Abstract
Activation of the noncanonical NF-κB signaling pathway involved in the proteolytic processing of NF-κB p100 to p52 is tightly regulated, and overproduction of p52 leads to lymphocyte hyperplasia and transformation. We have demonstrated that active but not latent Stat3, expressed in many types of human cancers involved in cell proliferation and survival, induces p100 processing to p52 by activation of IKKα and subsequent phosphorylation of p100. The Stat3-mediated p100 processing to p52 requires activation of Stat3 by the acetyltransferase activity of cAMP-response element-binding protein (CREB)-binding protein (CBP)/p300. A mutant of Stat3 defective in acetylation blocked Stat3-mediated p100 processing to p52 and acted as a dominant negative in blocking the production of p52. Furthermore, overexpression of p52 protected cells from apoptotic cell death. Thus, activation of the processing of p100 to p52 by Stat3 may represent one of the common pathways used by cancer cells to survive and escape therapy.
Keywords: apoptosis, cancer, cell signal, prostate
Stat3, a member of the signal transducers and activators of transcription (STAT) family, is a key signal transduction protein that mediates signaling by numerous cytokines, peptide growth factors, and oncoproteins (1). Upon stimulation, Stat3 can be activated by tyrosine or serine phosphorylation or acetylation (1–3). Accumulating evidence demonstrates that Stat3 activation plays important roles in cell differentiation, proliferation, development, apoptosis, and inflammation (4). Elevated activity of Stat3 has been found frequently in a wide variety of human tumors, including hematologic malignancies, head and neck cancer, breast cancer, and prostate cancer (4). Cell lines from multiple myelomas that have become growth-factor-independent require constitutively active Stat3 to protect against apoptosis (5). In addition, constitutively activated Stat3 induces cellular transformation in vitro and tumor formation in nude mice (6).
The NF-κB family of transcription factors consisting of RelA (p65), RelB, c-Rel, p50, and p52 plays a critical role in controlling expression of numerous genes that are involved in diverse processes, including inflammatory and immune responses, apoptosis, stress responses, malignant transformation, and tumor progression (7–11). Unlike RelA, RelB, and c-Rel proteins, which are directly synthesized as mature proteins, p50 and p52 are generated by proteolytic processing from their larger precursors NF-κB1 p105 and NF-κB2 p100, respectively (12, 13). The classical pathway of NF-κB activation involves the phosphorylation, subsequent ubiquitination, and degradation of inhibitors of NF-κB proteins (IκBs), which sequester p65 in the cytoplasm. Upon degradation of IκBs, the p65:p50 heterodimer translocates into the nucleus and activates target genes like IL-2, IL-6, and Bcl-2 (9, 11). The noncanonical NF-κB pathways that are involved in the processing of p100 to p52 require the recruitment of NF-κB-inducing kinase and subsequent activation of IκB kinase α (IKKα). IKKα phosphorylates p100 at two C-terminal serines, and, after ubiquitination and degradation of the ankyrin repeats, the subunit p52 is released. Whereas the processing of p105 to p50 is constitutive, the processing of p100 to p52 is a tightly controlled event in many cells and tissues (14–17).
Although processing of p100 to p52 is usually tightly regulated, this proteolytic processing can be activated by lymphotoxin β (18, 19), B cell-activating factor (20, 21), CD40 ligand (19, 22), and its cis-acting domain (17). Constitutive processing of p100 protein resulting in overexpression of p52 leads to lymphocyte hyperplasia and transformation (9, 22, 23). Here we show that active but not latent Stat3, expressed in many types of human cancers involved in cell proliferation and survival, induces p100 processing to p52 by activation of IKKα and subsequent phosphorylation of p100. Thus, activation of the processing of p100 to p52 by Stat3 may represent one of the common pathways used by cancer cells to survive and escape therapy.
Results
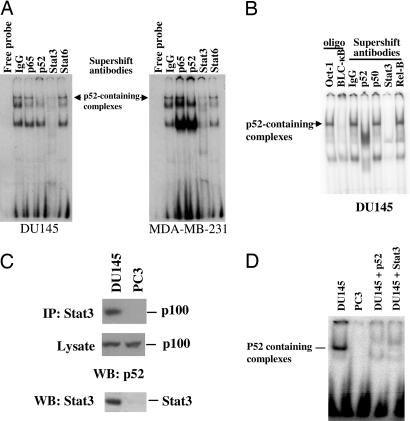
Stat3 Binds to p52 Complexes.
Once p52 is generated by proteolytic processing, the p52:RelB dimers undergo nuclear translocation and bind to the specific binding sites in the promoters of target genes. Such binding can be detected by EMSAs by using the consensus oligonucleotides for p52-containing dimers of NF-κB derived from the promoter region of the Blc gene, designated Blc-κB (24). To determine the transactivation ability of p52 in cancer cells, we performed EMSAs using radiolabeled Blc-κB with nuclear extracts from androgen-independent DU145 human prostate cancer cells. The p52 DNA-binding complex was detected in DU145 cells (Fig. 1A). Supershift analysis showed that antibodies against p52 could shift the complex, whereas antibodies against NF-κB p65, p50, or RelB failed to shift the complex (Fig. 1 A and B). Surprisingly, we found that Stat3 was present in the complex, as evidenced by the shifting of the p52-specific complex with anti-Stat3 antibodies (Fig. 1 A and B). In contrast, antibodies against Stat6 were unable to shift the complex (Fig. 1A). These results suggest that Stat3 but not Stat6 is associated with the NF-κB p52 DNA-binding complex. Similar results were observed in human MDA-MB-231 breast cancer cells. The p52 DNA-binding complex was detected in MDA-MB-231 cells by EMSA using Blc-κB as a probe. Antibodies against either p52 or Stat3 could shift the complex, whereas antibodies against p65 or Stat6 were not able to shift the complex (Fig. 1A), indicating that the presence of Stat3 in the p52 DNA-binding complex is not a cell-type-specific phenomenon.
