Abstract
Histone post-translational modifications occur, not only in the N-terminal tail domains, but also in the core domains. While modifications in the N-terminal tail function largely through the regulation of the binding of non-histone proteins to chromatin, based on their location in the nucleosome, core domain modifications may also function through distinct mechanisms involving structural alterations to the nucleosome. This article reviews the recent developments in regards to these novel histone modifications and discusses their important role in the regulation of chromatin structure.
INTRODUCTION
The study of histone modifications began over 40 years ago when Murray reported the identification of lysine methylation in calf thymus histones (1). Shortly thereafter, lysine acetylation and serine phosphorylation were also discovered in histones from such widely ranging sources as human lymphocytes, rat liver, peas and calf thymus (2–6). Later, in 1975, ubiquitylation and ADP-ribosyation were added to the list of modifications that can be affixed to histones (7,8). As recently as 2003, novel histone modifications were still being discovered, specifically, the addition of the small ubiquitin-related modifier, SUMO, onto histone lysine residues (9).
For nearly four decades the study of histone modifications focused exclusively on those that occurred on the tail domains of the core histones. While the core histones do not share any primary sequence similarity, they have a common domain structure. The central portion of each histone consists of a globular core domain that folds into the characteristic histone fold. These histone fold-containing regions associate with each other to constitute the bulk of the histone octamer around which DNA wraps. Flanking the core domains are the relatively unstructured tail domains. The histone tails extend out from the face of the nucleosome and through the DNA gyres into the area surrounding the nucleosome (10).
The N-terminal tail domains of the core histones contain an extraordinary number of sites that can be subjected to post-translational modification (11–13). Some tail modifications, such as acetylation and phosphorylation, can alter the charge of the tails and, therefore, have the potential to influence chromatin through electrostatic mechanisms. However, the primary mechanism by which tail modifications act appears to be through altering the ability of non-histone proteins to interact with chromatin (14–17).
The study of modifications in the N-terminal tail domains dominated research in this field for a number of reasons. The foremost being that the primary method for discovering histone modifications, Edman degradation, favored the analysis of the first 20–30 amino acids. The monopoly of the tail domains ended in 2002 with the application of mass spectrometry to the study of histone modifications. Mass spectrometry is a technique which takes advantage of the physical properties of ions to determine mass to charge ratios (m/z). This is particularly useful when the primary amino acid sequence is known because differences between predicted and observed masses, termed ‘mass shifts’, can be used to identify post-translational modifications. The application of mass spectrometry to the study of histone modifications allowed for the discovery of the first novel site of histone modification located outside of the tail, methylation of histone H3 lysine 79 (18,19). With this innovative application of mass spectrometry, a new door was opened for the study of histone modifications.
In the short time since the discovery of H3 lysine 79 methylation, over 30 histone modifications have been identified and/or quantified by mass spectrometry with the majority of these newly identified modifications localized to the core domains (20–22). Mapping of the precise locations of these core modifications onto the nucleosome crystal structure reveals that these modifications fall into clusters that can be organized into three distinct classes: (i) the solute accessible face, (ii) the histone lateral surface and (iii) the histone–histone interfaces (23,24). It is likely that modifications in these classes will have unique effects on chromatin structure and act through mechanisms that are distinct from those observed with tail domain modifications. The locations and the evolutionary conservation of these modifications predict that they may be of great physiological relevance. The limited data available concerning these modifications support this idea and suggest that histone core domain modifications may turn out to play as significant a role as modifications within the histone tails.
Similar to the situation observed with histone tail modifications, modifications located on the solute accessible face of the nucleosome have the ability to alter higher-order chromatin structure and chromatin–protein interactions. Histone lateral surface modifications are uniquely capable of affecting histone–DNA interaction and modifications on the histone–histone interface have the exclusive ability to disrupt intra-nucleosomal interactions thereby altering nucleosome stability. Mutations that alter sites of histone tail modifications have been shown to affect processes such as transcription, heterochromatic silencing and DNA damage repair; however, the effects in many cases were minor (25–31). Single amino acid substitutions of modifiable residues within the histone core have been shown to dramatically affect transcription, DNA damage repair, chromatin structure, chromatin assembly and heterochromatic gene silencing (18,19,32–36). In many instances, the locations of these residues help to explain why these phenotypes are observed. In this review, we will examine the literature available to date concerning histone core domain modifications and discuss how the locations of these modifications can, in many incidences, predict their function. In discussing modifications we will refer to the residue numbers in bovine histones, as the largest number have been identified in this organism. However, in discussing results of genetic analyses in yeast, we will refer to the residue numbers in yeast.
MODIFICATIONS OF THE NUCLEOSOMAL FACE
The solvent accessible face of the nucleosome provides a large surface on which interactions can occur that impact the regulation of chromatin structure. For example, specific regions of the nucleosome surface are critical for the assembly of silent chromatin structure in yeast and contacts between surface residues of histones H2A and H2B may mediate the inter-nucleosome interactions involved in the formation of higher order chromatin structures (29,37).
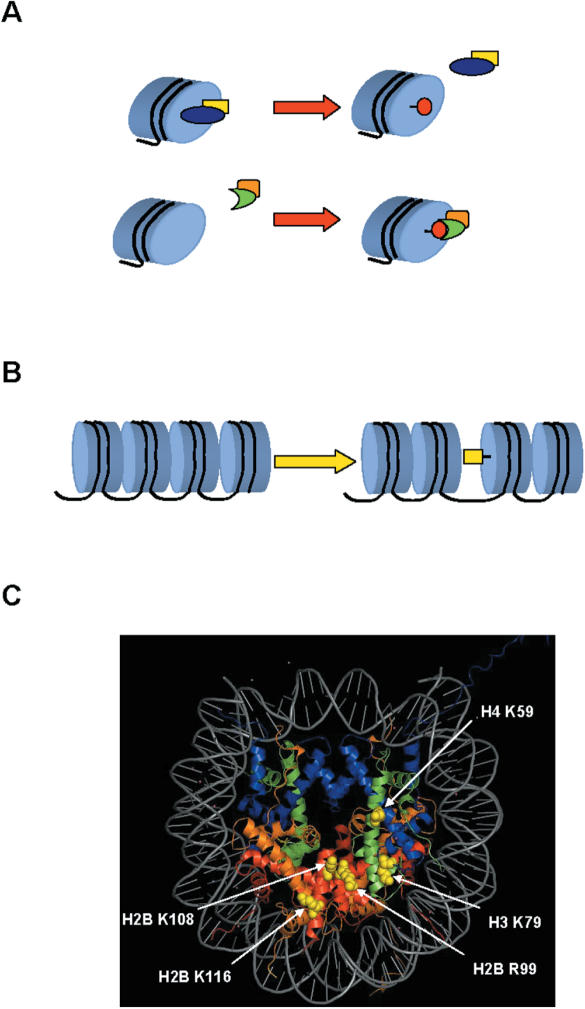
Therefore, modifications to this surface may function through a number of mechanisms to regulate chromatin structure. First, they may be functioning in much the same way as N-terminal tail modifications by controlling the ability of non-histone proteins to bind to the nucleosome (Figure 1A). Additionally, modifications to the nucleosome face may have more direct structural effects by influencing nucleosome–nucleosome interactions that are thought to occur during the formation of 30 nM filaments (Figure 1B).
Figure 1.
Modifications of the nucleosome face. Histone post-translational modifications located on the nucleosome face may influence chromatin structure through multiple mechanisms. (A) These modifications might regulate chromatin–protein interactions in either a positive (top) or negative (bottom) fashion. (B) Alternatively, these modifications could affect higher order chromatin structure by altering interactions between neighboring nucleosomes. (C) Surface modifications are modeled on the crystal structure of the nucleosome (pdb file 1AOI) (10). The histone proteins are shown in a ribbon diagram with histone H2A shown in red, H2B in orange, H3 in blue and H4 in green. The DNA helix is shown in gray. Modified residues are depicted as spheres and colored yellow. Image was generated using the program Pymol.
