Abstract
Galactolipids represent the most abundant lipid class in thylakoid membranes, where oxygenic photosynthesis is performed. The identification of galactolipids at specific sites within photosynthetic complexes by x-ray crystallography implies specific roles for galactolipids during photosynthetic electron transport. The preference for galactose and not for the more abundant sugar glucose in thylakoid lipids and their specific roles in photosynthesis are not understood. Introduction of a bacterial glucosyltransferase from Chloroflexus aurantiacus into the galactolipid-deficient dgd1 mutant of Arabidopsis thaliana resulted in the accumulation of a glucose-containing lipid in the thylakoids. At the same time, the growth defect of the dgd1 mutant was complemented. However, the degree of trimerization of light-harvesting complex II and the photosynthetic quantum yield of transformed dgd1 plants were only partially restored. These results indicate that specific interactions of the galactolipid head group with photosynthetic protein complexes might explain the preference for galactose in thylakoid lipids of higher plants. Therefore, galactose in thylakoid lipids can be exchanged with glucose without severe effects on growth, but the presence of galactose is crucial to maintain maximal photosynthetic efficiency.
Keywords: chloroplast, galactose, glucose, lipid, glucosylgalactosyldiacylglycerol
Directly or indirectly, almost all life on earth depends on the photosynthetic conversion of water, CO2, and sunlight into chemical energy and oxygen. In cyanobacteria, green algae, and plants, the primary reactions of oxygenic photosynthesis are executed in thylakoid membranes. They harbor a set of multimeric protein complexes embedded into a lipid matrix of unique composition. The structure of the protein complexes and the lipid composition of thylakoid membranes are highly conserved in all organisms performing oxygenic photosynthesis. Thylakoid lipids comprise the two galactolipids monogalactosyldiacylgly-cerol (βGalDG) and digalactosyldiacylglycerol (αGalβGalDG), a sulfolipid, and phosphatidylglycerol as the only phospholipid (1–3). Based on their high proportion in thylakoid membranes and the abundance of plants and algae, galactolipids represent the most abundant lipid class in the biosphere (4, 5). Galactolipids are crucial to establish the proton- and ion-impermeable matrix of chloroplast membranes. An appropriate ratio of βGalDG to αGalβGalDG is required to maintain the intricate bilayer characteristics required for insertion, folding, movement, and conformational changes of membrane proteins (6). Crystalline chlorophyll–protein complexes contain galactolipid molecules firmly bound to specific sites, some of them in close proximity to the electron transfer chain. This association implies important roles of galactolipids in photosynthetic exciton and electron transfer within and between the different complexes (7–9). Mutants and transgenic plants of Arabidopsis thaliana are the basis for our current understanding of the role of thylakoid lipids in photosynthesis and stress physiology (10, 11). Arabidopsis mutants with reduced proportions of βGalDG or αGalβGalDG were most informative, because their analysis revealed essential in vivo functions of galactolipids in photosynthesis and growth (12–14). However, the severe galactolipid reduction and thus the general shortage of lipid building blocks for thylakoid membrane assembly prevent the identification of specific galactolipid functions in photosynthetic complexes. Higher plants synthesize βGalDG from diacylglycerol and UDP-galactose as sugar donor (15, 16). This sugar nucleotide is used for thylakoid lipid synthesis despite the fact that UDP-glucose is more abundant in Arabidopsis, and an additional epimerization step of the sugar C4 carbon is required to convert UDP-glucose into UDP-galactose (17). Cyanobacteria have established a different pathway for βGalDG synthesis, because they first produce a glucolipid, βGlcDG (18), in a UDP-glucose-dependent reaction, which in a second step is converted into the galactolipid βGalDG by the unique epimerization of the lipid-bound sugar head group (19). The requirement for these additional epimerization steps in plants and cyanobacteria and the fact that many nonphotosynthetic bacteria contain glucolipids instead of galactolipids suggest that galactolipids have specific functions in photosynthesis that cannot be fulfilled by glucolipids. Therefore, galactolipids were maintained throughout evolution and were resistant toward replacement by corresponding glucolipids. However, galactolipid functions that rely on the C4 stereochemistry of their head groups are not known. Here we address the question of why oxygenic photosynthesis in higher plants relies on the presence of galactose, and not glucose, in thylakoid lipids.
Results
Introduction of a Bacterial Glycolipid into Arabidopsis.
The in situ exchange of plant galactolipids by glucolipids can be realized by replacing the galactosyltransferase activities in chloroplasts by glucosyltransferases of appropriate specificity (Fig. 1). For this purpose, glycosyltransferases were selected from bacterial origin that had previously been characterized by heterologous expression in prokaryotic or eukaryotic hosts (Escherichia coli, Pichia pastoris, Saccharomyces cerevisiae, and Synechococcus) and subsequent analysis of newly formed glycolipids (20–26). Additional putative glycosyltransferases were identified in bacteria based on sequence similarity to known genes (Table 1) (26), and by this 20 bacterial genes were selected for expression in plants. Because the enzymes should become active in chloroplasts, all sequences had to be fused with an N-terminal leader sequence for import into chloroplasts. These constructs were transferred into WT plants of Arabidopsis followed by lipid analyses of leaves. Only a single glycosyltransferase (gene chlo02003783, abbreviated as βGlcT and originating from the bacterium Chloroflexus aurantiacus) passed this functional selection. The plants expressing βGlcT (WT-βGlcT) produced a new dihexosyldiacylglycerol that was separated from the endogenous plant αGalβGalDG by TLC and accumulated in significant proportions (Table 2). The presence of the bacterial lipid did not result in any obvious growth phenotype of Arabidopsis (data not shown).