Fig. 1.
NF-κB p52 DNA binding activity in DU145 and MDA-MB-231 cells that contains Stat3 protein in p52-containing complexes. (A) NF-κB p52 EMSAs were carried out with nuclear fractions generated from DU145 and MDA-MB-231 cells by using a probe (BLC-κB, 5′-GGGAGATTTG-3′) specifically bound by RelB:p52 dimers. Specificity of p52 DNA binding was confirmed by supershift with antibodies specifically against p52 or control antibodies (IgG). A complex formed by nuclear extracts from both DU145 and MDA-MB-231 cells was supershifted with antibodies against Stat3 but not with antibodies against either NF-κB p65 or Stat6. (B) NF-κB p52 EMSA was carried out with nuclear fractions generated from DU145 cells by using the BLC-κB probe. Specificity of p52 DNA binding was confirmed by competition with excess unlabeled Oct-1 oligonucleotides (Oligo, Oct-1) or BLC-κB oligonucleotides (Oligo, BLC-κB) or supershift with antibodies against p52, p50, Stat3, RelB, or control antibodies (IgG). (C) Stat3 is associated with p100 protein. Cell extracts from DU145 and PC3 cells were subjected to immunoprecipitation (IP) with antibodies against Stat3. Precipitates or lysates were separated by SDS/PAGE and analyzed by immunoblotting (WB) with antibodies against p100. Expression of Stat3 was determined by immunoblotting (WB) the cell extracts with antibodies against Stat3. (D) Stat3 is associated with p52. Cell extracts from DU145 and PC3 cells were subjected to immunoprecipitation with antibodies against Stat3. The eluted immunoprecipitants were used for EMSAs using radiolabeled Blc-κB probe. Specificity of p52 DNA binding was confirmed by supershift with the antibodies specifically against p52 and Stat3 in DU145 cells.
These results suggest that Stat3 is present in the DNA-binding complex formed by p52-containing NF-κB dimers. The p52 is generated by proteolytic processing from NF-κB p100. To determine whether Stat3 is directly associated with p100, coimmunoprecipitation assays were performed. Immunoprecipitation of protein extracts with anti-Stat3 antibody and subsequent Western blotting demonstrated that Stat3 is associated with p100 in human DU145 cells that express endogenous Stat3, but not in PC3 cells that lack endogenous Stat3 (Fig. 1C). To determine whether Stat3 is also associated with p52, protein extracts were immunoprecipitated with anti-Stat3 antibody and eluted and probed for the presence of p52 by EMSA by using radiolabeled Blc-κB probe. The p52 DNA-binding complex was detected in immunoprecipitants with anti-Stat3 antibodies in DU145 cells but not in PC3 cells (Fig. 1D). Collectively, these results suggest that Stat3 may bind to the precursor p100 protein and induce its processing. Stat3 then binds to the product p52 to assist in the activation of target genes.
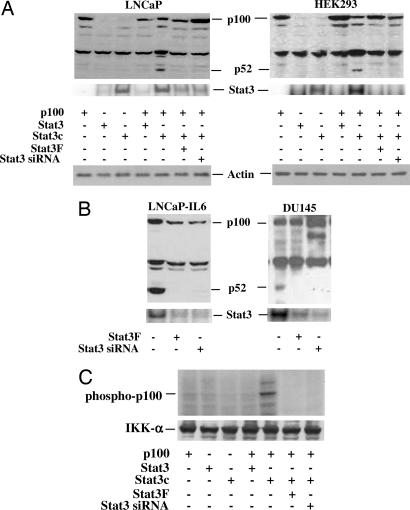
Stat3 Induces Processing of p100.
The presence of Stat3 in the p52 DNA-binding complex suggests that Stat3 may play a role in the induction of processing of p100 to p52 in cancer cells. We cotransfected plasmids expressing Stat3, a constitutively active form of Stat3 (Stat3c) (6), and a plasmid expressing WT p100 into LNCaP and HEK293 cells that lack endogenous p52 to determine whether Stat3 can induce the processing of p100. We found that constitutively active Stat3 (Stat3c) induced the processing of p100 to p52 in both LNCaP and HEK293 cells, whereas Stat3 by itself could not, suggesting that activated Stat3 is required to induce the processing of p100 to p52 (Fig. 2A). The activated Stat3-induced processing of p100 to p52 was blocked by using either Stat3 small interfering RNA (siRNA) (25) or a dominant-negative mutant of Stat3, Stat3F (26, 27) (Fig. 2 A and B).
Fig. 2.
