Abstract
Bunyamwera virus (family Bunyaviridae, genus Bunyavirus) contains a tripartite negative-sense RNA genome. The smallest RNA segment, S, encodes the nucleocapsid protein N and a nonstructural protein, NSs, in overlapping reading frames. We have generated a mutant virus lacking NSs, called BUNdelNSs, by reverse genetics. Compared with the wild-type (wt) virus, BUNdelNSs exhibited a smaller plaque size and generated titers of virus approximately 1 log lower. In mammalian cells, the mutant expressed greatly increased levels of N protein; significantly, the marked inhibition of host cell protein synthesis shown by wt virus was considerably impaired by BUNdelNSs. When inoculated by the intracerebral route BUNdelNSs killed BALB/c mice with a slower time course than wt and exhibited a reduced cell-to-cell spread, and titers of virus in the brain were lower. In addition, the abrogation of NSs expression changed Bunyamwera virus from a noninducer to an inducer of an interferon-β promoter. These results suggest that, although not essential for growth in tissue culture or in mice, the bunyavirus NSs protein has several functions in the virus life cycle and contributes to viral pathogenesis.
Keywords: reverse genetics, host cell shutoff, interferon antagonist
The family Bunyaviridae contains more than 300 mainly arthropod-borne viruses that share certain morphological and biochemical characteristics. Virus particles are spherical, enveloped, and contain a genome comprising three segments of single-stranded RNA. Virus multiplication occurs in the cytoplasm, and virions mature by budding primarily at membranes of the Golgi apparatus. Several members of the family cause encephalitis or hemorrhagic fevers in humans (e.g., Hantaan, Rift Valley fever, La Crosse, and Crimean–Congo hemorrhagic fever viruses) and are recognized as posing an increasing threat to human health, examples of the so-called “emerging infections” (1). The family is divided into five genera: Bunyavirus, Hantavirus, Nairovirus, Phlebovirus, and Tospovirus. Bunyamwera virus (BUN) is the prototype of both the family Bunyaviridae and the Bunyavirus genus. The Bunyavirus genus includes three human pathogens of note: La Crosse virus, which causes severe pediatric encephalitis in the United States; Oropouche virus, which has been responsible for repeated epidemics of a debilitating febrile illness in South America, involving thousands of patients; and Tahyna virus, which causes an influenza-like illness in Central Europe (1).
Bunyaviruses contain four structural polypeptides: two glycoproteins, G1 and G2, which are present on the surface of the virion, and two internal proteins, N (nucleocapsid protein) and L (RNA polymerase), which are associated with the three negative-sense virion RNA segments to form helical nucleocapsids. In infected cells, two virus-specified nonstructural proteins, NSs and NSm, have been identified. Genetic and biochemical analyses have shown that the largest RNA segment, L, encodes the L protein; the medium-sized RNA segment, M, encodes G1, G2, and NSm as a polyprotein; and the smallest RNA segment, S, encodes N and NSs in overlapping reading frames (reviewed in refs. 2–5).
Little is known about the function of the bunyavirus NSs protein. The protein varies in length from 83 to 109 aa among those members of the Bunyavirus genus, representing three serogroups, whose S genome segments have been sequenced (6–8). Variation in the length of the NSs proteins occurs at the carboxy terminus (with the exception of Guaroa bunyavirus NSs, which is also truncated at the amino terminus), and the NSs proteins display greater sequence variation within a serogroup (43–95% identity) than that found between the N proteins (62–96% identity; ref. 6).
Like other negative-strand RNA viruses, bunyaviruses replicate their genomes via full-length positive-strand RNA molecules (antigenomes), which are synthesized in a primer-independent manner by the viral RNA polymerase. In contrast, the synthesis of viral mRNAs (transcription) is a primer-dependent event; the primers are cleaved from the capped 5′ ends of host-cell mRNAs by a viral endonuclease activity (presumably a function of the L protein) similar to the cap snatching of influenza virus (2–5).