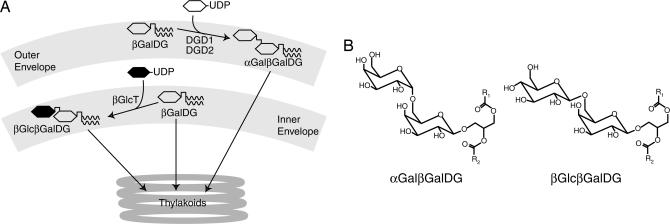
Fig. 1.
Transfer of a bacterial glycolipid into Arabidopsis. (A) Simplified scheme for the synthesis of αGalβGalDG and βGlcβGalDG in chloroplasts. In WT Arabidopsis most αGalβGalDG is formed from βGalDG in the outer envelope by one of the two galactosyltransferases DGD1 or DGD2. In transgenic plants expressing the glucosyltransferase βGlcT from Chloroflexus, βGlcβGalDG is mainly formed in the inner envelope. Galactose and glucose are depicted as open or filled hexagons, respectively, and diacylglycerol is depicted by a vertical line with two waved lines. (B) Structures of αGalβGalDG and βGlcβGalDG.
Table 1.
Bacterial glycosyltransferase and glycosyltransferase-like genes expressed in Arabidopsis
| Genus | Glycosyltransferase | Specificity in bacteria or yeast | Ref. | Expression in Arabidopsis |
|---|---|---|---|---|
| Acholeplasma | alMGS | αGlc | 22 | ? |
| Staphylococcus | ugt106B1 | βGlc, processive | 21 | − |
| Bacillus | ypfP | βGlc, processive | 20 | − |
| Agrobacterium | AGR_C_3323 | βGlc/βGal, processive | 25 | − |
| Mesorhizobium | mlr5650 | βGlc/βGal, processive | 25 | − |
| Deinococcus | dr1225 | αGlc | 26 | − |
| dr1076 | Unknown | − | ||
| Thermotoga | tm0744 | αGlc | 26 | − |
| Chlorobium | ct1882 | Unknown | − | |
| ct0548 | Unknown | − | ||
| ct0225 | Unknown | − | ||
| ct0226 | Unknown | − | ||
| Chloroflexus | chlo02003783 | βGlc | 26 | + |
| chlo02003782 | βGal | 26 | − | |
| chlo02003037 | Unknown | − | ||
| chlo02003249 | Unknown | − | ||
| chlo02003283 | Unknown | − | ||
| chlo02003531 | Unknown | − | ||
| chlo02004251 | Unknown | − | ||
| chlo02001411 | Unknown | − |
Genes from different bacteria known to be involved in glycolipid synthesis or showing sequence similarity to glycolipid synthase genes were fused with a chloroplast-targeting sequence, transferred into Arabidopsis WT plants, and tested for glycolipid accumulation. +, new glycolipid detected; −, no change in glycolipid content; ?, no transformant obtained.
Table 2.
Leaf lipid composition of Arabidopsis WT and dgd1 mutant plants expressing βGlcT from Chloroflexus
| Lipid | WT | WT-βGlcT | dgd1 | dgd1-βGlcT | dgd1-DGD1 |
|---|---|---|---|---|---|
| βGalDG | 52.5 ± 2.2 | 46.6 ± 4.0 | 42.3 ± 1.4 | 45.3 ± 1.1 | 50.7 ± 5.8 |
| PG | 6.8 ± 0.6 | 7.6 ± 0.6 | 13.7 ± 0.9 | 9.8 ± 0.6 | 7.8 ± 1.7 |
| βGlcβGalDG | —* | 8.7 ± 0.2 | —* | 17.7 ± 1.2 | —* |
| αGalβGalDG | 16.4 ± 0.3 | 18.2 ± 1.7 | 1.5 ± 0.3 | 2.7 ± 0.7 | 15.3 ± 1.1 |
| SQDG | 2.9 ± 0.5 | 4.6 ± 0.8 | 3.6 ± 0.6 | 6.5 ± 1.8 | 2.9 ± 2.0 |
| PE | 7.6 ± 0.9 | 6.3 ± 1.9 | 14.6 ± 1.6 | 5.4 ± 2.0 | 8.1 ± 1.1 |
| PC | 13.8 ± 1.5 | 8.1 ± 2.9 | 24.4 ± 1.5 | 12.6 ± 0.8 | 16.1 ± 0.8 |
Lipids were separated by TLC, and fatty acids were quantified by GC of methyl esters. Data (in mol%) represent means ± SD of three measurements. Data for dgd1-DGD1 (line R376) were taken from ref. 27. PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; SQDG, sulfoquinovosyldiacylglycerol.
*Below detection limit.
Characterization of the Novel Dihexosyldiacylglycerol Lipid Accumulating in Arabidopsis Plants Expressing Chloroflexus βGlcT.