Stat3 induces p52 production. (A) Active Stat3 induces p52 production in LNCaP and HEK293 cells. Cells were cotransfected with plasmids containing p100, Stat3, constitutively active Stat3 mutant (Stat3c), dominant-negative Stat3 mutant (Stat3F), or Stat3 siRNA. All transfections contained 2.0 μg of total plasmid DNA. After 48 h cells were lysed in RIPA buffer, and Western blot analyses were performed by using antibodies against p52. (B) Inhibition of p52 production by blocking Stat3 activation in LNCaP-IL6 and DU145 cells. LNCaP-IL6 and DU145 cells were transfected with dominant-negative Stat3 mutant (Stat3F), Stat3 siRNA, or control plasmid. All transfections contained 2 μg of total plasmid DNA. Cells were lysed after 48 h, and Western blot analyses were performed by using antibodies against p52. Stat3 activity was analyzed by EMSAs by using radiolabeled probe containing consensus Stat3 DNA binding sequence (5′-GATCCTTCTGGGAATTCCTAGATC). (C) Kinase assay to show the phosphorylation of p100 by IKKα. Lysates from Stat3, Sta3c, Stat3F, and Stat3 siRNA-transfected LNCaP cells were immunoprecipitated with anti-IKKα antibodies, and the immunoprecipitated enzyme was used to phosphorylate p100 in vitro. Reactions were stopped with SDS loading buffer after 30 min, loaded for SDS/PAGE, and transferred to a nitrocellulose membrane. The phosphorylated p100 was visualized by autoradiography. The membrane was reprobed with anti-IKK-α antibodies to normalize for equal amounts of kinase in each reaction.
The constitutively activated Stat3c was generated by point mutation of cysteine residues at A662 and N664 of the Stat3 molecule (6). This mutant Stat3c is a constitutively activated molecule that dimerizes spontaneously and binds to DNA and activates transcription (6). To determine whether endogenously activated Stat3 is able to induce the processing of p100 to p52, LNCaP cells stably transfected with IL-6 that express constitutively activated Stat3 were analyzed (28). The p52 protein was detected in IL-6-expressing and Stat3 active cells (Fig. 2B). Production of the p52 protein was abolished by using either Stat3 siRNA or the dominant-negative Stat3 mutant Stat3F (Fig. 2B). Similar results were observed in DU145 cells that express constitutively active Stat3 and p52, in which p52 levels were abolished by either Stat3 siRNA or Stat3F (Fig. 2B). These results demonstrate that activated Stat3 may play a direct role in the processing of p100 to p52.
Stat3 Induces p100 Phosphorylation by IKKα.
Because the processing of p100 to p52 requires the phosphorylation of p100 at the two C-terminal serines by activated IKKα (15), we determined whether Stat3 can induce the activation of IKKα and induce the phosphorylation of p100. We used an in vitro kinase assay in which the IKKα kinase is immunoprecipitated with specific antibodies against IKKα and used to phosphorylate p100 protein in vitro using [γ-32P]ATP. We found that Stat3c was able to induce the phosphorylation of p100 (Fig. 2C). To determine whether the activation of Stat3 was essential for the phosphorylation of p100, a dominant-negative mutant of Stat3 (Stat3F) and Stat3 siRNA were cotransfected with Stat3c into HEK293 cells. Stat3F and Stat3 siRNA nearly abolished the ability of Stat3c to induce phosphorylation of p100 (Fig. 2C), concordant with reduced Stat3c-induced p52 production by Stat3F and Stat3 siRNA (Fig. 2A). These results suggest that Stat3-induced p100 processing to p52 requires the phosphorylation of p100 by IKKα.
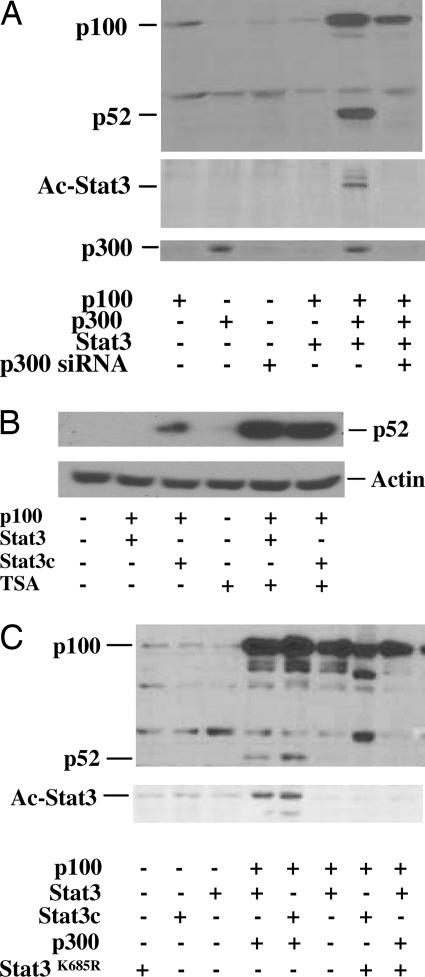
cAMP-Response Element-Binding Protein (CREB)-Binding Protein (CBP)/p300 Enhances Stat3-Mediated Processing of p100.