It seems unlikely that NSs plays a direct role in bunyavirus RNA synthesis. Transcriptase activity can be detected in detergent-disrupted virions that do not contain NSs (9–14), although Vialat and Bouloy (14) have suggested that NSs may be involved in mRNA transcription termination. By using a reverse genetic system with recombinant expressed bunyavirus proteins to drive reporter-gene expression from a chimeric bunyavirus-like RNA template, Dunn et al. (15) showed that transcription and replication of the RNA required only the N and L proteins, although the possibility of a regulatory role for NSs could not be discounted. To investigate the function of NSs further, we have exploited our ability to rescue infectious bunyavirus entirely from cloned cDNAs (16) to produce a mutant bunyavirus, BUNdelNSs, that does not synthesize the NSs protein. Among the characteristics of the mutant virus are a small plaque phenotype, production of increased levels of N protein, impaired capacity to shut off host cell protein synthesis, and the ability to induce an interferon-β promoter.
Materials and Methods
Cells and Viruses.
BHK-21 and Aedes albopictus C6/36 cells were maintained as described previously (16). Murine BF cells (cloned from a primary cell culture of a BALB/c mouse embryo; ref. 17) were kindly provided by R. Randall (University of St. Andrews, Scotland) and were grown as monolayers in DMEM supplemented with 10% FCS. Bunyaviruses were plaque purified in BHK-21 cells, and working stocks were grown in BHK-21 cells as described by Watret et al. (18).
Plasmids.
Plasmids were constructed by standard procedures (19) and confirmed by restriction enzyme analysis and/or nucleotide sequencing. Only the salient features of the plasmids are described here; details of the constructions are given in the supplementary Materials and Methods, published as supplemental data on the PNAS web site, www.pnas.org. Plasmid pT7riboBUNN (which encodes N but not NSs) was constructed from pT7riboBUNS (16) by using a PCR mutagenesis approach (19) to ablate the NSs ORF while retaining the coding region of N protein. pT7riboBUNNSs (which encodes NSs but not N) was also constructed from pT7riboBUNS and has a single base substitution to change the ATG initiation codon of the N ORF to TTG and has sequences downstream of the NSs ORF (bases 411 to 787) deleted. The expression of N and/or NSs proteins as appropriate was confirmed by coupled in vitro transcription/translation by using the Promega TNT kit (15).
Reporter plasmids for the interferon-promoter studies were kindly provided by S. Goodbourn (St. George's Hospital Medical School, University of London, U.K.). The IFN-β promoter-containing plasmid, pIFΔ(-125)lucter, contains IFN-β sequences from −125 to +72 fused to the firefly luciferase gene (20). The control plasmid, ptkΔ(-105)lucter, contains a fragment (bases −105 to −15) of the herpes simplex virus-thymidine kinase promoter fused to base −17 of the firefly luciferase gene (20).
Generation of the BUN NSs-Deleted Virus Mutant by Reverse Genetics.
Rescue experiments were performed in parallel as described previously (16), except that in one set, support plasmid pTF7–5BUNS (20 μg) was replaced by 15 μg of pTZBUNN and 1 μg of pTZBUNNSs (both described in ref. 15), and transcription plasmid pT7riboBUNS (1 μg) was replaced by 1.5 μg of pT7riboBUNN and 1.4 μg of pT7riboBUNNSs. Briefly, cells were infected with a recombinant vaccinia virus, which produces T7 RNA polymerase, followed by sequential transfections with mixtures of support plasmids to express all bunyavirus proteins and then transcription plasmids that produce full-length antigenome sense bunyavirus RNAs. After 48 h, the cells were lysed, and after passage in A. albopictus C6/36 mosquito cells, transfectant viruses were isolated by plaque formation on BHK cell monolayers as described previously (16, 18).
Analysis of Viral RNA.
Virion RNA from wild-type (wt) and BUNdelNSs viruses was prepared as described previously (16). To analyze the S genome segment, viral RNA was reverse transcribed by using primer NS2 (5′-CTCTTTATTGATTGAGTTGGAATTTCATGAT-3′), and the resultant cDNA was amplified by PCR by using primers S4 (ref. 16; 5′-AGTAGTGTGCTCCACCT-3′) and NS2. To determine the sequence of the S segment, viral RNA was reverse transcribed by using primer S3 (ref. 16; 5′-AGTAGTGTACTCCACAC-3′), and the resultant cDNA was amplified by PCR by using primers ab40 (5′-GTATGTGGTCATAACTAAGCC-3′) and S3. PCR products were sequenced directly and also after cloning into pBluescript; identical results were obtained from both templates.