By expression in yeast and bacteria (26) the Chloroflexus βGlcT was shown to encode a β-glucosyltransferase involved in dihexosyldiacylglycerol synthesis. The novel glycolipid accumulating in WT-βGlcT plants was isolated by TLC, and the composition of the head group and of the acyl groups was determined. Glucose and galactose as quantified by GC of alditol acetates were found in equal amounts in the novel glycolipid of WT-βGlcT plants (50% glucose and 50% galactose), in contrast to the endogenous αGalβGalDG, which almost exclusively contained galactose (2% glucose and 98% galactose; mean; n = 3; SD always <1%). For a detailed analysis of its structure, the newly formed dihexosyldiacylglycerol was acetylated and subjected to1H-NMR spectroscopy. This analysis revealed the same structure as deduced before in experiments on the expression of the bacterial βGlcT in cyanobacteria (26). Therefore, also in higher plants the expression of the βGlcT results in the formation of glucosylgalactosyldiacylglycerol (βGlcβGalDG). Most likely, the bacterial βGlcT forms this new glycolipid by adding a β-glucopyranose to the C6 position of βGalDG, thus showing the same specificity as in cyanobacteria (26). The new glycolipid deviates from the native plant αGalβGalDG by the epimeric C4 configuration (glucose) and the anomeric linkage (β instead of α) of the second hexose residue (Fig. 1B). βGlcβGalDG was localized in chloroplasts, as demonstrated by lipid analysis of isolated organelles (data not shown). The high proportion of hexadecatrienoic acid (abbreviated as 16:3, where 16 indicates the number of carbon atoms and 3 the number of double bonds in the acyl chain) in βGlcβGalDG of WT-βGlcT indicates that it was preferentially derived from the prokaryotic/chloroplast pathway of lipid synthesis (Table 3). As shown in Tables 3 and 4, the predominant fraction of 16:3 was found in the sn-2 position of the glycerol backbone of βGlcβGalDG, suggesting that it was derived from prokaryotic βGalDG, which is known to contain large amounts of 16:3 at sn-2. In contrast, the native plant αGalβGalDG lipid was enriched in α-linolenic acid (18:3) and contained little 16:3. Taken together, these results indicate that the bacterial glucosyltransferase βGlcT represents a suitable tool for in planta formation of a novel dihexosyldiacylglycerol lipid in chloroplasts in which the terminal α-galactosyl residue is replaced by β-glucose.
Table 3.
Fatty acid composition of leaf glycolipids
| Fatty acid | βGalDG |
βGlcβGalDG |
αGalβGalDG |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| WT | WT-βGlcT | dgd1 | dgd1-βGlcT | WT-βGlcT | dgd1-βGlcT | WT | WT-βGlcT | dgd1 | dgd1-βGlcT | |
| 16:0 | 1.1 | 0.7 | 5.1 | 3.6 | 2.0 | 3.7 | 12.1 | 5.6 | 25.5 | 24.4 |
| 16:1 | 0.9 | 0.1 | 3.7 | 1.0 | 0.7 | 0.5 | 0.3 | 0.2 | 4.5 | 0.8 |
| 16:2 | 0.1 | 0.5 | 0.6 | 0.1 | 0.6 | 0.1 | 0.6 | 0.2 | 1.7 | 0.1 |
| 16:3 | 35.1 | 31.7 | 15.6 | 22.2 | 14.6 | 19.0 | 3.4 | 2.1 | 3.5 | 5.7 |
| 18:0 | 0.3 | 0.1 | 0.6 | 0.3 | 0.5 | 0.4 | 0.8 | 0.4 | 3.8 | 3.7 |
| 18:1 | 1.0 | 0.1 | 1.0 | 0.8 | 0.2 | 0.4 | 0.6 | 0.1 | 5.0 | 1.6 |
| 18:2 | 1.9 | 1.3 | 2.6 | 1.9 | 2.4 | 1.6 | 4.1 | 2.8 | 9.0 | 12.5 |
| 18:3 | 59.5 | 65.6 | 70.1 | 70.1 | 78.9 | 74.3 | 77.9 | 88.5 | 47.2 | 51.0 |
Lipids were isolated by TLC, and fatty acid composition was determined by GC of methyl esters. Data are given in mol% and represent means of three experiments. SD was always <2.0 mol%. Fatty acids are abbreviated as X:Y, where X and Y depict the number of carbon atoms and double bonds, respectively.
Table 4.
Prokaryotic and eukaryotic molecular species of leaf glycolipids
| Fatty acid composition | βGalDG |
βGlcβGalDG |
αGalβGalDG |
|||
|---|---|---|---|---|---|---|
| WT | WT-βGlcT | WT | WT-βGlcT | WT | WT-βGlcT | |
| C16 at sn-2 | 73.0 ± 1.3 | 68.1 ± 2.4 | — | 31.9 ± 2.0 | 11.8 ± 0.5 | 13.2 ± 1.5 |
| C18 at sn-2 | 26.7 ± 1.6 | 31.8 ± 2.4 | — | 67.6 ± 1.7 | 88.1 ± 0.5 | 86.6 ± 2.5 |
Glycolipids isolated by TLC were digested with R. arrhizus lipase, and reaction products were separated by TLC. The fatty acids of lysoglycolipids were measured by GC after transmethylation and indicate the fatty acid composition (total C16/prokaryotic species or total C18/eukaryotic species) at the sn-2 position. Data (in mol%) represent means ± SD of three experiments.