It has been demonstrated that the Stat3 C terminus can function as a transactivator capable of recruiting CBP/p300 coactivator, resulting in the acetylation and dimerization of Stat3 and activation of sequence-specific DNA binding and transcription (2, 3, 29, 30). Acetylation of Stat3 is important for Stat3 activation, nuclear translocation, and transactivational ability (2, 3). To determine whether CBP/p300 was critical for Stat3 to induce p100 processing to p52, LNCaP cells were cotransfected with p100, Stat3, and CBP/p300. Stat3 alone did not induce p100 processing to p52. However, cotransfection with Stat3 and CBP/p300 induced p100 processing to p52 that was abolished by CBP/p300 siRNA (Fig. 3A). CBP/p300 are important transcriptional coactivators exhibiting histone acetyltransferase activity that activates Stat3 by acetylation (3, 31, 32). To determine whether acetylation is required for the Stat3 activity of inducing p100 processing, the cell lysates were immunoprecipitated with antibody specifically against acetylated lysine and probed with antibody specifically against Stat3. Cotransfection of CBP/p300 with Stat3 was able to acetylate Stat3, resulting in the processing of p100 to p52 (Fig. 3A), indicating that Stat3-induced p100 processing to p52 requires CBP/p300-mediated acetylation.
Fig. 3.
Stat3 acetylation by CBP/p300 induces processing of p100 to p52 in LNCaP cells. (A) Cells were cotransfected with plasmids containing p100, Stat3, p300, or p300 siRNA (5′-GAGGATATTTCAGAGTCTA-3′). All transfections contained 2 μg of total plasmid DNA. Cells were lysed after 48 h, and Western blot analyses were performed by using antibodies against p52. Stat3 acetylation (Ac-Stat3) was detected by immunoprecipitation of the lysates with anti-Stat3 and Western blot with antibodies against acetylated lysine. (B) TSA enhances Stat3-induced processing of p100 to p52. HEK293 cells were cotransfected with p100 and Stat3 or Stat3c, and the cells were treated with or without 0.2 μM TSA. The cell extracts were used for Western blot analysis by using antibody against p52. (C) Stat3 defective for acetylation at lysine (Stat3K685R) lost the ability to induce processing of p100 to p52. LNCaP cells were cotransfected with plasmids containing p100, Stat3, Stat3c, p300, or Stat3K685R. Cells were lysed after 48 h, and Western blot was performed by using antibodies against p52. Stat3 acetylation (Ac-Stat3) was detected by immunoprecipitation of the lysates with anti-Stat3 and Western blot with antibodies against acetylated lysine.
Acetylation of Stat3 Enhances Processing of p100 to p52.
To examine whether acetylation enhances the processing of p100 to p52, HEK293 cells were cotransfected with p100 and Stat3 or Stat3c. The cells were treated with or without trichostatin A (TSA), a broad inhibitor of histone deacetylases. Whereas only Stat3c induced the processing of p100 to p52 in the absence of TSA, both Stat3 and Stat3c were able to induce the processing of p100 to p52 in the presence of TSA (Fig. 3B), suggesting that acetylation enhances the processing of p100 to p52. To confirm whether acetylation is essential for Stat3-mediated p100 processing to p52, we used a mutant of Stat3 that is defective for acetylation at lysine-685 (Stat3K685R) to examine whether it affects the production of p52 from p100 (2). When Stat3 acetylation was blocked by cotransfection of Stat3K685R with both CBP/p300 and p100 into LNCaP cells, there was no detectable production of p52 protein, corresponding to decreased levels of acetylated Stat3 (Fig. 3C). However, WT Stat3 cotransfected with p100 and CBP/p300 produced p52 protein. We cotransfected Stat3K685R with Stat3, CBP/p300, and p100 into LNCaP cells and found that Stat3K685R can inhibit p100 processing to p52, indicating that the Stat3K685R mutant might be capable of acting as a dominant negative for Stat3-induced production of p52. These results further confirm that acetylation of Stat3 is required for p100 processing to p52.
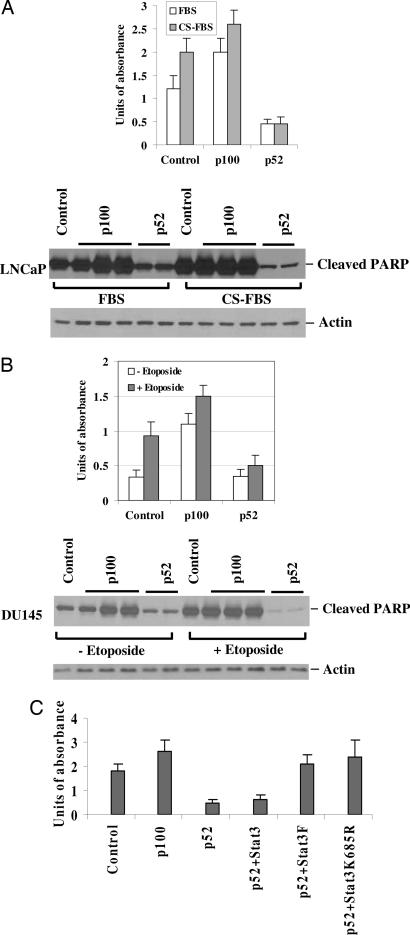
p52 Is an Antiapoptotic Factor.