For Northern blot analysis, infected cell RNA, prepared by using Trizol reagent, was separated by formaldehyde agarose gel electrophoresis (19) and transferred to Hybond-N+ (Amersham Pharmacia) membrane. The membrane was hybridized overnight at 50°C in 50% formamide (19) with a mixture of [32P]CTP RNA probes transcribed from pT7riboBUNS, pT7riboBUNM, and pT7riboBUNL DNAs (16), followed by extensive washing and exposure to x-ray film.
Metabolic Labeling of Viral Proteins.
BHK cells in 35-mm Petri dishes were infected with 5 plaque-forming units (pfu) per cell of the different viruses and labeled with 30 μCi per dish of [35S]methionine for 2 h at 24 h or 48 h after infection. Cell lysates were prepared and analyzed by SDS/PAGE, as described previously (18).
Transient Transfections and Reporter Assays.
Subconfluent monolayers of BF cells were incubated with 3-μg plasmid DNAs in 500 μl of OptiMEM containing 15-μl liposomes, prepared as described by Rose et al. (21). After 6 h at 37°C, the liposome–DNA mixture was removed, and cells were mock infected or infected with wt BUN or BUNdelNSs at 5 pfu per cell. Cells were harvested 15 h after infection and were lysed in 200 μl of passive lysis buffer (Promega). An aliquot of 20 μl of cell lysate was used to measure firefly luciferase activity as described by the manufacturer (Promega).
Pathogenicity Studies.
Five-week-old specific pathogen-free female BALB/c mice were inoculated either i.p. or intracerebrally (i.c.) with virus. During these studies, any mice that were moribund or severely paralyzed were killed. For the pathogenesis studies, brains were removed and bisected sagittally along the midline. One-half was immediately frozen at −70°C and used to assay for infectious virus; the brains were thawed on ice and homogenized in 1.35 ml of PBS containing 0.75% (wt/vol) BSA (PBS/BSA) to give an initial 1:10 dilution. Serial 10-fold dilutions were made in PBS/BSA, and 100 μl of each dilution was assayed on confluent BHK cells in 60-mm dishes as described (18). The other half of each brain was fixed by immersion in 4% formaldehyde in PBS, embedded in paraffin wax, and 5-mm sections were cut onto Biobond-coated glass microscope slides. The sections were immunostained as previously described (22) to detect viral proteins by using a polyclonal anti-BUN antiserum (18).
Results
Generation of Transfectant Bunyamwera Virus.
Because the N and NSs ORFs overlap, opportunities to introduce specific mutations into the NSs ORF would be severely limited, as such mutations may also affect the N ORF. Thus, it would be difficult to ascribe any observed phenotype to a particular amino acid change. In passing, this may be a reason for the paucity of temperature-sensitive mutations that map to the bunyavirus S segment compared with the hundreds mapping to the M and L segments (reviewed in ref. 23). We planned, therefore, to produce a four-segment bunyavirus possessing two S-like segments, in which one would encode the N protein and the other would encode the NSs protein, and both segments would maintain the S segment noncoding sequences. The rationale for this is that, first, bunyaviruses are probably diploid for the S segment (24–26) and could package an additional S-like segment; second, analogous experiments have been conducted with influenza A virus, in which a virus with nine instead of the usual complement of eight segments has been constructed (27).
The established rescue system described previously (16) requires two types of plasmid for each genome segment, a “support” plasmid, containing essentially the protein coding sequence under control of a T7 promoter to allow transient expression of the protein in transfected cells, and a “transcription” plasmid, which contains a full-length cDNA copy of a genome segment flanked by T7 promoter and hepatitis δ ribozyme sequences. These transcription plasmids yield full-length antigenome RNAs of each segment, which are subsequently encapsidated by N protein, replicated, and packaged into virions.