Histone H3 lysine 79 methylation is the most well-characterized modification of the nucleosome face. This modification has been observed in a number of organisms including yeast, calf thymus, human and chicken (18,19,38–40). This evolutionary conservation in such a wide variety of eukaryotes is a strong indication that it plays a fundamental role in the regulation of chromatin structure.
Histone H3 lysine 79 is located within the globular core domain in the loop 1 region of the H3 histone fold (Figure 1C). This area does not contact DNA and is located on the solvent-accessible nucleosome surface. Within a single nucleosome, the two copies of histone H3 are oriented such that the two H3 lysine 79 residues lie on opposite sides of the ‘disk’. This location is within a region that was genetically identified as being important for silent chromatin structure (41).
In budding yeast, silent chromatin is found in several localized regions of the genome. These regions are found at the telomeres, the silent mating loci (HML and HMR) and the rDNA repeats. For both telomeres and the silent mating loci, the Sir2p, Sir3p and Sir4p proteins are structural components of silent chromatin structure (42). These proteins are directed to the proper position through associations with other factors that bind to either telomeric repeats or silencer sequences at HML and HMR. The SIR proteins then spread throughout the region of chromatin to be silenced through interactions with the core histones (43–45). These SIR/histone interactions are sensitive to histone post-translational modification status and require the histone deacetylase activity of Sir2p (46–49). The rDNA repeat locus is packaged in a fundamentally distinct type of silent chromatin structure that also requires Sir2p but is independent of Sir3p and Sir4p (50–52).
Yeast is an excellent model system for the study of silent chromatin as sensitive in vivo assays have been developed for monitoring the formation and maintenance of silent chromatin structure. These systems are based on the ability to place reporter genes in regions of silent chromatin. The most frequently used reporter gene is URA3 for which both positive and negative growth selections exist (53).
Although histone H3 lysine 79 itself is not methylated in silent chromatin, these in vivo assays revealed lysine 79 is required for the proper formation of silent chromatin at telomeres and the silent mating loci (38). Lysine 79 also appears to be required for rDNA silencing because its mutation to alanine or arginine disrupted rDNA silencing (18,41). The fact that deletion of DOT1, the gene encoding the histone methyltransferase responsible for this modification, did not disrupt rDNA silencing argues that the non-methylated state of lysine 79 is needed for normal rDNA silencing (54).
The role of H3 lysine 79 methylation in heterochromatic gene silencing is likely to involve the regulation of the association of non-histone proteins with chromatin. However, this modification does not function by directly promoting the association of structural components of silent chromatin structure with nucleosomes as is seen with the methylation of H3 lysine 9 and the binding of HP1 in higher eukaryotes. Rather, H3 lysine 79 methylation appears to act by preventing the binding of SIR proteins to regions of euchromatin and thereby ensuring that these factors are available for the assembly of silent chromatin at the proper locations (55). This model was first suggested by the observation that ∼90% of the histone H3 molecules in yeast are methylated on lysine 79. This percentage corresponds very well with the fraction of the yeast genome that is present as euchromatin (18). Indeed, H3 lysine 79 methylation is found throughout the euchromatic regions of the yeast genome (as well as euchromatic regions in higher eukaryotes) but is significantly under-represented in silent chromatin (38). Also consistent with this idea, H3 K79A point mutants displayed a dramatic decrease in binding of Sir2p and Sir3p to yeast telomeres and silent mating loci with a concomitant increase in SIR protein occupancy at adjacent euchromatic sites. Similar ChIP analysis revealed a more modest decrease in Sir2p and Sir3p binding to telomere and HMRa regions in a dot1 deletion strain (18,19).
The mechanism underlying the influence of H3 lysine 79 methylation on SIR protein binding to chromatin is not known. This modification may directly block a surface on the nucleosome face that is necessary for one or more of the SIR proteins to bind. Alternatively, H3 lysine 79 methylation may interfere with internucleosomal interactions that are necessary for forming a higher-order structure that is a prerequisite for stable SIR protein association. Resolution of this issue is likely to require a detailed biochemical analysis of SIR protein–nucleosome interactions.
Dot1p is unique among histone lysine methyltransferases in that it is the only enzyme in this class that lacks a SET [Su(var), Enhancer of zeste, Trithorax] domain (56). SET is a highly conserved domain shown to be essential to the activity of all other histone lysine methyltransferases identified (57,58). Crystal structures have been determined for fragments of the human and yeast Dot1p enzymes revealing that they share some structural properties of the SET domain proteins and also have features similar to those found in protein arginine methyltransferases (59,60). Modeling studies suggest that Dot1p makes extensive contacts with the face of the nucleosome, including the surface genetically identified as being critical for yeast silent chromatin structure (41). In addition, positively charged patches away from the active site may mediate interactions between Dot1p and nucleosomal DNA (59,60).
Although lysine 79 methylation levels do not change in response to locus-specific gene activation or repression, hDOT1L, has been implicated in the misregulation of mixed lineage leukemia (MLL) target genes in cases of leukemia. hDOT1L was found to interact with AF10, a putative transcription factor and frequent MLL fusion partner responsible for upregulation of Hox genes in cases of acute myeloid leukemia (AML) (61). Furthermore, an artificially constructed hDOT1L-MLL fusion protein was capable of inducing leukemogenesis in murine progenitor bone marrow cells. This leukemic transformation was dependent on the histone methyltransferase activity of hDOT1L. Interestingly, transformation with either MLL-AF10 or MLL-hDOT1L resulted in upregulation of Hox genes and hypermethylation of H3 lysine 79 (61). Based on their experimental results, Okada et al. (61) have proposed that AF10 may function as a bridge that links hDOT1L to MLL-AF10 target genes. If this proves to be the case, hDOT1L may serve as a new therapeutic target.
While histone H3 lysine 79 methylation was first characterized with respect to its influence on gene expression, this modification also has important functions in the cell that appear to be largely independent of its effects on transcription. This modification plays a key role in checkpoint signaling pathways as deletion of DOT1 in budding yeast results in defects in both the pachytene and DNA damage checkpoints (G1/S and intra-S) (62–64).
The involvement of H3 lysine 79 methylation in the DNA damage checkpoint appears to be mediated through effects on the binding of checkpoint proteins to chromatin. The human protein 53BP1, whose homologs in budding and fission yeast are Rad9p and Crb2, respectively, contains a conserved tandem tudor domain. This protein domain was originally identified in the SMN protein as a methylated arginine-binding module (65–68). Huyen et al. (69) followed up on this observation by demonstrating that 53BP1 only binds to histones that are post-translationally modified. Subsequent analyses identified histone H3 lysine 79 methylation as the specific post-translational modification recognized by 53BP1. Curiously, unlike many histone modifications involved in the DNA repair process, H3 lysine 79 methylation is not induced by the presence of DNA damage suggesting that some aspect of chromatin structure is altered by the presence of DNA damage such that this modification can be accessed by 53BP1 (69).
A second site of methylation on the solute accessible nucleosome face, histone H4 lysine 59, was first observed by mass spectroscopic analysis of bovine histones (Figure 1C). As expected from the relative proximity of this modification to H3 lysine 79, mutations that alter this residue in yeast have significant effects on silent chromatin structure (22). Although H4 K59 mutations are phenotypically similar to H3 K79 alleles it is not known whether H4 lysine 59 methylation acts through similar mechanisms.
Three additional sites of modification on the nucleosome face were identified via mass spectrometry. Intriguingly, these modifications, which include acetylation of histone H2B lysines 108 and 116 and methylation of H2B arginine 99, form a tight cluster on the surface of the nucleosome suggesting the possibility that they may function in a coordinated fashion (Figure 1C) (22). However, these modifications have not been examined in any detail and, therefore, their in vivo significance is not yet known.
NUCLEOSOME LATERAL SURFACE MODIFICATIONS
A nucleosome consists of 147 bp of DNA wrapped around the histone octamer making direct contact at 14 distinct locations on the superhelix (10). Using mass spectrometry, Zhang et al. (22) recently identified >20 novel modifications within the histone core. A striking number of these modifications were mapped to the nucleosome lateral surface following the path of the DNA as it wraps around the histone octamer (23,24). Several of these newly identified core modifications were mapped to residues that directly bind the DNA, while others were positioned in close proximity to the DNA. Although the latter group does not make direct contact with DNA, these residues have the potential to indirectly influence the histone–DNA interface.