p100 is a proapoptotic protein and acts as an IκB-like protein because of the presence of ankyrin repeats at the C terminus (33). To determine the potential significance of overexpression of p52 in cancer cells, we infected the androgen-sensitive LNCaP prostate cancer cells with adenoviruses encoding p100 and p52 and measured the effect on apoptosis. Expression of p100 considerably increased apoptotic cell death, whereas overexpression of p52 was able to reduce apoptotic cell death (Fig. 4A). Furthermore, overexpression of p52 could rescue LNCaP cells from androgen deprivation (charcoal-stripped serum condition)-induced apoptosis (Fig. 4A). To examine whether overexpression of p52 increases resistance to apoptotic cell death induced by chemotherapeutic agents, DU145 cells were treated with 50 μM etoposide for 24 h after infection with adenoviruses encoding p100 and p52. Etoposide induced massive apoptosis in DU145 cells, as indicated by the amount of poly(ADP-ribose)polymerase cleavage and cell death (Fig. 4B). Expression of p52 reduced the extent of apoptosis induced by etoposide (Fig. 4B). These results demonstrate that, whereas p100 is a proapoptotic protein, p52 is an antiapoptotic protein for prostate cancer cells. To examine whether Stat3 activation is required for p52-mediated antiapoptotic activity in prostate cancer cells, DU145 cells were transfected with p52 or cotransfected with latent Stat3, Stat3F, or Stat3K685R and treated with etoposide. Cotransfection of Stat3F or Stat3K685R almost abolished p52-induced antiapoptotic activity, whereas expression of latent Stat3 had no effect (Fig. 4C). These results suggest that the antiapoptotic activity of p52 is mediated by active Stat3.
Fig. 4.
p52 protects cells from apoptotic death. (A) p52 protects LNCaP cells from apoptotic death induced by androgen deprivation. LNCaP cells were infected with adenoviruses expressing p100 or p52. The cells were cultured in RPMI medium 1640 supplemented with 10% FBS or 10% charcoal-stripped FBS. The degree of apoptosis was measured by cell death detection ELISA according to the manufacturer's instructions (catalog no. 1544675, Roche, Indianapolis, IN). Protein fractions were prepared and used for Western blot analysis by using antibody against cleaved poly(ADP-ribose)polymerase. (B) p52 increases the resistance of DU145 cells to etoposide-induced apoptosis. DU145 cells were infected with adenoviruses containing p100 or p52 and then treated with or without 50 μM etoposide for 24 h. The degree of apoptosis was measured by cell death detection ELISA, and cleavage of poly(ADP-ribose)polymerase was confirmed by Western blot analysis. (C) Dominant-negative Stat3 and the Stat3 mutant defective in acetylation abolish the antiapoptotic activity of p52. DU145 cells were transfected with p100 or p52 or cotransfected with Stat3F or Stat3K685R as indicated. The cells were then treated with 50 μM etoposide for 24 h. The degree of apoptosis was measured by cell death detection ELISA.
Discussion
The p100 processing event is highly regulated in normal cells and is dysregulated in transformed cells, even though the factors contributing to constitutive processing are still unknown. Here we show that p100 is processed to p52 by activated Stat3 in both prostate and breast cancer cells. Our findings provide evidence for a mechanism of p100 processing in which activated Stat3 induces the processing of p100 to p52 that requires CBP/p300-mediated acetylation. Furthermore, we show that p52 protein is antiapoptotic in prostate cancer cells and promotes survival upon treatment with androgen deprivation or with chemotherapeutic agents.
In this study we demonstrate that active but not latent Stat3 induces p100 processing. This finding is based on the observation that a dominant-negative Stat3 mutant (Stat3F), a non-tyrosine-phosphorylatable Stat3 (Y705F), blocked active Stat3-induced p100 processing (Fig. 2), suggesting that this process requires active Stat3 by tyrosine phosphorylation. Furthermore, Stat3-mediated p100 processing depends on Stat3 acetylation. Two lines of evidence support this observation. (i) CBP/p300 activated Stat3 by acetylation and induced p100 processing (Fig. 3A). (ii) A mutant of Stat3 that is defective for acetylation at lysine-685 blocked active Stat3-induced p100 processing (Fig. 3C). Therefore, Stat3-mediated p100 processing requires Stat3 activation in a tyrosine phosphorylation- and acetylation-dependent manner.
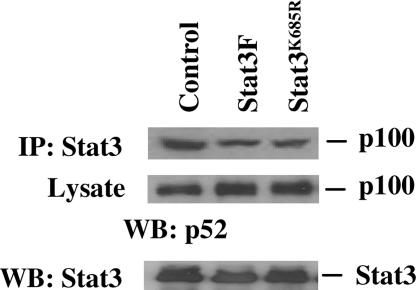
Coimmunoprecipitation assays demonstrated that Stat3 and p100 form a complex (Fig. 1C), whereas EMSAs suggested that Stat3 is also associated with p52 (Fig. 1D). Formation of the Stat3–p100 complex seems to depend on neither tyrosine phosphorylation nor acetylation of Stat3, because neither Stat3F nor Stat3K685R abolishes the association of Stat3 with p100 in DU145 cells (Fig. 5). These results suggest that, although both the latent and active forms of Stat3 interact with p100, only the active Stat3 induces the processing of p100 to p52 (Fig. 2).
Fig. 5.
Effect of phosphorylation and acetylation of Stat3 on the formation of Stat3–p100 complexes. DU145 cells were transfected with control vector, Stat3F, or Stat3K685R. Cell extracts were subjected to immunoprecipitation (IP) with antibodies against Stat3. Precipitates or lysates were separated by SDS/PAGE and analyzed by immunoblotting (WB) with antibodies against p100. Expression of Stat3 was determined by immunoblotting (WB) the cell extracts with antibodies against Stat3.
