatal rat cardiomyocytes, inhibits protein synthesis and collagen production when it is added to cultured cardiomyocytes (24) or cardiac fibroblasts (25). In line with these findings, our observations suggest that absence of Adm in the developing heart leads to abnormally enhanced growth. However, in apparent contrast, our studies also show that the absence of Adm results in less than normal vascular smooth muscle cell development in large arteries, which is unlikely to be secondary to low blood pressure caused by a poorly functioning heart because it does not occur in other mice with cardiac failure.
The extreme hydrops of Adm−/− embryos requires comment. For the above reasons, it is unlikely to be solely because of cardiac or vascular defects. Likewise, it is unlikely that abnormal vascular permeability could account for the hydrops, because gene disruption models with vascular permeability defects do not show the same degree of edema (26–28). Abnormalities in lymphatic vessel development could theoretically cause the severe edema, and lymphatic vessels sprout from veins around E12.5 in mice, which is the time of the hydrops onset in our mutants. Furthermore, we find that Adm is expressed in the thoracic duct of Adm+/− embryos (Fig. 2d). However, current data are not yet sufficient to allow a chain of events to be specified that leads to the hydrops of Adm−/− embryos. Nevertheless, our overall data suggest that a genetically determined absence of Adm could be one cause of nonimmune hydrops fetalis in humans.
Acknowledgments
We thank J. Cross, N. Freedman, N. Maeda, H. Rockman, W. Samson, K. Sulik, and members of the Smithies and Maeda laboratories for helpful advice and discussions, and R. Bagnell, H. S. Kim, N. Shaw, and X. Chen for assistance with our experiments. This work was supported by grants from the National Institutes of Health to O.S. (GM20069 and HL49277) and to K.M.C. (HL10344).
Abbreviations
- Adm
Adrenomedullin
- EGFP
enhanced green fluorescent protein
- CGRP
calcitonin gene-related peptide
- En
embryonic day n
- H&E
hematoxylin/eosin
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.021548898.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.021548898
References
- 1.Sakata J, Shimokubo T, Kitamura K, Nakamura S, Kangawa K, Matsuo H, Eto T. Biochem Biophys Res Commun. 1993;195:921–927. doi: 10.1006/bbrc.1993.2132. [DOI] [PubMed] [Google Scholar]
- 2.Hinson J P, Kapas S, Smith D M. Endocr Rev. 2000;21:138–167. doi: 10.1210/edrv.21.2.0396. [DOI] [PubMed] [Google Scholar]
- 3.McLatchie L M, Fraser N J, Main M J, Wise A, Brown J, Thompson N, Solari R, Lee M G, Foord S M. Nature (London) 1998;393:333–339. doi: 10.1038/30666. [DOI] [PubMed] [Google Scholar]
- 4.Koller B H, Hagemann L J, Doetschman T, Hagaman J R, Huang S, Williams P J, First N L, Maeda N, Smithies O. Proc Natl Acad Sci USA. 1989;86:8927–8931. doi: 10.1073/pnas.86.22.8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kapas S, Martinez A, Cuttitta F, Hinson J P. J Endocrinol. 1998;156:477–484. doi: 10.1677/joe.0.1560477. [DOI] [PubMed] [Google Scholar]
- 6.Satoh F, Takahashi K, Murakami O, Totsune K, Sone M, Ohneda M, Sasano H, Mouri T. Neurosci Lett. 1996;203:207–210. doi: 10.1016/0304-3940(95)12294-x. [DOI] [PubMed] [Google Scholar]
- 7.Paszty C, Mohandas N, Stevens M E, Loring J F, Liebhaber S A, Brion C M, Rubin E M. Nat Genet. 1995;11:33–39. doi: 10.1038/ng0995-33. [DOI] [PubMed] [Google Scholar]
- 8.Sucov H M, Dyson E, Gumeringer C L, Price J, Chien K R, Evans R M. Genes Dev. 1994;8:1007–1018. doi: 10.1101/gad.8.9.1007. [DOI] [PubMed] [Google Scholar]