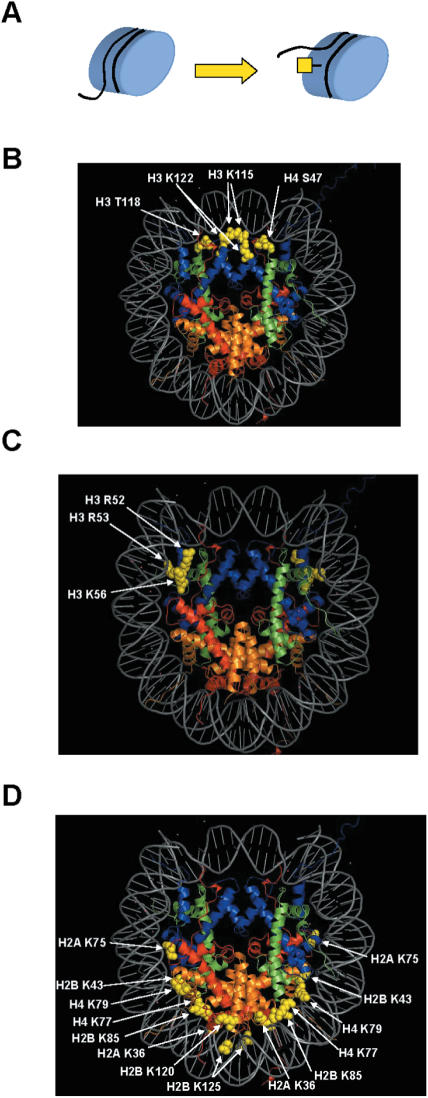
The position of modifications on the lateral surface of the nucleosome immediately suggested that their primary function would be through the regulation of histone–DNA interactions (Figure 2A) (23). Cosgrove et al. (24) proposed a detailed model, termed regulated nucleosome mobility, to describe how these modifications could be involved in altering the fluidity of chromatin structure. In this model, a chromatin remodeling activity (either an ATP-dependent chromatin remodeller or nucleosome assembly/disassembly activity) acts on a nucleosome to alter histone–DNA contacts such that sites of modification on the lateral surface are exposed. The exposed sites can then be acted on by histone modifying activities to either add or remove post-translational modifications which, in turn, lead to nucleosomes with altered mobility. This altered mobility can then lead to changes in the accessibility of specific sequences of DNA or changes in higher order chromatin structure.
Figure 2.
Nucleosome lateral surface modifications. (A) Modifications to the lateral surface of the nucleosome are likely to influence chromatin structure through the modulation of histone–DNA interactions. (B) Modifications located at the nucleosome dyad axis are modeled on the nucleosome crystal structure. (C) Modifications near the DNA entry–exit points are highlighted on the nucleosome crystal structure. (D) Additional modifications near site of histone–DNA contact are shown on the nucleosome crystal structure. Crystal structure was rendered as described in the legend to Figure 1.
While the regulated nucleosome mobility model provides a mechanistic foundation for understanding the cooperativity that exists between histone modifying enzymes and chromatin remodeling activities, the modification of residues on the lateral surface may not be obligatorily linked to the action of chromatin remodeling activities. The work of Widom and co-workers have elegantly demonstrated that there is spontaneous unwrapping of the DNA helix from the surface of the nucleosome (70–79). This unwrapping not only makes the DNA accessible but also exposes residues on the lateral face of the nucleosome to histone modifying activities. Changes to the modification state of the nucleosome lateral surface might then impact chromatin structure by altering the equilibrium constants governing this unwrapping.
Structural studies indicate that the interactions between DNA and histones are at their greatest in the region of the nucleosome dyad (80). It is perhaps significant then that several lateral surface modifications cluster in this region of the nucleosome (Figure 2B) (20,22). Included in this group, along with histone H3 lysine residues 115 and 122, are histone H3 threonine 118 and histone H4 serine 47 which are sites of SIN alleles in yeast (81,82). SIN alleles of the histones were originally isolated as point mutations that partially alleviated the requirement for the SWI/SNF ATP-dependent chromatin remodeling complex for the transcriptional activation of a subset of yeast genes (83). Nucleosomes reconstituted in vitro with histones harboring specific SIN alleles (H4 R45C and R45H) displayed similar nucleosome positioning as seen with wild-type histones, but these Sin mutant nucleosomes were completely incapable of forming fully compacted chromatin fibers (84). Muthurajan et al. analyzed crystal structures of several Sin mutant nucleosomes and reported disruptions in local protein–DNA interactions (85). The effects observed on higher order chromatin structure could well be a direct result of decreases in histone–DNA interactions, but this possibility requires further investigation. Consistent with a role for these residues in modulating histone–DNA contacts, nucleosomes reconstituted with histones containing mutations at the sites of SIN alleles (including H3 threonine 118) show disruption to specific histone–DNA contacts and increased nucleosome mobility (85–87). While none of the H3 T118 alleles tested mimicked the presence of a phosphate group at this position, it is likely that a negatively charged modification at a histone–DNA contact point would have a significant impact on nucleosome stability. Consistent with this idea, substituting H3 T118 or H4 serine 47 with a glutamic acid residue generates lethal and slow growth phenotypes, respectively, in yeast (35). While it is not known whether mutations that alter histone H3 lysines residues 115 and 122 generate SIN alleles, they do result in a number of interesting phenotypes. Mutations that alter H3 lysine 122 to either alanine of glutamine are among the few histone point mutants that have a general slow growth phenotype. In addition, the H3 K122Q allele is defective for silent chromatin structure at the rDNA repeats while the H3 K122R allele shows increased rDNA silencing (35). These results suggest that rDNA chromatin structure is sensitive to the acetylation state of H3 lysine 122. Histone H3 K115 mutations also affect silent chromatin structure with the genetic analyses indicating that this residue needs to be in the deacetylated state for proper transcriptional silencing. In addition, alteration of H3 K115 led to hydroxyurea (HU) sensitivity (35).
Spontaneous unwrapping of the DNA from a nucleosome appears to initiate at the DNA entry–exit point on the nucleosome dyad (78). The most well-characterized lateral surface modification, acetylation of histone H3 lysine 56, is positioned at this location on the nucleosome (Figure 2C). H3 lysine 56 acetylation is conserved in yeast and flies, but it has not been shown definitively to occur in mammals (22,32–35). Evidence for the methylation of arginine 52, arginine 53 or lysine 56 has been reported. Although the exact site of the detected methylation was not identified, it raises the possibility that lysine 56 might be methylated in mammals (22). This does not, however, rule out the possibility that a fraction of mammalian H3 molecules, that have thus far eluded detection, are acetylated at lysine 56. In yeast, H3 lysine 56 acetylation is abundant with roughly a quarter of the histone H3 molecules isolated from asynchronous cultures bearing this modification (32,34). This is likely an underestimate of its prevalence as H3 lysine 56 acetylation appears to be highly cell cycle regulated although there is some conflicting data in the literature (32,33,88). H3 lysine 56 acetylation is prominent during S phase, but then is rapidly lost during G2 (32–34).
Consistent with the S-phase appearance of H3 lysine 56 acetylation, several lines of evidence indicate that this modification is incorporated into chromatin during histone deposition. First, Masumoto et al. (32,88) used inducible forms of H3 to demonstrate that H3 lysine 56 acetylation occurs largely on newly synthesized histones. In addition, histone H3 molecules that co-purify with the chromatin assembly factor CAF-1 are acetylated on lysine 56. Also, changing H3 lysine 56 to glutamine (H3 K56Q), which mimics the acetylated state, resulted in chromatin with an increased sensitivity to MNase digestion and led to a decrease in endogenous plasmid supercoiling (32). These are two common assays used to detect abnormalities in chromatin formation. The H3 K56Q allele was also defective in telomeric and rDNA silencing (35). Areas of heterochromatic gene silencing are thought to be more sensitive to defects in chromatin formation. The K56R point mutant, which mimics the unacetylated state, displayed normal MNase resistance, plasmid supercoiling, and telomeric/rDNA silencing (32,35). Taken together these data are consistent with the proposed role for H3 lysine 56 acetylation in chromatin assembly, with the default status for condensed chromatin being unacetylated at this residue.