NF-κB p100 is a proapoptotic protein with antioncogenic function and is the only NF-κB family member known to be mutated in human cancer (33, 34). Our finding that p52 is an antiapoptotic protein and that overexpression of p52 increases resistance to chemotherapy in prostate cancer cells is in agreement with the role of p52 in tumor development (35–37). In view of the fact that Stat3 is activated in many types of human cancers (4, 38), it is likely that the induction of processing of p100 by Stat3 resulting in generation of p52 protein may commonly be present in many types of human cancers. This phenomenon may result in the simultaneous loss of p100 and the generation of p52 protein in cancer cells, which may disrupt the balance between the proapoptotic protein p100 with an antioncogenic function and the antiapoptotic protein p52 with an oncogenic property. It is possible that the effects of Stat3 on cell proliferation, differentiation, survival, and immune evasion may be magnified by the induction of p52 production, which regulates the transcription of genes that control cell proliferation, survival, and transformation. Stat3 is activated in many types of cancers, and it is likely that activation of the processing of p100 to p52 by Stat3 is one of the common pathways used by cancer cells to survive and escape therapy.
Materials and Methods
Cell Lines, Plasmids, and Antibodies.
The prostate cancer cell lines LNCaP, PC3, and DU145, the breast cancer cell line MDA-MB-231, and HEK293 cells were purchased from the American Type Culture Collection. The antibodies against Stat3, p100/p52, p65, p50, and RelB were purchased from Santa Cruz Biotechnology. Anti-phospho-Y705-Stat3 and anti-acetyl lysine antibodies were purchased from Cell Signaling Technology (Beverly, MA). The acetylation defective mutant of Stat3, Stat3K685R, was generously provided by Eugene Chin (Brown University Medical School, Providence, RI). All other reagents were of analytical grade from Sigma.
Preparation of Cell Lysates and Immunoblotting.
Cells were lysed in a high-salt buffer containing 10 mM Hepes (pH 7.9), 0.25 M NaCl, 1% Nonidet P-40, and 1 mM EDTA, and total protein in the lysates was determined with the Coomassie blue Protein Assay Reagent (Pierce). Equal amounts of protein were electrophoresed by 10% SDS/PAGE and transferred to a nitrocellulose membrane. The membranes were blocked for 1 h at room temperature in 5% milk in 1× PBS plus 0.1% Tween 20 and incubated with primary antibody diluted in 1% BSA overnight. After washing, the membranes were incubated for 1 h in secondary antibody conjugated to horseradish peroxidase diluted in 5% milk in 1× PBS plus 0.1% Tween 20. After washing, the membranes were incubated in a 1:1 ratio of reagents A and B (ECL, Amersham Pharmacia) for 1 min and exposed to film.
EMSAs.
Stat3, Stat3c, p100, and p300 plasmids were cotransfected into LNCaP or HEK293 cells, and cytoplasmic and nuclear extracts were made from the cells by using low-salt and high-salt buffers, respectively, as described previously (26). Ten micrograms of nuclear protein was incubated with binding buffer containing 10 mM Hepes (pH 7.9), 400 mM NaCl, 1 mM EDTA, 40% glycerol, and 1 μg of poly(dI-dC) per reaction with 105 cpm of the [γ-32P]ATP-labeled consensus oligonucleotides (Stat3, NF-κB, and Blc-κB) for 20 min at room temperature. The reactions were stopped with the addition of 6× DNA-loading buffer and electrophoresed on a 5% nondenaturing polyacrylamide gel. The gel was dried and exposed to a phosphorimager screen. For supershift assays, the reaction mixtures were incubated with the appropriate antibodies for 45 min after the initial 20-min reaction, and the reactions were stopped with loading buffer and run on a 5% nondenaturing polyacrylamide gel.
Immunoprecipitation.
Du145 and PC3 cells were lysed in high-salt buffer. Equal amounts of the lysates were immunoprecipitated with 1 μg of the Stat3 antibodies with 30 μl of protein A/G-agarose with constant rotation overnight and washed with 10 mM Hepes (pH 7.9), 1 mM EDTA, 150 mM NaCl, and 1% Nonidet P-40 twice with 400 μl each. The precipitated proteins were eluted with 30 μl of SDS/PAGE sample buffer and boiled for 10 min. The eluted proteins were electrophoresed by 10% SDS/PAGE, transferred to nitrocellulose membranes, and probed with anti-p100/p52 antibodies.
In Vitro Kinase Assays.
Stat3, Stat3c, p100, Stat3F, and Stat3 siRNA plasmids were cotransfected into LNCaP cells, and cells were lysed in high-salt buffer as described earlier. Equal amounts of lysates were immunoprecipitated overnight with anti-IKKα antibodies, and the protein A/G-agarose beads were incubated with 5 μCi (1 Ci = 37 GBq) of [γ-32P]ATP in the presence of 1× kinase assay buffer (20 mM Hepes, pH 7.5/10 mM MgCl2/20 mM β-glycerophosphate/50 μM Na-orthovanadate/1 mM DTT/20 μM ATP) for 30 min at 30°C. HEK293 cells were transfected with pCDNA3.1-HA-p100 (WT) expression vector, and 50 μg of total protein with overexpressed p100 was used as the substrate in each reaction. The reaction was stopped by the addition of 20 μl of 4× SDS/PAGE sample buffer and boiling for 10 min. The reaction mixtures were electrophoresed by 10% SDS/PAGE and transferred to nitrocellulose membranes, and the p100 phosphorylated by the kinase IKKα was visualized by autoradiography. The membrane was probed with anti-IKKα antibodies to normalize for equal amounts of kinase in each reaction.