The Bacterial Lipid βGlcβGalDG Complements Growth Deficiency of the Arabidopsis dgd1 Mutant.
To study the capacity of the bacterial βGlcβGalDG lipid in supporting photosynthesis in thylakoid membranes, the Chloroflexus βGlcT gene was introduced into the dgd1 mutant of Arabidopsis, in which the reduced activity of the αGalβGalDG synthase DGD1 leads to the loss of 90% of αGalβGalDG (1.5 mol% versus 16.4 mol%; Table 2) (12). In the dgd1 mutant, thylakoid ultrastructure and efficiency of photosynthetic light reactions are affected, and this is accompanied by strongly reduced growth (Fig. 2A) (12). The changes observed for the dgd1 mutant are clearly caused by αGalβGalDG deficiency, because complementation with the native DGD1 gene product fully restored photosynthetic efficiency and growth (line dgd1-DGD1; Fig. 2) (15, 27). Introduction of the βGlcT gene from Chloroflexus into the dgd1 mutant resulted in the accumulation of βGlcβGalDG in several complemented lines, but in the following the results of only one representative line are shown. The level of total dihexosyldiacylglycerols in line dgd1-βGlcT was in the range of WT proportions because it contained 17.7 mol% of βGlcβGalDG in addition to 2.7 mol% of the native αGalβGalDG (Table 2 and Fig. 2B). In parallel, the chlorophyll content of the transformant was increased to almost WT levels (Table 5), and the dwarf growth phenotype was rescued (Fig. 2A). These results suggest that βGlcβGalDG can replace αGalβGalDG as a building block for thylakoid membrane assembly.
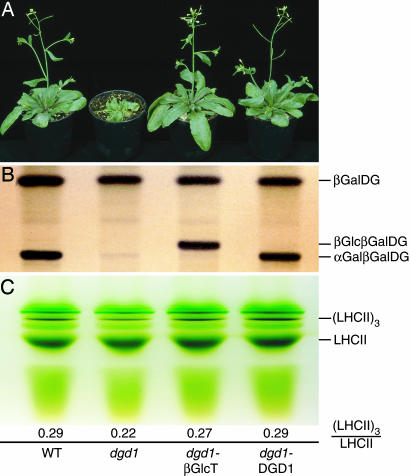
Fig. 2.
Complementation of galactolipid deficiency by expression of the Chloroflexus glucosyltransferase βGlcT in the dgd1 mutant of Arabidopsis. (A) Growth phenotype of 5-week-old WT, dgd1 (deficient in αGalβGalDG biosynthesis), dgd1-βGlcT (expressing the Chloroflexus gene βGlcT), or dgd1-DGD1 plants (transformed with the authentic Arabidopsis DGD1 cDNA). (B) Leaf lipids of WT, dgd1, dgd1-βGlcT, and dgd1-DGD1 plants were separated by TLC and stained with α-naphthol. (C) Separation of pigment–protein complexes. Chloroplasts isolated from the four lines were solubilized with octyl glucoside, and pigment–protein complexes were separated by green gel electrophoresis. Numbers indicate the ratio of trimeric to monomeric LHCII complexes.
Table 5.
Chlorophyll contents in Arabidopsis dgd1 plants expressing the Chloroflexus glycosyltransferase βGlcT
| Plant | Chlorophyll a + b, mg/g of fresh weight | Chlorophyll a/b ratio |
|---|---|---|
| WT | 1.38 ± 0.16 | 2.84 ± 0.12 |
| dgd1 | 0.91 ± 0.11* | 2.27 ± 0.17* |
| dgd1-βGlcT | 1.05 ± 0.16* | 2.42 ± 0.22* |
| dgd1-DGD1 | 1.21 ± 0.14 | 2.70 ± 0.13 |
Data represent means ± SD of five measurements.
*Significantly different from WT according to Student’s t test (P < 0.05).
Stability of Light-Harvesting Complex (LHC) II Trimers in dgd1-βGlcT Plants Is Largely Restored.
To address the question of specific galactolipid functions we studied the trimerization state of the major form of chlorophyll antenna proteins, LHCII. This pigment–protein crystallizes as trimers in vitro with individual molecules of phosphatidylglycerol and αGalβGalDG firmly bound at specific sites of the contact zones (8, 28). It is assumed that in vivo the trimers are the functionally relevant structures and that the two lipids are required for association and stabilization in thylakoid membranes. Therefore, we subjected thylakoid membranes from WT, dgd1, dgd1-βGlcT, and dgd1-DGD1 (15, 27) to detergent extraction and separation of pigment–protein complexes by gel electrophoresis. The ratio of LHCII trimers to monomers as calculated from green gels is considered to reflect the in vivo stability of the trimers (Fig. 2C). This ratio is strongly reduced for dgd1 (0.22) as compared with the values found for WT and dgd1-DGD1 (0.29) (Fig. 2C) (12). On the other hand, the increased ratio of LHCII trimers to LHCII monomers in dgd1-βGlcT (0.27) points to an increased stability of the trimers almost reaching WT values.
Photosynthetic Efficiency in Transgenic dgd1 Plants Accumulating βGlcβGalDG.