A role for lysine 56 acetylation in modulating histone–DNA interacts at the DNA entry–exit point is likely to underlie the observation that lysine 56 acetylation is important for transcription of a subset of SNF5-regulated genes including the core histone genes and SUC2 (34). ChIP analysis showed that chromatin in the vicinity of the HTA1 and HTA2 genes (encoding histone H2A) is hyperacetylated on H3 lysine 56. This hyperacetylation takes place in a cell cycle regulated fashion, peaking just prior to S phase when the histone genes are transcribed. The acetylation of H3 lysine 56 at the HTA1 and HTA2 loci is functionally relevant as the loss of this modification compromises the ability of Snf5p to bind to and activate these genes. However, contrary to the prediction of the regulated nucleosome mobility model, the converse is not true as the acetylation of H3 lysine 56 is independent of SNF5. Therefore, at these loci, H3 lysine 56 acetylation occurs prior to the action of the SWI/SNF chromatin remodeling activity.
A critical step in furthering our understanding of H3 lysine 56 acetylation is the identification of the histone acetyltransferase responsible for this modification. Ozdemir et al. (33) used an antibody specific for H3 lysine 56 acetylation to test deletion strains for 16 known yeast protein acetyltransferases and found that H3 lysine 56 acetylation levels were unaffected by the loss of any single enzyme. Using a different approach, Xu et al. (34) performed ChIP from strains deleted for a number of known histone acetyltransferases and found that loss of the protein known as Spt10p abolished the cell cycle regulated acetylation of H3 lysine 56 at the histone genes. In addition, an spt10Δ resulted in changes in histone gene expression similar to those of H3 K56 mutants. Spt10 is a site-specific DNA-binding protein that binds to histone gene promoters in vivo (89). However, there is currently no evidence that Spt10 can directly acetylate histone H3 lysine 56 at histone gene promoters or other loci.
H3 lysine 56 has also been shown to play a role in DNA damage repair (32,33,35). The K56R point mutant is sensitive to a variety of DNA damaging agents including camptothecin (CPT), methyl methanesulfonate (MMS) and Bleomycin. However, these cells are not sensitive to UV-induced damage. H3 lysine 56 acetylation has been shown to persist much longer into G2 following exposure to camptothecin (CPT) in a Rad9p-dependent manner (32). This suggests that one function of the DNA damage checkpoint is to maintain H3 lysine 56 acetylation near sites of DNA damage until repair is complete. Interestingly, the H3 K56Q mutant was also sensitive to MMS, Bleomycin and CPT indicating that the ability to modulate between the acetylated and unacetylated states may be important for DNA damage repair.
In addition to lysine 56, mass spectrometric analysis also identified histone H3 arginine 52 and arginine 53 as possible sites of mono-methylation located near the DNA entry–exit point (Figure 2C) (22). Hyland et al. (35) analyzed point mutants of these residues for effects on cell viability, heterochromatic gene silencing and DNA damage repair. Intriguingly, changing H3 arginine 52 to either alanine, lysine or glutamine resulted in lethality suggesting that this residue plays a critical role in chromatin structure. When expressed in combination with wild-type histone alleles, R52 mutants caused defects in telomeric and, to a lesser degree, rDNA silencing and sensitivity to HU. While mutations that altered R53 were tolerated, R53Q and R53K alleles displayed mild defects in telomeric and rDNA silencing, respectively.
A number of other residues near points of histone–DNA contact are found around the circumference of the nucleosome (Figure 2D). These include residues modified in bovine histones, such as histone H2A lysine residues 36, 74, 75 and 77, histone H2B lysines 43, 85, 120 and 125 and histone H4 lysines 77 and 79 (22). Analysis of point mutants revealed that H4 lysine 79 affected both telomeric and rDNA silencing. The phenotypes of H4 K79 point mutants suggest that the unacetylated state is important for the compact chromatin structure within these regions. The impact of H4 lysine 77 modification was less clear as mutations that mimic the constutively acetylated state at this residue did not disrupt silent chromatin structure at either telomeres or the rDNA repeats (35).
With the exception of histone H4 lysine 77, in vivo analyses of modification sites on the lateral surface of the nucleosome are consistent with the regulated nucleosome mobility model and its suggestion that the precise nature of histone–DNA contacts on this surface are critical determinants of higher order chromatin structure dynamics. However, the mechanistic link between lateral surface modifications and chromatin remodeling factors remains to be clarified. There is clearly much that needs to be learned about the mechanisms employed by eukaryotic cells to control the modification status of this vital surface of the nucleosome.
MODIFICATIONS WITHIN HISTONE–HISTONE INTERFACES
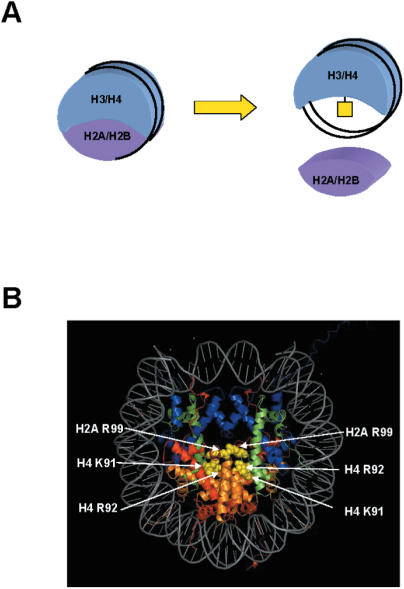
At a very basic level, chromatin structure is dependent upon specific histone–histone interactions that lead to the formation of the histone octamer. These histone–histone interactions include those that mediate the formation of the H3/H4 and H2A/H2B histone fold pairs, those that allow the formation of the H3/H4 tetramer and the H2A/H2B dimer and those between tetramers and dimers that result in completion of the histone octamer. Given the critical nature of these interactions, the use of post-translational modifications for the modulation of intranucleosomal histone–histone contacts may be an important mechanism for regulating chromatin structure. In this model, the modification of residues at points of histone–histone contact would influence chromatin structure by directly impacting the structure of the histone octamer (Figure 3A).
Figure 3.
Modifications to regions of histone–histone interface. (A) Modifications located at points of contact between histone proteins may control chromatin structure through physical alterations in the nucleosome such as destabilization of tetramer–dimers interactions. (B) Locations of modifications in regions of histone–histone interface are depicted on the crystal structure of the nucleosome. Nucleosome was rendered as described in the legend to Figure 1.
There are a number of modifiable residues that are within regions of the histone proteins that are involved in intranucleosomal interactions (20,22). Several of these modifications may not be involved in modulating these interactions. For example, histone H3 lysine residues 115 and 122 and threonine 118 are within the 4 helix bundle that holds together the H3/H4 tetramer. However, these residues are oriented toward the DNA and away from the histone–histone interface (Figure 2C). Therefore, as described above, these modifications are much more likely to regulate histone–DNA contacts than histone–histone interactions.
The best example of a post-translational modification that functions through structural effects on the histone octamer is the acetylation of histone H4 lysine 91. H4 lysine 91 acetylation was first identified by mass spectrometry on bovine histones (22,90). Lysine 91 is in the region of histone H4 that interacts with histone H2B and helps to stabilize the tetramer–dimer interaction necessary for the formation of the histone octamer (Figure 3B) (10,91). In fact, histone H4 lysine 91 is closely juxtaposed with a glutamic acid residue in histone H2B with which it likely forms a salt bridge (24). Therefore, histone H4 lysine 91 is well positioned to be a point at which octamer structure could be regulated through post-translational modification.