Construction of Adenoviruses.
cDNAs encoding full-length p100 (WT), p100 (SS/AA), and p52 were cut from pCDNA3.1 vectors encoding HA-tagged p100 (WT), p100 (SS/AA), and p52, respectively (HinDIII/XbaI), and cloned into the shuttle vector pDC316 (Microbix Biosystems, Toronto, ON, Canada) under a mouse CMV promoter. The shuttle vectors and the pBHGlox(delta)E1,3Cre vector containing the adenoviral genome (Microbix Biosystems) were cotransfected into the packaging (HEK293) cells by using Superfect reagent (Qiagen). The supernatants containing the adenoviruses were collected and used to infect HEK293 cells. Adenoviral recombinants containing the cloned genes as well as homologous recombinants were selected from the initial heterogeneous pool and plaque-purified by infecting HEK293 cells at different dilutions and picking up single plaques formed by single infectious units. Viral recombinants expressing optimal amounts of the proteins of interest were purified, amplified in HEK293 cells, and used to infect either LNCaP or DU145 cells.
Apoptosis Assays and Cell Death Detection ELISA.
LNCaP or DU145 cells were treated with 50–150 μM etoposide for 24–48 h to induce apoptosis after infection with adenoviruses encoding p100 or p52. LNCaP cells were cultured under androgen-deprivation conditions (10% charcoal-stripped serum) for 3–7 days after infection with the adenoviruses. The degree of apoptosis was measured by cell death detection ELISA according to the manufacturer's instructions. Briefly, floating and attached cells were collected and homogenized in 400 μl of incubation buffer. Five microliters of the supernatant diluted in 95 μl of incubation buffer was used in the ELISA. The wells were coated with anti-histone antibodies and then incubated with the lysates, horseradish peroxidase-conjugated anti-DNA antibodies, and the substrate subsequently, and absorbance was read at 620 nm.
Acknowledgments
We thank Dr. James Darnell, Jr. (Rockefeller University, New York) for Stat3c, Dr. Koichi Nakajima (Osaka University, Osaka) for Stat3F, Dr. Sergio Onate (Roswell Park Cancer Institute) for p300, Dr. Eugene Chin for Stat3K685R, and Dr. John Isaacs for critical reading of this manuscript. This work was supported by National Institutes of Health Grants CA90271 and CA109441, U.S. Army Medical Research Materiel Command Prostate Cancer Research Program Grant DAMD17-017-17-0089, and the Roswell Park Alliance Foundation.
Abbreviations
- STAT
signal transducer and activator of transcription
- siRNA
small interfering RNA
- CBP
cAMP-response element-binding protein (CREB)-binding protein
- TSA
trichostatin A.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office. V.M.R. is a guest editor invited by the Editorial Board.
References
- 1.Darnell J. E., Jr. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 2.Yuan Z. L., Guan Y. J., Chatterjee D., Chin Y. E. Science. 2005;307:269–273. doi: 10.1126/science.1105166. [DOI] [PubMed] [Google Scholar]
- 3.Wang R., Cherukuri P., Luo J. J. Biol. Chem. 2005;280:11528–11534. doi: 10.1074/jbc.M413930200. [DOI] [PubMed] [Google Scholar]
- 4.Bowman T., Garcia R., Turkson J., Jove R. Oncogene. 2000;19:2474–2488. doi: 10.1038/sj.onc.1203527. [DOI] [PubMed] [Google Scholar]
- 5.Catlett-Falcone R., Landowski T. H., Oshiro M. M., Turkson J., Levitzki A., Savino R., Ciliberto G., Moscinski L., Fernandez-Luna J. L., Nunez G., et al. Immunity. 1999;10:105–115. doi: 10.1016/s1074-7613(00)80011-4. [DOI] [PubMed] [Google Scholar]
- 6.Bromberg J. F., Wrzeszczynska M. H., Devgan G., Zhao Y., Pestell R. G., Albanese C., Darnell J. E., Jr. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 7.Lin X., Mu Y., Cunningham E. T., Jr., Marcu K. B., Geleziunas R., Greene W. C. Mol. Cell. Biol. 1998;18:5899–5907. doi: 10.1128/mcb.18.10.5899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghosh S., May M. J., Kopp E. B. Annu. Rev. Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 9.Sun S. C., Xiao G. Cancer Metastasis Rev. 2003;22:405–422. doi: 10.1023/a:1023733231406. [DOI] [PubMed] [Google Scholar]
- 10.Karin M., Greten F. R. Nat. Rev. Immunol. 2005;5:749–759. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- 11.Karin M., Cao Y., Greten F. R., Li Z. W. Nat. Rev. Cancer. 2002;2:301–310. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- 12.Fan C. M., Maniatis T. Nature. 1991;354:395–398. doi: 10.1038/354395a0. [DOI] [PubMed] [Google Scholar]