The functional performance of photosystems can be assessed more closely by measuring parameters of photosynthetic electron flow. Leaves from the four different Arabidopsis lines were used to compare the effective photosystem (PS)II quantum yield, a measure of photosynthetic efficiency. As shown in Fig. 3, quantum yields at low and medium light intensities (up to 500 μmol quanta·m−2·s−1) were reduced in dgd1 as compared with WT and dgd1-DGD1, whereas dgd1-βGlcT showed intermediate quantum yields. In contrast, quantum yields of dgd1 and dgd1-βGlcT were very similar at high light (>500 μmol·m−2·s−1), but both were lower than in WT and dgd1-DGD1. Therefore, the compensatory effect of an exchange of αGalβGalDG by βGlcβGalDG is restricted to low and medium light intensities. Because the WT and the complemented line dgd1-βGlcT contain similar proportions of dihexosyldiacylglycerol (16.4 mol% of αGalβGalDG and 20.4 mol% of αGalβGalDG plus βGlcβGalDG, respectively; Table 2), the alterations in photosynthetic efficiency can be attributed to differences in the anomeric and epimeric configuration of the sugar head groups.
Fig. 3.
Light-response curves of PSII quantum yield for WT, dgd1, and complemented plants. Dark-adapted plants, exposed to different light conditions, were used for chlorophyll fluorescence measurements to determine effective PSII quantum yield, (Fm′ − F)/Fm′. Data represent means and SE of four measurements. PAR, photosynthetically active radiation.
Discussion
Expression of the Chloroflexus βGlcT gene in Arabidopsis resulted in the accumulation of βGlcβGalDG in chloroplasts, which represents the first example of the synthesis of a foreign membrane lipid in significant proportions in a higher plant. Previous work on the transfer of genes of lipid metabolism into higher plants was focused on the formation of fatty acids and reserve lipids (reviewed in ref. 29). Overexpression of genes involved in phospholipid synthesis (e.g., aminoalcoholphoshotransferase; ref. 30) or of galactolipid synthesis (e.g., DGD1 and DGD2) (14, 15) in Arabidopsis had only minor effects on membrane lipid composition, suggesting that the amounts of phospholipids and glycolipids are subject to strict control. Phosphate deprivation results in the replacement of phospholipids by αGalβGalDG and by sulfolipid (14, 31) and thus represents the only known condition under which plant galactolipid content is severely altered. Interestingly, the sum of total dihexosyldiacylglycerol in WT-βGlcT (βGlcβGalDG and αGalβGalDG, 26.9 mol%; Table 2) is much higher than in WT (αGalβGalDG, 16.4 mol%). This finding suggests that, in contrast to endogenous plant αGalβGalDG synthases (DGD1 and DGD2), the heterologous Chloroflexus enzyme may not be subject to homeostatic regulation. In addition, βGlcβGalDG may resist to some extent the lipid turnover processes catalyzed by specific lipases and glycosidases.
The accumulation of hexadecatrienoic acid (16:3) in the sn-2 position of βGlcβGalDG indicates that it is synthesized from 16:3-rich βGalDG via the prokaryotic/chloroplast pathway of lipid synthesis (32, 33). The preferred biosynthesis of βGlcβGalDG from chloroplast-type βGalDG can be explained by the colocalization of βGlcT with the βGalDG synthase MGD1 in the inner chloroplast envelope membrane (34). βGlcβGalDG still contains large amounts of eukaryotic molecular species (Table 4), indicating that the βGlcT has access to both prokaryotic and eukaryotic substrates. Native plant αGalβGalDG, however, is low in 16:3, because it is largely synthesized from endoplasmic reticulum-derived/eukaryotic lipid. DGD1, the major αGalβGalDG synthase in Arabidopsis, is localized to the outer chloroplast envelope, where it mostly converts endoplasmic reticulum-derived βGalDG originating from the βGalDG synthases MGD2 and MGD3 into αGalβGalDG (35). This can explain why αGalβGalDG in Arabidopsis contains low amounts of 16:3. Futhermore, it is possible that the plant αGalβGalDG synthases DGD1 and DGD2 discriminate against 16:3-rich βGalDG as substrate for galactosylation, whereas the bacterial enzyme βGlcT is less selective for βGalDG with specific fatty acid composition. From the fact that βGlcβGalDG is localized in thylakoids, and assuming a restriction of glycolipid biosynthesis to chloroplast envelopes, it has to be concluded that βGlcβGalDG is accepted by the lipid trafficking machinery required for glycolipid transport from envelope membranes to thylakoids. This system is not discriminating between native and heterologous glycolipids.