Subsequently, acetylation of histone H4 lysine 91 was identified in the population of molecules that co-purified with the nuclear Hat1p/Hat2p/Hif1p complex isolated from yeast (36). This complex is composed of the type B histone acetyltransferase subunits Hat1p and Hat2p (which are thought to be responsible for the acetylation of newly synthesized histone H4 molecules) in association with a histone H3/H4 specific chaperone that possesses in vitro chromatin assembly activity (Hif1p) (92,93). This observation was important for a number of reasons. First, identification of H4 lysine 91 acetylation in yeast indicated that this is a highly conserved modification. Second, the association of histone H4 acetylated on lysine 91 with proteins involved in histone deposition suggested that this modification occurs prior to chromatin assembly. This led Ye et al. (36) to propose that the dynamic nature of acetylation at this residue might modulate dimer–tetramer interactions during chromatin assembly. Whereby acetylation of H4 lysine 91 would prevent salt-bridge formation and weaken octamer stability, removal of the acetyl group from this site following histone deposition would allow salt bridge formation and stabilization of the histone octamer.
To investigate this possibility, an H4 K91A point mutant was generated and studied for various phenotypic effects. H4 K91A mutants were shown to be deficient in repair of MMS, CPT and HU-induced double-strand DNA damage as well as UV-induced single-strand DNA damage. By combining the H4 K91A allele with the deletion of proteins involved in cell cycle checkpoint (Mec1p and Mec3p), NHEJ (Yku70p) or homologous recombination (Rad52p), they concluded that K91A was not directly involved in the process of repairing the DNA lesion. However, the H4 K91A allele did not increase the DNA damage sensitivity of mutants lacking the histone chaperones Asf1p and Cac1p consistent with a role for H4 lysine 91 in DNA repair linked chromatin assembly (36). In a separate study, Hyland et al. (35) reported that H4 K91 mutants did not display sensitivity to MMS or CPT, but were slightly sensitive to HU. It is not known whether comparable concentrations of DNA damaging agents were tested in these studies. As DNA damage sensitivity phenotypes can be highly concentration dependent, this may explain the conflicting results. In addition, strain specific differences in DNA damage sensitivity could also be a factor.
Areas of heterochromatin are often sensitive to defects in chromatin assembly (94–102). For example, many mutants that are deleted for one or more assembly factors display defects in telomeric silencing. Consistent with this, H4 K91A mutants displayed a general up regulation of telomere-proximal genes with the telomeric chromatin acquiring euchromatic features such as loss of Sir2p, increased H3 lysine 79 methylation and histone H4 hyperacetylation (36).
Biochemical evidence also supports the model that histone H4 lysine 91 acetylation functions by physically altering the structure of the nucleosome by interfering with dimer–tetramer interactions. For example, chromatin isolated from H4 K91A cells was more rapidly digested by micrococcal nuclease (MNase), which normally cuts DNA that is not fully protected by the octamer. In addition, H2A/H2B dimers were more easily displaced by salt from chromatin with a mutation of histone H4 lysine 91 (36).
Taken together, the data suggest that histone H4 lysine 91 acetylation occurs on molecules prior to histone deposition. The presence of the acetyl group on H4 lysine 91 (and the concomitant charge neutralization) would interfere with the stable association of H2A/H2B dimers with the H3/H4 tetramer. At the appropriate time, deacetylation of H4 lysine 91 would permit completion of the histone octamer and allow chromatin assembly to proceed. In addition to chromatin assembly, histone H4 lysine 91 acetylation might also play a role in gene expression by preserving the open chromatin structure that can be generated by the release of H2A/H2B dimers during transcription. Transcriptional repression would then be accompanied by lysine 91 deacetylation and reassembly of the histone octamer.
Histone H4 arginine 92 was identified by mass spectroscopy as a site of methylation in bovine histones (22). Located adjacent to histone H4 lysine 91, arginine 92 is also closely juxtaposed to a glutamic acid residue on histone H2B (E73) but, intriguingly, on the other dimer (Figure 3B). In yeast, H4 R92K and R92A point mutants were defective in telomeric silencing and repair of HU-induced DNA damage (35). This suggests that either the arginine residue itself or the ability to modulate between the modified and unmodified states is important for these processes. A more complete characterization may reveal that, like lysine 91, arginine 92 plays a role in nucleosome stability by mediating dimer–tetramer interactions.
The final core domain modification that has the potential to influence histone–histone interactions is histone H2A lysine 99 methylation. This modification was identified by mass spectrometry in bovine histones (22). While the residue at this position is an arginine (also a potential methylation site) in some organisms, it is not conserved in budding yeast. Hence, there are no in vivo clues as to the function of this modification. This modification occurs at an intriguing site at the center of the histone octamer with the two lysine 99 residues oriented toward each other (Figure 3B). Hence, they are unlikely to be accessible from the surface of the nucleosome and are clearly not in a position to influence histone–DNA interactions. Therefore, one possible mechanism by which this modification might act is through altering histone–histone packing at the center of the octamer.
SUMMARY AND OUTLOOK
The application of mass spectrometry to the study of histone post-translational modifications has allowed for the discovery of a new class of histone modifications located outside of the unstructured N-terminal tails. These core domain modifications are conserved across a wide range of organisms. Two residues among these newly identified sites of modification, H3 arginine 52 and T118, are required for viability in yeast and other sites of histone core modification have been shown to have dramatic affects on DNA-dependent processes such as transcription, chromatin assembly, and DNA damage repair.
For the purpose of this review, we divided these core modifications into three classes based their locations on the nucleosome. The experimental data available to date surrounding these modifications supports the idea that the location of a modification can be used to predict functional roles. Modifications on the solute accessible face appear to exert their affects on transcription, heterochromatic gene silencing, and DNA repair by modulating histone–protein interactions and higher order chromatin structure.
Modifications located on the histone lateral surface affect transcription, chromatin assembly, and DNA damage repair by mediating histone–DNA interactions. Among the lateral surface modifications, there are several residues positioned at the DNA entry–exit point that have been proposed to play a role in DNA breathing. It is clear that the residues directly involved in DNA binding are important for histone–DNA interactions. The role of modifiable residues that do not directly contact the DNA is slightly less obvious. Though not in direct contact with the DNA, these residues could mediate histone–DNA interactions indirectly, perhaps through intervening water molecules (103). Alternatively, these residues could be important for protein recruitment to the lateral surface. In general, lateral surface modifications may function to keep chromatin association with histones weakened in order to allow access by transcriptional or DNA damage repair machinery.
Core modifications located within the histone–histone interfaces are distinctly capable of regulating nucleosome stability. These modifications affect chromatin assembly, heterochromatic silencing and DNA damage repair, by modulating nucleosome structure. Specifically, the modified states may weaken intranucleosomal interactions to allow access by DNA damage repair machinery and to allow orderly assembly of nucleosomes onto the DNA following transcription, DNA repair or replication.
The first core modification to be discovered, H3 lysine 79, has also been the most extensively characterized. Histone H3 lysine 79 methylation is unique in that the enzyme responsible for this modification, Dot1, has been clearly demonstrated both in vivo and in vitro. The next major advance in our understanding of histone core domain modifications will be identification of the modifying enzymes that act on the other sites in the histone core domains. It will be interesting to see whether novel enzymes will be uncovered or whether the same enzymes that act on the N-terminal tails will also be able to accommodate the very different substrate topology found in the histone core domains.
The existence of histone core domain modifications was first reported in 2002. In the short time since their discovery, both their abundance and importance have increased significantly. Based on the data available to date, these modifications appear to play diverse roles in the regulation of chromatin structure. As their characterization continues, the tale of these modifications is certain to make for interesting reading.