- 13.Betts J. C., Nabel G. J. Mol. Cell. Biol. 1996;16:6363–6371. doi: 10.1128/mcb.16.11.6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao G., Harhaj E. W., Sun S. C. Mol. Cell. 2001;7:401–409. doi: 10.1016/s1097-2765(01)00187-3. [DOI] [PubMed] [Google Scholar]
- 15.Xiao G., Fong A., Sun S. C. J. Biol. Chem. 2004;279:30099–30105. doi: 10.1074/jbc.M401428200. [DOI] [PubMed] [Google Scholar]
- 16.Qing G., Xiao G. J. Biol. Chem. 2005;280:9765–9768. doi: 10.1074/jbc.C400502200. [DOI] [PubMed] [Google Scholar]
- 17.Qing G., Qu Z., Xiao G. J. Biol. Chem. 2005;280:18–27. doi: 10.1074/jbc.M406619200. [DOI] [PubMed] [Google Scholar]
- 18.Dejardin E., Droin N. M., Delhase M., Haas E., Cao Y., Makris C., Li Z. W., Karin M., Ware C. F., Green D. R. Immunity. 2002;17:525–535. doi: 10.1016/s1074-7613(02)00423-5. [DOI] [PubMed] [Google Scholar]
- 19.Coope H. J., Atkinson P. G., Huhse B., Belich M., Janzen J., Holman M. J., Klaus G. G., Johnston L. H., Ley S. C. EMBO J. 2002;21:5375–5385. doi: 10.1093/emboj/cdf542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caamano J. H., Rizzo C. A., Durham S. K., Barton D. S., Raventos-Suarez C., Snapper C. M., Bravo R. J. Exp. Med. 1998;187:185–196. doi: 10.1084/jem.187.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franzoso G., Carlson L., Poljak L., Shores E. W., Epstein S., Leonardi A., Grinberg A., Tran T., Scharton-Kersten T., Anver M., et al. J. Exp. Med. 1998;187:147–159. doi: 10.1084/jem.187.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishikawa H., Carrasco D., Claudio E., Ryseck R. P., Bravo R. J. Exp. Med. 1997;186:999–1014. doi: 10.1084/jem.186.7.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ciana P., Neri A., Cappellini C., Cavallo F., Pomati M., Chang C. C., Maiolo A. T., Lombardi L. Oncogene. 1997;14:1805–1810. doi: 10.1038/sj.onc.1201015. [DOI] [PubMed] [Google Scholar]
- 24.Bonizzi G., Bebien M., Otero D. C., Johnson-Vroom K. E., Cao Y., Vu D., Jegga A. G., Aronow B. J., Ghosh G., Rickert R. C., Karin M. EMBO J. 2004;23:4202–4210. doi: 10.1038/sj.emboj.7600391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee S. O., Lou W., Qureshi K. M., Mehraein-Ghomi F., Trump D. L., Gao A. C. Prostate. 2004;60:303–309. doi: 10.1002/pros.20072. [DOI] [PubMed] [Google Scholar]
- 26.Ni Z., Lou W., Leman E. S., Gao A. C. Cancer Res. 2000;60:1225–1228. [PubMed] [Google Scholar]
- 27.Nakajima K., Yamanaka Y., Nakae K., Kojima H., Ichiba M., Kiuchi N., Kitaoka T., Fukada T., Hibi M., Hirano T. EMBO J. 1996;15:3651–3658. [PMC free article] [PubMed] [Google Scholar]
- 28.Lee S. O., Lou W., Hou M., de Miguel F., Gerber L., Gao A. C. Clin. Cancer Res. 2003;9:370–376. [PubMed] [Google Scholar]
- 29.Paulson M., Pisharody S., Pan L., Guadagno S., Mui A. L., Levy D. E. J. Biol. Chem. 1999;274:25343–25349. doi: 10.1074/jbc.274.36.25343. [DOI] [PubMed] [Google Scholar]
- 30.Nakashima K., Yanagisawa M., Arakawa H., Kimura N., Hisatsune T., Kawabata M., Miyazono K., Taga T. Science. 1999;284:479–482. doi: 10.1126/science.284.5413.479. [DOI] [PubMed] [Google Scholar]
- 31.Zhang J. J., Vinkemeier U., Gu W., Chakravarti D., Horvath C. M., Darnell J. E., Jr. Proc. Natl. Acad. Sci. USA. 1996;93:15092–15096. doi: 10.1073/pnas.93.26.15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korzus E., Torchia J., Rose D. W., Xu L., Kurokawa R., McInerney E. M., Mullen T. M., Glass C. K., Rosenfeld M. G. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y., Cui H., Schroering A., Ding J. L., Lane W. S., McGill G., Fisher D. E., Ding H. F. Nat. Cell Biol. 2002;4:888–893. doi: 10.1038/ncb872. [DOI] [PubMed] [Google Scholar]
- 34.Rayet B., Gelinas C. Oncogene. 1999;18:6938–6947. doi: 10.1038/sj.onc.1203221. [DOI] [PubMed] [Google Scholar]
- 35.Perkins N. D. Oncogene. 2003;22:7553–7556. doi: 10.1038/sj.onc.1207139. [DOI] [PubMed] [Google Scholar]
- 36.Nonaka M., Horie R., Itoh K., Watanabe T., Yamamoto N., Yamaoka S. Oncogene. 2005;24:3976–3986. doi: 10.1038/sj.onc.1208564. [DOI] [PubMed] [Google Scholar]
- 37.Cogswell P. C., Guttridge D. C., Funkhouser W. K., Baldwin A. S., Jr. Oncogene. 2000;19:1123–1131. doi: 10.1038/sj.onc.1203412. [DOI] [PubMed] [Google Scholar]
- 38.Lee S. O., Gao A. C. Vitam. Horm. 2005;70:333–357. doi: 10.1016/S0083-6729(05)70011-X. [DOI] [PubMed] [Google Scholar]