The restoration of WT-like growth in dgd1-βGlcT indicates that a replacement of the outer α-galactose by a β-glucose residue in dihexosyldiacylglycerol is not affecting its role as a building block for thylakoid membrane assembly. Nevertheless, photosynthetic measurements revealed differences among WT, dgd1-DGD1, and dgd1-βGlcT plants, demonstrating that βGlcβGalDG, in comparison to αGalβGalDG, does not support maximal photosynthetic efficiency. αGalβGalDG was localized to the periphery of LCHII trimers as observed by x-ray crystallography, whereas another thylakoid lipid, phosphatidylglycerol, was found within the trimeric complex (8). Green gel electrophoresis of dgd1 thylakoids revealed that the intensity of the LHCII band previously designated LHCP1 and containing oligomeric/trimeric complexes was decreased, whereas the monomeric LHCII band (LHCP3) was increased (Fig. 2C) (12, 28, 36). This result indicates that αGalβGalDG is important for the stability of LHCII trimers. The amount of trimeric complexes in dgd1-βGlcT plants was increased as compared with dgd1 but did not reach WT or dgd1-DGD1 levels. In this regard it is interesting to note that the hydroxyl groups of the sugars of αGalβGalDG can interact with the polypeptide chains of LHCII by means of hydrogen bonding at its binding sites (8, 28) and that these interactions presumably are different when αGalβGalDG is replaced with βGlcβGalDG in dgd1-βGlcT plants. This might be one explanation for the fact that αGalβGalDG is the preferred dihexosyldiacylglycerol lipid for the functional stabilization of pigment–protein complexes. In addition, αGalβGalDG deficiency in the dgd1 mutant was previously shown to affect the integrity of PSI, PSII, and the oxygen evolving complex (37, 38). Furthermore, αGalβGalDG was recently identified in the crystal structure of PSII, and it is possible that this lipid is involved in PSII dimerization (39). Therefore, the decrease in dgd1 quantum yield that could not be fully complemented with βGlcβGalDG in dgd1-βGlcT plants might be caused by the fact that αGalβGalDG is required for the functional integrity of LHCII and of PSI, PSII, or the oxygen evolving complex.
The Arabidopsis dgd1-βGlcT plants still contain residual amounts of αGalβGalDG. Thus, it is possible that αGalβGalDG in these plants fulfills functions that cannot be complemented by βGlcβGalDG. Therefore, the expression of βGlcT in the dgd1 dgd2 double mutant, which is totally devoid of αGalβGalDG (14), may reveal additional galactose-specific functions of αGalβGalDG. Our approach led to the exchange of the outer α-galactose by a β-glucose residue in diglycosyldiacylglycerol and thus represents only the first step toward a complete C4 epimerization of the galactolipid matrix. Thus, the next step would be the replacement of βGalDG by βGlcDG, which at the same time would lead to an exchange of the inner galactose of dihexosyldiacylglycerol.
In conclusion, the superior performance of galactolipids in photosynthesis as compared with glucolipids might be ascribed to specific interactions of galactolipid head groups with glycolipid binding sites in photosynthetic protein complexes. These interactions were conserved throughout evolution and most likely involved an optimization of the protein domains of glycolipid binding sites. This conclusion refers to both cyanobacteria, where this glycolipid/protein interaction was established for the first time and has been maintained since, and eukaryotic plants, where the pathway of the galactolipid biosynthesis has been altered without affecting the C4 stereochemistry of the glycolipid head groups.
Materials and Methods
Plant Material and Growth Conditions.
A. thaliana was grown in growth chambers at 60% humidity at 21°C with 16 h of light per day (120 μmol quanta m−2 s−1). The dgd1 mutant (ecotype Columbia) and dgd1 plants complemented with the authentic Arabidopsis DGD1 cDNA (line R376) were described previously (12, 15, 27).
Origin of Genes and Plant Transformation.
The chloroplast signal sequence of βGalDG synthase (type A) was amplified by PCR by using the primers GH69LeaderntF (GGG CCC ATG ATG CAG CAT TCT TCT TC) and GH69LeaderntR2 (ACC TAG GAT AAG CAC CTT TTT CGG AGG) from genomic tobacco DNA and subcloned into pUC18. The signal sequence was released with ApaI/AvrII and ligated into a modified binary vector derived from pCAMB35SOCS12 containing a cauliflower mosaic virus 35S promoter and an octopine synthase terminator (A. Abbadi and E.H., unpublished observations). The βGlcT ORF chlo02003783 (GenBank accession no. ZP_00356752) amplified from genomic Chloroflexus DNA by PCR was subcloned into pUC18 (24) and inserted into the AvrII/BamHI sites, C-terminal to the tobacco αGalβGalDG synthase (type A) signal sequence of the modified pCAMB35SOCS12 vector. The binary vector was transferred into Agrobacterium tumefaciens GV3101 and used for Arabidopsis (WT Columbia and dgd1) transformation by means of infiltration (40).
Lipid Analysis.
Lipids were isolated from leaves, separated by TLC, and stained with iodine vapor or α-naphthol (12, 25). Chloroplasts for glycolipid analysis were obtained from leaves after homogenization and centrifugation through a 50%/80% Percoll step gradient (41). Individual lipids isolated from TLC plates were used to prepare fatty acid methyl esters, which were quantified by GC with pentadecanoic acid (15:0) as internal standard (42). Positional analysis of acyl groups in glycolipids was done by GC quantification of fatty acids after digestion with Rhizopus arrhizus lipase (43). The sugar composition of glycolipid head groups isolated from leaves by TLC was determined after hydrolysis and conversion of the monosaccharides to alditol acetates by GC (44). The proton NMR spectrum of acetylated βGlcβGalDG was recorded in CDCl3 at 600 MHz and yielded essentially the same signals and coupling constants for all structurally relevant protons as described in refs. 25 and 26.
Chlorophyll and Chlorophyll Fluorescence Measurements.