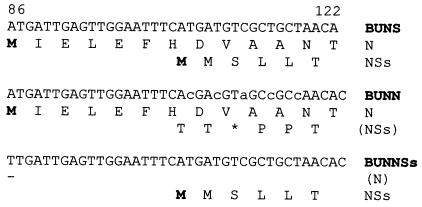
Support plasmids encoding either the N or NSs proteins were already available (15). Two new transcription plasmids, pT7riboBUNN and pT7riboBUNNSs, were made that encode the N and NSs ORFs, respectively, each flanked by the complete 5′ and 3′ noncoding sequences of the S segment. pT7riboBUNN contains five-point mutations that abrogate NSs expression: the tandem ATG initiation codons were both changed to ACG codons, codon 3 was changed from TCG (Ser) to TAG (a stop codon), and codons 4 and 5 were changed from Leu to Pro codons. All changes are silent with respect to the N reading frame (Fig. 1). Multiple mutations were made to minimize the chance of reversion to restore the NSs ORF. pT7riboBUNNSs contains a single point mutation to ablate the N protein initiation codon (ATG to TTG) and a 377-nt deletion at the 3′ end of the N ORF. Hence, pT7riboBUNN will generate a full-length S segment (961 bases) and pT7riboBUNNSs a smaller S-like RNA of 584 bases.
Figure 1.
Nucleotide sequences from bases 86 to 122 of BUN S segment in pT7riboBUNS, pT7riboBUNN, and pT7riboBUNNSs showing the N and NSs start codons and mutations made to ablate either the N or NSs reading frames. Amino acid translations are given below the nucleotide sequences. Initiating methionine residues are indicated in bold lettering and stop codons by an asterisk.
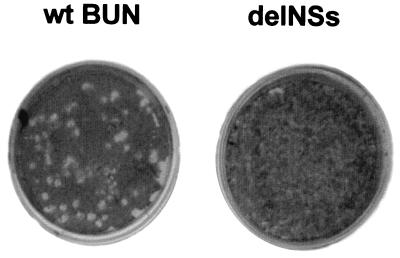
Rescue experiments were performed as described previously (16), except that four support plasmids were used in the first transfection, followed by the four transcription plasmids in the second round. Four plasmids were used at each transfection stage to avoid the potential of vaccinia virus-induced recombination (28) between a wt S segment cDNA (encoding NSs) and the modified NSs-deleted construct. Thus, transfection with the support plasmids pTZBUNN, pTZBUNNSs (15), pTF7–5BUNM, and pTF7–5BUNL was followed by transfection with transcription plasmids pT7riboBUNN, pT7riboBUNNSs, pT7riboBUNM, and pT7riboBUNL(+)Xho. The last plasmid contains an XhoI site introduced within the L coding sequence as a genetic marker (16). The progeny from the transfection experiment was passaged in A. albopictus C6/36 mosquito cells (to remove the vaccinia virus), and the supernatant was plaqued on BHK cells. A few plaques were observed that were markedly smaller than those given by wt virus, obtained from a rescue experiment carried out in parallel but by using wt plasmids (Fig. 2). Wild-type BUN plaques were around 3–5 mm in diameter, whereas those of the transfectant viruses were typically 1–2 mm in diameter. Seven plaques from the rescue experiment were grown up, and two of these (designated 1a and 9a) were passaged 10 times in BHK cells. The yields of these viruses were about 10-fold lower than wt, giving titers approximately 1–5 × 106 pfu/ml.
Figure 2.
Comparison of plaques produced by wt BUN (Left) and the BUNdelNSs mutant (Right) in BHK cells. Cell monolayers were fixed with 4% formaldehyde and stained with Giemsa's solution.
Analysis of Viral RNAs.
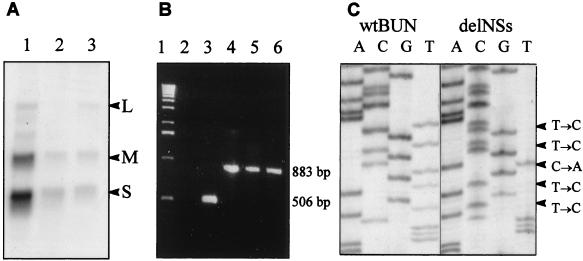
To verify that the transfectant viruses contained four genome segments as expected, viral RNAs were analyzed by Northern blots and reverse transcriptase–PCR (RT-PCR). However, Northern blots of RNA extracted from cells infected with the two transfectant viruses 1a or 9a revealed only three size classes of viral RNAs, corresponding to the L, M, and S segments, with no evidence for an RNA smaller than authentic S (Fig. 3A). This was confirmed by RT-PCR of virion RNA (Fig. 3B). Primers were designed such that the full-length S RNA (encoding just N protein) and the smaller S-like RNA, encoding NSs, would give products of different sizes (883 and 506 bases, respectively). By using RNA prepared from both transfectant viruses 1a and 9a, only the 883-bp product was observed. Subsequent nucleotide sequence analysis of the S RNA from both 1a and 9a viruses showed it to be identical in sequence to the transfected pT7riboBUNN plasmid (Fig. 3C). Thus, the transfectant viruses apparently contained three segments of RNA in their genomes, of which the S segment encoded only the N protein and not NSs.