Acknowledgments
Work in the authors laboratory is supported by the National Institutes of Health (R01 GM62970) and the V Foundation for Cancer Research. Funding to pay the Open Access publication charges for this article was waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Murray K. The occurrence of epsilon-N-methyl lysine in histones. Biochemistry. 1964;127:10–15. doi: 10.1021/bi00889a003. [DOI] [PubMed] [Google Scholar]
- 2.Kleinsmith L.J., Allfrey V.G., Mirsky A.E. Phosphoprotein metabolism in isolated lymphocyte nuclei. Proc. Natl Acad. Sci. USA. 1966;55:1182–1189. doi: 10.1073/pnas.55.5.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ord M.G., Stocken L.A. Phosphate and thiol groups in histone f3 from rat liver and thymus nuclei. Biochem. J. 1967;102:631–636. doi: 10.1042/bj1020631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeLange R.J., Fambrough D.M., Smith E.L., Bonner J. Calf and pea histone IV. I. Amino acid compositions and the identical COOH-terminal 19-residue sequence. J. Biol. Chem. 1968;243:5906–5913. [PubMed] [Google Scholar]
- 5.Gershey E.L., Vidali G., Allfrey V.G. Chemical studies of histone acetylation. The occurrence of epsilon-N-acetyllysine in the f2a1 histone. J. Biol. Chem. 1968;243:5018–5022. [PubMed] [Google Scholar]
- 6.Vidali G., Gershey E.L., Allfrey V.G. Chemical studies of histone acetylation. The distribution of epsilon-N-acetyllysine in calf thymus histones. J. Biol. Chem. 1968;243:6361–6366. [PubMed] [Google Scholar]
- 7.Goldknopf I.L., Taylor C.W., Baum R.M., Yeoman L.C., Olson M.O., Prestayko A.W., Busch H. Isolation and characterization of protein A24, a ‘histone-like’ non-histone chromosomal protein. J. Biol. Chem. 1975;250:7182–7187. [PubMed] [Google Scholar]
- 8.Ueda K., Omachi A., Kawaichi M., Hayaishi O. Natural occurrence of poly(ADP-ribosyl) histones in rat liver. Proc. Natl Acad. Sci. USA. 1975;72:205–209. doi: 10.1073/pnas.72.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shiio Y., Eisenman R.N. Histone sumoylation is associated with transcriptional repression. Proc. Natl Acad. Sci. USA. 2003;100:13225–13230. doi: 10.1073/pnas.1735528100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luger K., Mader A.W., Richmond R.K., Sargent D.F., Richmond T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 11.Grant P.A. A tale of histone modifications. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-4-reviews0003. REVIEWS0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goll M.G., Bestor T.H. Histone modification and replacement in chromatin activation. Genes Dev. 2002;16:1739–1742. doi: 10.1101/gad.1013902. [DOI] [PubMed] [Google Scholar]
- 13.Turner B.M. Cellular memory and the histone code. Cell. 2002;111:285–291. doi: 10.1016/s0092-8674(02)01080-2. [DOI] [PubMed] [Google Scholar]
- 14.Martin C., Zhang Y. The diverse functions of histone lysine methylation. Nature Rev. Mol. Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 15.Grewal S.I., Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- 16.Iizuka M., Smith M.M. Functional consequences of histone modifications. Curr. Opin. Genet. Dev. 2003;13:154–160. doi: 10.1016/s0959-437x(03)00020-0. [DOI] [PubMed] [Google Scholar]
- 17.Jenuwein T., Allis C.D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 18.van Leeuwen F., Gafken P.R., Gottschling D.E. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–756. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- 19.Ng H.H., Feng Q., Wang H., Erdjument-Bromage H., Tempst P., Zhang Y., Struhl K. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 2002;16:1518–1527. doi: 10.1101/gad.1001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cocklin R.R., Wang M. Identification of methylation and acetylation sites on mouse histone H3 using matrix-assisted laser desorption/ionization time-of-flight and nanoelectrospray ionization tandem mass spectrometry. J. Protein Chem. 2003;22:327–334. doi: 10.1023/a:1025334006014. [DOI] [PubMed] [Google Scholar]
- 21.Zhang K., Tang H., Huang L., Blankenship J.W., Jones P.R., Xiang F., Yau P.M., Burlingame A.L. Identification of acetylation and methylation sites of histone H3 from chicken erythrocytes by high-accuracy matrix-assisted laser desorption ionization-time-of-flight, matrix-assisted laser desorption ionization-postsource decay, and nanoelectrospray ionization tandem mass spectrometry. Anal. Biochem. 2002;306:259–269. doi: 10.1006/abio.2002.5719. [DOI] [PubMed] [Google Scholar]
- 22.Zhang L., Eugeni E.E., Parthun M.R., Freitas M.A. Identification of novel histone post-translational modifications by peptide mass fingerprinting. Chromosoma. 2003;112:77–86. doi: 10.1007/s00412-003-0244-6. [DOI] [PubMed] [Google Scholar]
- 23.Freitas M.A., Sklenar A.R., Parthun M.R. Application of mass spectrometry to the identification and quantification of histone post-translational modifications. J. Cell Biochem. 2004;92:691–700. doi: 10.1002/jcb.20106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cosgrove M.S., Boeke J.D., Wolberger C. Regulated nucleosome mobility and the histone code. Nature Struct. Mol. Biol. 2004;11:1037–1043. doi: 10.1038/nsmb851. [DOI] [PubMed] [Google Scholar]
- 25.Ma X.J., Wu J., Altheim B.A., Schultz M.C., Grunstein M. Deposition-related sites K5/K12 in histone H4 are not required for nucleosome deposition in yeast. Proc. Natl Acad. Sci. USA. 1998;95:6693–6698. doi: 10.1073/pnas.95.12.6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Megee P.C., Morgan B.A., Mittman B.A., Smith M.M. Genetic analysis of histone H4: essential role of lysines subject to reversible acetylation. Science. 1990;247:841–845. doi: 10.1126/science.2106160. [DOI] [PubMed] [Google Scholar]
- 27.Megee P.C., Morgan B.A., Smith M.M. Histone H4 and the maintenance of genome integrity. Genes Dev. 1995;9:1716–1727. doi: 10.1101/gad.9.14.1716. [DOI] [PubMed] [Google Scholar]
- 28.Thompson J.S., Ling X., Grunstein M. Histone H3 amino terminus is required for telomeric and silent mating locus repression in yeast. Nature. 1994;369:245–247. doi: 10.1038/369245a0. [DOI] [PubMed] [Google Scholar]
- 29.Park E.C., Szostak J.W. Point mutations in the yeast histone H4 gene prevent silencing of the silent mating type locus HML. Mol. Cell. Biol. 1990;10:4932–4934. doi: 10.1128/mcb.10.9.4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kelly T.J., Qin S., Gottschling D.E., Parthun M.R. Type B histone acetyltransferase Hat1p participates in telomeric silencing. Mol. Cell. Biol. 2000;20:7051–7058. doi: 10.1128/mcb.20.19.7051-7058.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qin S., Parthun M.R. Histone H3 and the histone acetyltransferase Hat1p contribute to DNA double-strand break repair. Mol. Cell. Biol. 2002;22:8353–8365. doi: 10.1128/MCB.22.23.8353-8365.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Masumoto H., Hawke D., Kobayashi R., Verreault A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature. 2005;436:294–298. doi: 10.1038/nature03714. [DOI] [PubMed] [Google Scholar]
- 33.Ozdemir A., Spicuglia S., Lasonder E., Vermeulen M., Campsteijn C., Stunnenberg H.G., Logie C. Characterization of lysine 56 of histone H3 as an acetylation site in Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:25949–25952. doi: 10.1074/jbc.C500181200. [DOI] [PubMed] [Google Scholar]