Chlorophyll content in leaves was measured photometrically after extraction with 80% acetone (45). In vivo chlorophyll fluorescence was determined with a pulse amplitude modulation fluorimeter (Imaging PAM; Heinz Walz, Effeltrich, Germany). Plants were dark-adapted before fluorescence measurements for 60 min. Fluorescent light-response curves were recorded after a 5-min exposure of the plants to the photosynthetically active radiation as indicated. Effective PSII quantum yield was calculated following the equation (Fm′ − F)/Fm′, where Fm′ and F are the fluorescence emission of a light-adapted plant under measuring light or after application of a saturating light pulse, respectively (46).
Pigment–Protein Electrophoresis.
Chloroplasts were isolated from leaves and pigment–protein complexes solubilized with a detergent mixture of SDS and octyl glucoside (n-octyl-β-d-glucopyranoside) in a ratio of chlorophyll/SDS/octyl glucoside of 1:1:9 (36). After PAGE, green gels were scanned, and relative band intensities were calculated (12).
Acknowledgments
We thank Regina Wendenburg (Max Planck Institute of Molecular Plant Physiology) for green gel electrophoresis. This work was supported by grants from Deutsche Forschungsgemeinschaft (Sonderforschungsbereiche 429 and 470) and Bundesministerium für Bildung und Forschung (Project Napus 2000, Part FK 0312252 F).
Abbreviations
- βGalDG
monogalactosyldiacylglycerol
- αGalβGalDG
digalactosyldiacylglycerol
- βGlcβGalDG
glucosylgalactosyldiacylglycerol
- βGlcT
β-glucosyltransferase from Chloroflexus
- LHC
light-harvesting complex
- PS
photosystem.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Benson A. A. In: Structure and Function of Chloroplasts. Gibbs M., editor. Berlin: Springer; 1971. pp. 129–148. [Google Scholar]
- 2.Block M. A., Maréchal E., Joyard J. In: Regulation of Photosynthesis. Aro E.-M., Andersson B., editors. Dordrecht, The Netherlands: Kluwer; 2001. pp. 195–218. [Google Scholar]
- 3.Dörmann P., Benning C. Trends Plant Sci. 2002;7:112–118. doi: 10.1016/s1360-1385(01)02216-6. [DOI] [PubMed] [Google Scholar]
- 4.Carter H. E., McCluer R. H., Slifer E. D. J. Am. Chem. Soc. 1956;78:3735–3738. [Google Scholar]
- 5.Benson A. A., Wiser R., Ferrari R. A., Miller J. A. J. Am. Chem. Soc. 1958;80:4740. [Google Scholar]
- 6.Webb M. S., Green B. R. Biochim. Biophys. Acta. 1991;1060:133–158. [Google Scholar]
- 7.Jordan P., Fromme P., Witt H. T., Klukas O., Saenger W., Krauss N. Nature. 2001;411:909–917. doi: 10.1038/35082000. [DOI] [PubMed] [Google Scholar]
- 8.Liu Z., Yan H., Wang K., Kuang T., Zhang J., Gui L., An X., Chang W. Nature. 2004;428:287–292. doi: 10.1038/nature02373. [DOI] [PubMed] [Google Scholar]
- 9.Fyfe P. K., Hughes A. V., Heathcote P., Jones M. R. Trends Plant Sci. 2005;10:275–282. doi: 10.1016/j.tplants.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 10.Browse J., McCourt P., Somerville C. R. Science. 1985;227:763–765. doi: 10.1126/science.227.4688.763. [DOI] [PubMed] [Google Scholar]
- 11.Murata N., Ishizaki-Nishizawa O., Higashi S., Hayashi H., Tasaka Y., Nishida I. Nature. 1992;356:710–713. [Google Scholar]
- 12.Dörmann P., Hoffmann-Benning S., Balbo I., Benning C. Plant Cell. 1995;7:1801–1810. doi: 10.1105/tpc.7.11.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jarvis P., Dörmann P., Peto C. A., Lutes J., Benning C., Chory J. Proc. Natl. Acad. Sci. USA. 2000;97:8175–8179. doi: 10.1073/pnas.100132197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelly A. A., Froehlich J. E., Dörmann P. Plant Cell. 2003;15:2694–2706. doi: 10.1105/tpc.016675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dörmann P., Balbo I., Benning C. Science. 1999;284:2181–2184. doi: 10.1126/science.284.5423.2181. [DOI] [PubMed] [Google Scholar]
- 16.Shimojima M., Ohta H., Iwamatsu A., Masuda T., Shioi Y., Takamiya K.-I. Proc. Natl. Acad. Sci. USA. 1997;94:333–337. doi: 10.1073/pnas.94.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dörmann P., Benning C. Plant J. 1998;13:641–652. doi: 10.1046/j.1365-313x.1998.00067.x. [DOI] [PubMed] [Google Scholar]