Figure 3.
Characterization of the transfectant virus genome. (A) Northern blot analysis. BHK cells were infected with wt BUN (lane 1) or with transfectant viruses 1a (lane 2) or 9a (lane 3). Total cell RNA was extracted at 31 h after infection and separated on a 1.2% agarose gel containing 3.7% formaldehyde. Blotted RNA was hybridized with a mixture of 32P-labeled riboprobes that detected the L, M, and S RNA segments, as indicated. (B) RT-PCR analysis. Virion RNA was reverse transcribed by using primer NS2, and cDNA products were amplified by PCR by using primers S4 and NS2. PCR products from plasmid constructs pT7riboBUNN and pT7riboBUNNSs are included as markers for the expected product sizes. Lane 1, 1-kbp ladder; lane 2, PCR negative control; lane 3, PCR product from pT7riboBUNNSs; lane 4, PCR product from pT7riboBUNN; lane 5, RT-PCR product from transfectant virus 1a RNA; lane 6, RT-PCR product from transfectant virus 9a RNA. (C) Nucleotide sequence comparison. The nucleotide sequences of cloned RT-PCR products from the S segment RNA of wt BUN (Left) or transfectant virus 9a (Right) were determined in the region of the NSs start codon. The nucleotide changes from the wt to the mutant sequence are indicated (Right).
Protein Synthesis in Virus-Infected Cells.
To analyze the proteins synthesized by the transfectant viruses, BHK cells were infected at 5 pfu per cell with wt BUN or transfectant virus, labeled with [35S]methionine at 24 h after infection, and cell extracts were analyzed by PAGE followed by autoradiography (Fig. 4A). A related bunyavirus, Maguari virus, was included in this experiment to aid in identification of viral proteins. The NSs protein was clearly identified in wt BUN and Maguari virus-infected cells but was absent in the extracts of cells infected with the transfectant virus. Thus, the transfectant viruses were designated BUNdelNSs. It was also noticeable that the amount of N protein produced by BUNdelNSs, relative to the G1 protein, was increased compared with wt. Quantification of this gel by using a PhosphorImager indicated that approximately 10 times more N protein was produced in cells infected with BUNdelNSs compared with wt BUN.
Figure 4.
Protein profiles of wt and transfectant bunyaviruses. (A) Absence of NSs protein and overexpression of N protein in cells infected with the transfectant viruses. BHK cells were infected with wt BUN (lane 1), Maguari virus (lane 2), or with transfectant viruses BUNdelNSs 1a (lane 3) or BUNdelNSs 9a (lane 4) and labeled with [35S]methionine for 2 h at 24 h after infection. Cell lysates were separated by SDS/PAGE, and labeled proteins were visualized by autoradiography. Positions of bunyavirus proteins are indicated. (B) Shutoff of host cell protein synthesis. BHK cells were infected with 5 pfu per cell of wt BUN (lane 1), transfectant virus BUNdelNSs 1a (lane 2), and transfectant virus BUNdelNSs 9a (lane 3) or were mock infected (lane 4). Cells were labeled with [35S]methionine for 2 h at 48 h after infection, and then equal amounts of cell lysate were analyzed by SDS/PAGE.
NSs-Deleted Viruses Are Defective in Shutoff of Host Cell Protein Synthesis.