- 34.Xu F., Zhang K., Grunstein M. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell. 2005;121:375–385. doi: 10.1016/j.cell.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 35.Hyland E.M., Cosgrove M.S., Molina H., Wang D., Pandey A., Cottee R.J., Boeke J.D. Insights into the role of histone H3 and histone H4 core modifiable residues in Saccharomyces cerevisiae. Mol. Cell. Biol. 2005;25:10060–10070. doi: 10.1128/MCB.25.22.10060-10070.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ye J., Ai X., Eugeni E.E., Zhang L., Carpenter L.R., Jelinek M.A., Freitas M.A., Parthun M.R. Histone H4 lysine 91 acetylation a core domain modification associated with chromatin assembly. Mol. Cell. 2005;18:123–130. doi: 10.1016/j.molcel.2005.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schalch T., Duda S., Sargent D.F., Richmond T.J. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature. 2005;436:138–141. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
- 38.Ng H.H., Ciccone D.N., Morshead K.B., Oettinger M.A., Struhl K. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: a potential mechanism for position-effect variegation. Proc. Natl Acad. Sci. USA. 2003;100:1820–1825. doi: 10.1073/pnas.0437846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feng Q., Wang H., Ng H.H., Erdjument-Bromage H., Tempst P., Struhl K., Zhang Y. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 2002;12:1052–1058. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- 40.Lacoste N., Utley R.T., Hunter J.M., Poirier G.G., Cote J. Disruptor of telomeric silencing-1 is a chromatin-specific histone H3 methyltransferase. J. Biol. Chem. 2002;277:30421–30424. doi: 10.1074/jbc.C200366200. [DOI] [PubMed] [Google Scholar]
- 41.Park J.H., Cosgrove M.S., Youngman E., Wolberger C., Boeke J.D. A core nucleosome surface crucial for transcriptional silencing. Nature Genet. 2002;32:273–279. doi: 10.1038/ng982. [DOI] [PubMed] [Google Scholar]
- 42.Rusche L.N., Kirchmaier A.L., Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- 43.Hecht A., Laroche T., Strahl-Bolsinger S., Gasser S.M., Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- 44.Strahl-Bolsinger S., Hecht A., Luo K., Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- 45.Grunstein M. Yeast heterochromatin: regulation of its assembly and inheritance by histones. Cell. 1998;93:325–328. doi: 10.1016/s0092-8674(00)81160-5. [DOI] [PubMed] [Google Scholar]
- 46.Rusche L.N., Kirchmaier A.L., Rine J. Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol. Biol. Cell. 2002;13:2207–2222. doi: 10.1091/mbc.E02-03-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meijsing S.H., Ehrenhofer-Murray A.E. The silencing complex SAS-I links histone acetylation to the assembly of repressed chromatin by CAF-I and Asf1 in Saccharomyces cerevisiae. Genes Dev. 2001;15:3169–3182. doi: 10.1101/gad.929001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kimura A., Umehara T., Horikoshi M. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nature Genet. 2002;32:370–377. doi: 10.1038/ng993. [DOI] [PubMed] [Google Scholar]
- 49.Suka N., Luo K., Grunstein M. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nature Genet. 2002;32:378–383. doi: 10.1038/ng1017. [DOI] [PubMed] [Google Scholar]
- 50.Bryk M., Banerjee M., Murphy M., Knudsen K.E., Garfinkel D.J., Curcio M.J. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 1997;11:255–269. doi: 10.1101/gad.11.2.255. [DOI] [PubMed] [Google Scholar]
- 51.Smith J.S., Boeke J.D. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 1997;11:241–254. doi: 10.1101/gad.11.2.241. [DOI] [PubMed] [Google Scholar]
- 52.Fritze C.E., Verschueren K., Strich R., Easton Esposito R. Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J. 1997;16:6495–6509. doi: 10.1093/emboj/16.21.6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boeke J.D., Trueheart J., Natsoulis G., Fink G.R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 54.Singer M.S., Kahana A., Wolf A.J., Meisinger L.L., Peterson S.E., Goggin C., Mahowald M., Gottschling D.E. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics. 1998;150:613–632. doi: 10.1093/genetics/150.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Leeuwen F., Gottschling D.E. Genome-wide histone modifications: gaining specificity by preventing promiscuity. Curr. Opin. Cell Biol. 2002;14:756–762. doi: 10.1016/s0955-0674(02)00393-9. [DOI] [PubMed] [Google Scholar]
- 56.Dlakic M. Chromatin silencing protein and pachytene checkpoint regulator Dot1p has a methyltransferase fold. Trends Biochem. Sci. 2001;26:405–407. doi: 10.1016/s0968-0004(01)01856-4. [DOI] [PubMed] [Google Scholar]
- 57.Yeates T.O. Structures of SET domain proteins: protein lysine methyltransferases make their mark. Cell. 2002;111:5–7. doi: 10.1016/s0092-8674(02)01010-3. [DOI] [PubMed] [Google Scholar]
- 58.Jenuwein T., Laible G., Dorn R., Reuter G. SET domain proteins modulate chromatin domains in eu- and heterochromatin. Cell Mol. Life Sci. 1998;54:80–93. doi: 10.1007/s000180050127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Min J., Feng Q., Li Z., Zhang Y., Xu R.M. Structure of the catalytic domain of human DOT1L, a non-SET domain nucleosomal histone methyltransferase. Cell. 2003;112:711–723. doi: 10.1016/s0092-8674(03)00114-4. [DOI] [PubMed] [Google Scholar]
- 60.Sawada K., Yang Z., Horton J.R., Collins R.E., Zhang X., Cheng X. Structure of the conserved core of the yeast Dot1p, a nucleosomal histone H3 lysine 79 methyltransferase. J. Biol. Chem. 2004;279:43296–43306. doi: 10.1074/jbc.M405902200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Okada Y., Feng Q., Lin Y., Jiang Q., Li Y., Coffield V.M., Su L., Xu G., Zhang Y. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121:167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 62.San-Segundo P.A., Roeder G.S. Role for the silencing protein Dot1 in meiotic checkpoint control. Mol. Biol. Cell. 2000;11:3601–3615. doi: 10.1091/mbc.11.10.3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Giannattasio M., Lazzaro F., Plevani P., Muzi-Falconi M. The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6-Bre1 and H3 methylation by Dot1. J. Biol. Chem. 2005;280:9879–9886. doi: 10.1074/jbc.M414453200. [DOI] [PubMed] [Google Scholar]
- 64.Wysocki R., Javaheri A., Allard S., Sha F., Cote J., Kron S.J. Role of Dot1-dependent histone H3 methylation in G1 and S phase DNA damage checkpoint functions of Rad9. Mol. Cell. Biol. 2005;25:8430–8443. doi: 10.1128/MCB.25.19.8430-8443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Selenko P., Sprangers R., Stier G., Buhler D., Fischer U., Sattler M. SMN tudor domain structure and its interaction with the Sm proteins. Nature Struct. Biol. 2001;8:27–31. doi: 10.1038/83014. [DOI] [PubMed] [Google Scholar]
- 66.Sprangers R., Groves M.R., Sinning I., Sattler M. High-resolution X-ray and NMR structures of the SMN Tudor domain: conformational variation in the binding site for symmetrically dimethylated arginine residues. J. Mol. Biol. 2003;327:507–520. doi: 10.1016/s0022-2836(03)00148-7. [DOI] [PubMed] [Google Scholar]
- 67.Friesen W.J., Massenet S., Paushkin S., Wyce A., Dreyfuss G. SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets. Mol. Cell. 2001;7:1111–1117. doi: 10.1016/s1097-2765(01)00244-1. [DOI] [PubMed] [Google Scholar]
- 68.Brahms H., Meheus L., de Brabandere V., Fischer U., Luhrmann R. Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B′ and the Sm-like protein LSm4, and their interaction with the SMN protein. RNA. 2001;7:1531–1542. doi: 10.1017/s135583820101442x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huyen Y., Zgheib O., Ditullio R.A., Jr, Gorgoulis V.G., Zacharatos P., Petty T.J., Sheston E.A., Mellert H.S., Stavridi E.S., Halazonetis T.D. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- 70.Polach K.J., Widom J. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J. Mol. Biol. 1995;254:130–149. doi: 10.1006/jmbi.1995.0606. [DOI] [PubMed] [Google Scholar]
- 71.Polach K.J., Widom J. A model for the cooperative binding of eukaryotic regulatory proteins to nucleosomal target sites. J. Mol. Biol. 1996;258:800–812. doi: 10.1006/jmbi.1996.0288. [DOI] [PubMed] [Google Scholar]