- 18.Feige G. B., Heinz E., Wrage K., Cochems N., Ponzelar E. In: Biogenesis and Function of Plant Lipids. Douce R. S., editor. Amsterdam: Elsevier; 1980. pp. 135–140. [Google Scholar]
- 19.Sato N. Plant Physiol. Biochem. 1994;32:121–126. [Google Scholar]
- 20.Jorasch P., Wolter F. P., Zähringer U., Heinz E. Mol. Microbiol. 1998;29:419–431. doi: 10.1046/j.1365-2958.1998.00930.x. [DOI] [PubMed] [Google Scholar]
- 21.Jorasch P., Warnecke D. C., Lindner B., Zähringer U., Heinz E. Eur. J. Biochem. 2000;267:3770–3783. doi: 10.1046/j.1432-1327.2000.01414.x. [DOI] [PubMed] [Google Scholar]
- 22.Berg S., Edman M., Li L., Wikström M., Wieslander Å. J. Biol. Chem. 2001;276:22056–22063. doi: 10.1074/jbc.M102576200. [DOI] [PubMed] [Google Scholar]
- 23.Edman M., Berg S., Storm P., Wikström M., Vikström S., Öhman A., Wieslander Å. J. Biol. Chem. 2003;278:8420–8428. doi: 10.1074/jbc.M211492200. [DOI] [PubMed] [Google Scholar]
- 24.Knudsen E., Jantzen E., Bryn K., Ormerod J. G., Sirevåg R. Arch. Microbiol. 1982;132:149–154. [Google Scholar]
- 25.Hölzl G., Leipelt M., Ott C., Zähringer U., Lindner B., Warnecke D., Heinz E. Glycobiology. 2005;15:874–886. doi: 10.1093/glycob/cwi066. [DOI] [PubMed] [Google Scholar]
- 26.Hölzl G., Zähringer U., Warnecke D., Heinz E. Plant Cell Physiol. 2005;46:1766–1778. doi: 10.1093/pcp/pci189. [DOI] [PubMed] [Google Scholar]
- 27.Härtel H., Dörmann P., Benning C. J. Photochem. Photobiol. B. 2001;61:46–51. doi: 10.1016/s1011-1344(01)00144-0. [DOI] [PubMed] [Google Scholar]
- 28.Nussberger S., Dörr K., Wang N., Kühlbrandt W. J. Mol. Biol. 1993;234:347–356. doi: 10.1006/jmbi.1993.1591. [DOI] [PubMed] [Google Scholar]
- 29.Thelen J. J., Ohrogge J. B. Metab. Eng. 2002;4:12–21. doi: 10.1006/mben.2001.0204. [DOI] [PubMed] [Google Scholar]
- 30.Goode J. H., Dewey R. E. Plant Physiol. Biochem. 1999;37:445–457. [Google Scholar]
- 31.Härtel H., Dörmann P., Benning C. Proc. Natl. Acad. Sci. USA. 2000;97:10649–10654. doi: 10.1073/pnas.180320497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heinz E., Roughan P. G. Plant Physiol. 1983;72:273–279. doi: 10.1104/pp.72.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Browse J., Warwick N., Somerville C. R., Slack C. R. Biochem. J. 1986;235:25–31. doi: 10.1042/bj2350025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Awai K., Maréchal E., Block M. A., Brun D., Masuda T., Shimada H., Takamiya I.-i., Ohta H., Joyard J. Proc. Natl. Acad. Sci. USA. 2001;98:10960–10965. doi: 10.1073/pnas.181331498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Froehlich J., Benning C., Dörmann P. J. Biol. Chem. 2001;276:31806–31812. doi: 10.1074/jbc.M104652200. [DOI] [PubMed] [Google Scholar]
- 36.Andersson B., Anderson J. M., Ryrie I. J. Eur. J. Biochem. 1982;123:465–472. doi: 10.1111/j.1432-1033.1982.tb19790.x. [DOI] [PubMed] [Google Scholar]
- 37.Guo J., Zhang Z., Bi Y., Yang W., Xu Y., Zhang L. FEBS Lett. 2005;579:3619–3624. doi: 10.1016/j.febslet.2005.05.049. [DOI] [PubMed] [Google Scholar]
- 38.Reifarth F., Christen G., Seeliger A. G., Dörmann P., Benning C., Renger G. Biochemistry. 1997;36:11769–11776. doi: 10.1021/bi9709654. [DOI] [PubMed] [Google Scholar]
- 39.Loll B., Kern J., Saenger W., Zouni A., Biesiadka J. Nature. 2005;438:1040–1044. doi: 10.1038/nature04224. [DOI] [PubMed] [Google Scholar]
- 40.Bent A. F., Kunkel B. N., Dahlbeck D., Brown K. L., Schmidt R., Giraudat J., Leung J., Staskawicz B. J. Science. 1994;265:1856–1860. doi: 10.1126/science.8091210. [DOI] [PubMed] [Google Scholar]
- 41.Tietje C., Heinz E. Planta. 1998;206:72–78. [Google Scholar]
- 42.Browse J., McCourt P. J., Somerville C. R. Anal. Biochem. 1985;152:141–145. doi: 10.1016/0003-2697(86)90132-6. [DOI] [PubMed] [Google Scholar]
- 43.Siebertz H. P., Heinz E., Linscheid M., Joyard J., Douce R. Eur. J. Biochem. 1979;101:429–438. doi: 10.1111/j.1432-1033.1979.tb19736.x. [DOI] [PubMed] [Google Scholar]
- 44.Reiter W.-D., Chapple C., Somerville C. R. Plant J. 1997;12:335–345. doi: 10.1046/j.1365-313x.1997.12020335.x. [DOI] [PubMed] [Google Scholar]
- 45.Lichtenthaler H. K. Methods Enzymol. 1987;148:350–382. [Google Scholar]
- 46.Schreiber U., Schliwa U., Bilger W. Photosynth. Res. 1986;10:51–62. doi: 10.1007/BF00024185. [DOI] [PubMed] [Google Scholar]