Examination of the gel in Fig. 4A showed there was apparently a reduced shutoff of host cell protein synthesis in BUNdelNSs-infected cells compared with wt. To investigate this more fully, BHK cells were infected with 5 pfu per cell of each virus and radiolabeled under identical conditions, and equal amounts of cell lysate were analyzed by PAGE (Fig. 4B). At 48 h after infection, host cell protein synthesis was almost completely shut off in wt BUN-infected BHK cells. In contrast, in BUNdelNSs-infected cells, host cell protein synthesis was reduced compared with mock-infected cells but not inhibited to the same extent as in wt-infected cells.
Pathogenesis of BUNdelNSs in BALB/c Mice.
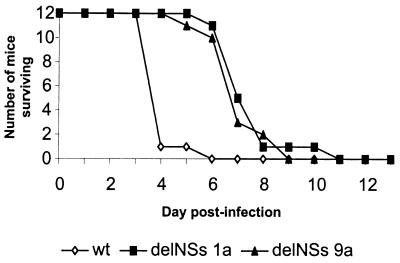
Groups of 12 5-week-old BALB/c mice were inoculated with wt or BUNdelNSs viruses either i.c. with 2,000 pfu or i.p. with 10,000 pfu and monitored for 30 days. With the exception of one mouse inoculated i.p. with wt BUN, which died at 7 days after infection, all mice inoculated i.p. survived (data not shown). In contrast, mice inoculated i.c. by either virus died, but there was a clear difference in disease progression (Fig. 5). Eleven of the 12 mice inoculated i.c. with wt BUN were dead or moribund and had to be killed by 4 days after infection, whereas most of the mice inoculated with BUNdelNSs survived for at least 3 days longer.
Figure 5.
Pathogenesis of bunyaviruses in mice. Three groups of 12 5-week-old female BALB/c mice were inoculated i.c. with 2,000 pfu of wt BUN or BUNdelNSs viruses 1a or 9a. The animals were monitored for 14 days. Any mice that were moribund or severely paralyzed were killed and scored as dead that day.
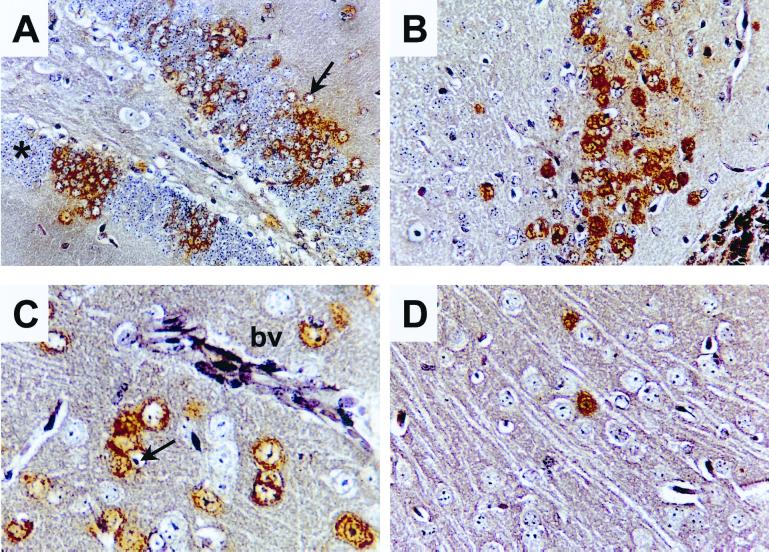
To examine this phenotypic difference in more detail, brains were taken from the mice, and virus titers were determined by plaque assay, whereas the distribution and tropism of the infection were observed by immunostaining. Brains were removed at days 4, 8, and 14 after infection from the i.p.-infected mice and at 4 and 8 days from the i.c.-infected mice. The results from these experiments are summarized in Table 1. At day 4, a low level of infectious virus was detected in the brain of one of three mice infected i.p. with wt BUN. Of the mice inoculated i.p. with BUNdelNSs, none had detectable infectious virus in the brain at 4 or 8 days. In contrast, all of the mice infected i.c. had infectious virus in the brain at day 4, with wt virus reaching a higher titer than the BUNdelNSs viruses. The tropism of both viruses was essentially similar in that neurones were infected (Fig. 6). However, wt virus had spread to many cells by 4 days after infection i.c. (A–C), whereas BUNdelNSs (D) was detected in only a few cells. In both cases, most of the infected cells had a normal neuronal morphology. The attenuation of BUNdelNSs virus in infected mice suggests a role for NSs as a virulence factor.