- 72.Protacio R.U., Polach K.J., Widom J. Coupled-enzymatic assays for the rate and mechanism of DNA site exposure in a nucleosome. J. Mol. Biol. 1997;274:708–721. doi: 10.1006/jmbi.1997.1440. [DOI] [PubMed] [Google Scholar]
- 73.Anderson J.D., Widom J. Sequence and position-dependence of the equilibrium accessibility of nucleosomal DNA target sites. J. Mol. Biol. 2000;296:979–987. doi: 10.1006/jmbi.2000.3531. [DOI] [PubMed] [Google Scholar]
- 74.Polach K.J., Lowary P.T., Widom J. Effects of core histone tail domains on the equilibrium constants for dynamic DNA site accessibility in nucleosomes. J. Mol. Biol. 2000;298:211–223. doi: 10.1006/jmbi.2000.3644. [DOI] [PubMed] [Google Scholar]
- 75.Anderson J.D., Lowary P.T., Widom J. Effects of histone acetylation on the equilibrium accessibility of nucleosomal DNA target sites. J. Mol. Biol. 2001;307:977–985. doi: 10.1006/jmbi.2001.4528. [DOI] [PubMed] [Google Scholar]
- 76.Anderson J.D., Thastrom A., Widom J. Spontaneous access of proteins to buried nucleosomal DNA target sites occurs via a mechanism that is distinct from nucleosome translocation. Mol. Cell. Biol. 2002;22:7147–7157. doi: 10.1128/MCB.22.20.7147-7157.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Miller J.A., Widom J. Collaborative competition mechanism for gene activation in vivo. Mol. Cell. Biol. 2003;23:1623–1632. doi: 10.1128/MCB.23.5.1623-1632.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li G., Widom J. Nucleosomes facilitate their own invasion. Nature Struct. Mol. Biol. 2004;11:763–769. doi: 10.1038/nsmb801. [DOI] [PubMed] [Google Scholar]
- 79.Li G., Levitus M., Bustamante C., Widom J. Rapid spontaneous accessibility of nucleosomal DNA. Nature Struct. Mol. Biol. 2005;12:46–53. doi: 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
- 80.Luger K., Richmond T.J. DNA binding within the nucleosome core. Curr. Opin. Struct. Biol. 1998;8:33–40. doi: 10.1016/s0959-440x(98)80007-9. [DOI] [PubMed] [Google Scholar]
- 81.Kruger W., Peterson C.L., Sil A., Coburn C., Arents G., Moudrianakis E.N., Herskowitz I. Amino acid substitutions in the structured domains of histones H3 and H4 partially relieve the requirement of the yeast SWI/SNF complex for transcription. Genes Dev. 1995;9:2770–2779. doi: 10.1101/gad.9.22.2770. [DOI] [PubMed] [Google Scholar]
- 82.Fleming A.B., Pennings S. Antagonistic remodelling by Swi-Snf and Tup1-Ssn6 of an extensive chromatin region forms the background for FLO1 gene regulation. EMBO J. 2001;20:5219–5231. doi: 10.1093/emboj/20.18.5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kruger W., Herskowitz I. A negative regulator of HO transcription, SIN1 (SPT2), is a nonspecific DNA-binding protein related to HMG1. Mol. Cell. Biol. 1991;11:4135–4146. doi: 10.1128/mcb.11.8.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Horn P.J., Crowley K.A., Carruthers L.M., Hansen J.C., Peterson C.L. The SIN domain of the histone octamer is essential for intramolecular folding of nucleosomal arrays. Nature Struct. Biol. 2002;9:167–171. doi: 10.1038/nsb762. [DOI] [PubMed] [Google Scholar]
- 85.Muthurajan U.M., Bao Y., Forsberg L.J., Edayathumangalam R.S., Dyer P.N., White C.L., Luger K. Crystal structures of histone Sin mutant nucleosomes reveal altered protein–DNA interactions. EMBO J. 2004;23:260–271. doi: 10.1038/sj.emboj.7600046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Flaus A., Rencurel C., Ferreira H., Wiechens N., Owen-Hughes T. Sin mutations alter inherent nucleosome mobility. EMBO J. 2004;23:343–353. doi: 10.1038/sj.emboj.7600047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kurumizaka H., Wolffe A.P. Sin mutations of histone H3: influence on nucleosome core structure and function. Mol. Cell. Biol. 1997;17:6953–6969. doi: 10.1128/mcb.17.12.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhou H., Madden B.J., Muddiman D.C., Zhang Z. Chromatin assembly factor 1 interacts with histone h3 methylated at lysine 79 in the processes of epigenetic silencing and DNA repair. Biochemistry. 2006;45:2852–2861. doi: 10.1021/bi0521083. [DOI] [PubMed] [Google Scholar]
- 89.Mendiratta G., Eriksson P.R., Shen C.H., Clark D.J. The DNA-binding domain of the yeast Spt10p activator includes a zinc finger that is homologous to foamy virus integrase. J. Biol. Chem. 2006;281:7040–7048. doi: 10.1074/jbc.M511416200. [DOI] [PubMed] [Google Scholar]
- 90.Zhang L., Freitas M. Comparison of peptide mass mapping and electron capture dissociation as assays for histone posttranslational modifications. Int. J. Mass Spectrom. 2004;234:213–225. [Google Scholar]
- 91.Santisteban M.S., Arents G., Moudrianakis E.N., Smith M.M. Histone octamer function in vivo: mutations in the dimer–tetramer interfaces disrupt both gene activation and repression. EMBO J. 1997;16:2493–2506. doi: 10.1093/emboj/16.9.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ai X., Parthun M.R. The nuclear Hat1p/Hat2p complex: a molecular link between type B histone acetyltransferases and chromatin assembly. Mol. Cell. 2004;14:195–205. doi: 10.1016/s1097-2765(04)00184-4. [DOI] [PubMed] [Google Scholar]
- 93.Poveda A., Pamblanco M., Tafrov S., Tordera V., Sternglanz R., Sendra R. Hif1 is a component of yeast histone acetyltransferase B, a complex mainly localized in the nucleus. J. Biol. Chem. 2004;279:16033–16043. doi: 10.1074/jbc.M314228200. [DOI] [PubMed] [Google Scholar]
- 94.Adams C.R., Kamakaka R.T. Chromatin assembly: biochemical identities and genetic redundancy. Curr. Opin. Genet. Dev. 1999;9:185–190. doi: 10.1016/S0959-437X(99)80028-8. [DOI] [PubMed] [Google Scholar]
- 95.Kaufman P.D., Cohen J.L., Osley M.A. Hir proteins are required for position-dependent gene silencing in Saccharomyces cerevisiae in the absence of chromatin assembly factor I. Mol. Cell. Biol. 1998;18:4793–4806. doi: 10.1128/mcb.18.8.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kaufman P.D., Kobayashi R., Stillman B. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev. 1997;11:345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
- 97.Enomoto S., Berman J. Chromatin assembly factor I contributes to the maintenance, but not the re-establishment, of silencing at the yeast silent mating loci. Genes Dev. 1998;12:219–232. doi: 10.1101/gad.12.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Enomoto S., McCune-Zierath P.D., Gerami-Nejad M., Sanders M.A., Berman J. RLF2, a subunit of yeast chromatin assembly factor-I, is required for telomeric chromatin function in vivo. Genes Dev. 1997;11:358–370. doi: 10.1101/gad.11.3.358. [DOI] [PubMed] [Google Scholar]
- 99.Monson E.K., de Bruin D., Zakian V.A. The yeast Cac1 protein is required for the stable inheritance of transcriptionally repressed chromatin at telomeres. Proc. Natl Acad. Sci. USA. 1997;94:13081–13086. doi: 10.1073/pnas.94.24.13081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sharp J.A., Fouts E.T., Krawitz D.C., Kaufman P.D. Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr. Biol. 2001;11:463–473. doi: 10.1016/s0960-9822(01)00140-3. [DOI] [PubMed] [Google Scholar]
- 101.Krawitz D.C., Kama T., Kaufman P.D. Chromatin assembly factor I mutants defective for PCNA binding require Asf1/Hir proteins for silencing. Mol. Cell. Biol. 2002;22:614–625. doi: 10.1128/MCB.22.2.614-625.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pirrotta V., Gross D.S. Epigenetic silencing mechanisms in budding yeast and fruit fly: different paths, same destinations. Mol. Cell. 2005;18:395–398. doi: 10.1016/j.molcel.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 103.Davey C.A., Sargent D.F., Luger K., Maeder A.W., Richmond T.J. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J. Mol. Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]