- 9.Huang W Y, Aramburu J, Douglas P S, Izumo S. Nat Med. 2000;6:482–483. doi: 10.1038/74914. [DOI] [PubMed] [Google Scholar]
- 10.Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. FEBS Lett. 1997;407:313–319. doi: 10.1016/s0014-5793(97)00313-x. [DOI] [PubMed] [Google Scholar]
- 11.Godwin A R, Stadler H S, Nakamura K, Capecchi M R. Proc Natl Acad Sci USA. 1998;95:13042–13047. doi: 10.1073/pnas.95.22.13042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barrow J R, Stadler H S, Capecchi M R. Development (Cambridge, UK) 2000;127:933–944. doi: 10.1242/dev.127.5.933. [DOI] [PubMed] [Google Scholar]
- 13.Fleischmann M, Bloch W, Kolossov E, Andressen C, Muller M, Brem G, Hescheler J, Addicks K, Fleischmann B K. FEBS Lett. 1998;440:370–376. doi: 10.1016/s0014-5793(98)01476-8. [DOI] [PubMed] [Google Scholar]
- 14.Xian M, Honbo N, Zhang J, Liew C C, Karliner J S, Lau Y F. J Mol Cell Cardiol. 1999;31:2155–2165. doi: 10.1006/jmcc.1999.1046. [DOI] [PubMed] [Google Scholar]
- 15.Sapin V, Dolle P, Hindelang C, Kastner P, Chambon P. Dev Biol. 1997;191:29–41. doi: 10.1006/dbio.1997.8687. [DOI] [PubMed] [Google Scholar]
- 16.Yang J T, Rayburn H, Hynes R O. Development (Cambridge, UK) 1995;121:549–560. doi: 10.1242/dev.121.2.549. [DOI] [PubMed] [Google Scholar]
- 17.Schatteman G C, Motley S T, Effmann E L, Bowen-Pope D F. Teratology. 1995;51:351–366. doi: 10.1002/tera.1420510602. [DOI] [PubMed] [Google Scholar]
- 18.Jaber M, Koch W J, Rockman H, Smith B, Bond R A, Sulik K K, Ross J, Jr, Lefkowitz R J, Caron M G, Giros B. Proc Natl Acad Sci USA. 1996;93:12974–12979. doi: 10.1073/pnas.93.23.12974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riley P, Anson-Cartwright L, Cross J C. Nat Genet. 1998;18:271–275. doi: 10.1038/ng0398-271. [DOI] [PubMed] [Google Scholar]
- 20.Firulli A B, McFadden D G, Lin Q, Srivastava D, Olson E N. Nat Genet. 1998;18:266–270. doi: 10.1038/ng0398-266. [DOI] [PubMed] [Google Scholar]
- 21.Barak Y, Nelson M C, Ong E S, Jones Y Z, Ruiz-Lozano P, Chien K R, Koder A, Evans R M. Mol Cell. 1999;4:585–595. doi: 10.1016/s1097-2765(00)80209-9. [DOI] [PubMed] [Google Scholar]
- 22.Mesaeli N, Nakamura K, Zvaritch E, Dickie P, Dziak E, Krause K H, Opas M, MacLennan D H, Michalak M. J Cell Biol. 1999;144:857–868. doi: 10.1083/jcb.144.5.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee Y, Song A J, Baker R, Micales B, Conway S J, Lyons G E. Circ Res. 2000;86:932–938. doi: 10.1161/01.res.86.9.932. [DOI] [PubMed] [Google Scholar]
- 24.Tsuruda T, Kato J, Kitamura K, Kuwasako K, Imamura T, Koiwaya Y, Tsuji T, Kangawa K, Eto T. Hypertension. 1998;31:505–510. doi: 10.1161/01.hyp.31.1.505. [DOI] [PubMed] [Google Scholar]
- 25.Horio T, Nishikimi T, Yoshihara F, Matsuo H, Takishita S, Kangawa K. Eur J Pharmacol. 1999;382:1–9. doi: 10.1016/s0014-2999(99)00559-2. [DOI] [PubMed] [Google Scholar]
- 26.Puri M C, Rossant J, Alitalo K, Bernstein A, Partanen J. EMBO J. 1995;14:5884–5891. doi: 10.1002/j.1460-2075.1995.tb00276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suri C, Jones P F, Patan S, Bartunkova S, Maisonpierre P C, Davis S, Sato T N, Yancopoulos G D. Cell. 1996;87:1171–1180. doi: 10.1016/s0092-8674(00)81813-9. [DOI] [PubMed] [Google Scholar]
- 28.Kim K, Drummond I, Ibraghimov-Beskrovnaya O, Klinger K, Arnaout M A. Proc Natl Acad Sci USA. 2000;97:1731–1736. doi: 10.1073/pnas.040550097. . (First Published February 4, 2000; 10.1073/pnas.040550097) [DOI] [PMC free article] [PubMed] [Google Scholar]