Table 1.
Pathogenesis of wt and delNSs viruses in BALB/c mice
| Virus | Route* | Day† | Titers‡ |
|---|---|---|---|
| wt | i.p. | 4 | 0, 0, 4 × 102 |
| 8 | 0, 0 | ||
| 1a | i.p. | 4 | 0, 0, 0 |
| 8 | 0, 0 | ||
| 9a | i.p. | 4 | 0, 0, 0 |
| 8 | 0, 0 | ||
| wt | i.c. | 4 | 1.8 × 107, 6.1 × 107 |
| 8 | ND§ | ||
| 1a | i.c. | 4 | 2.5 × 105, 2 × 106, 2 × 105 |
| 8 | 1 × 105, ND¶ | ||
| 9a | i.c. | 4 | 3 × 106, 5 × 106, 1.7 × 106 |
| 8 | 4.2 × 104, 1.3 × 105 |
ND, not determined.
Inoculations given by i.p. contained 10,000 pfu virus in 0.1 ml of PBSA, whereas those given by i.c. contained 2,000 pfu virus in 0.02 ml of PBSA.
Mice were killed and the brains halved and frozen.
The titer of virus in half of the brain homogenate was determined. Zero titer implies titer below the level of detectability.
All mice dead by 4 days after infection.
Second mouse died at 7 days after infection.
Figure 6.
Histology and immunostaining of brain sections from infected BALB/c mice. Animals were infected with 2,000 pfu of wt BUN (A–C) or BUNdelNSs 1a (D) by the intracerebral route. Mice were killed 4 days after infection, and brain sections were prepared. Micrographs show different areas of the brain immunostained for viral proteins (brown). (A) Foci of positive neurones in the hippocampal dentate gyrus. Note that staining is predominantly cytoplasmic and that several infected cells have enlarged pale staining nuclei (arrow). For comparison, unstained neurones with a normal morphology are indicated by an asterisk. (B) Positive neurones in the anterior olfactory nucleus. C and D show sections of the frontal cortex. (C) Positive neurones adjacent to a blood vessel (bv) in the cortex. Note again the enlarged pale staining nuclei (arrow) of infected cells. (D) Infection with BUNdelNSs virus results in fewer infected cells.
Induction of an IFN-β Promoter by BUNdelNSs Virus.
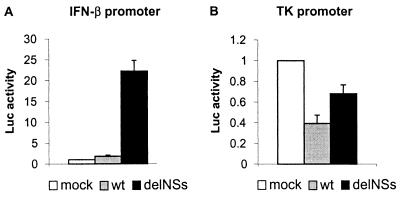
Type I interferons (IFN α and β) are important components of the innate immune system, and accessory proteins encoded by several negative-strand RNA viruses have been found to antagonize the IFN system (17, 29–32). In particular, the NS1 protein of influenza A virus counteracts the production of IFN in infected cells, resulting in an attenuated phenotype of NS1-deleted viruses (29, 33). To investigate whether the BUN NSs gene could play a similar role, we studied IFN induction in a transient transfection assay. Cells derived from BALB/c mice were transfected with a reporter plasmid containing the IFN-β promoter linked to a luciferase gene and subsequently mock infected or infected with wt BUN or BUNdelNSs. As shown in Fig. 7A, luciferase activity in wt BUN-infected cells was scarcely above the background level measured in mock-infected cells. In contrast, there was approximately 20-fold increase in luciferase activity in cells infected with BUNdelNSs, indicating strong activation of the IFN-β promoter. It is formally possible that the difference in IFN induction may have been because of the reduced ability of BUNdelNSs to shut off host cell protein synthesis. To address this question, cells were transfected with a plasmid containing the luciferase gene under control of the weak herpes simplex virus-thymidine kinase promoter and then infected as before. Fig. 7B shows that control promoter activity was slightly reduced when cells were infected with either wt BUN or BUNdelNSs viruses; wt BUN had a greater impact than BUNdelNSs. The difference between wt BUN and BUNdelNSs, however, was only about 2-fold at the observed time point, whereas the difference in activation of the IFN-β promoter was more than 20-fold. This indicates that activation of the IFN-β promoter was specific and independent of the reduced ability of BUNdelNSs to shut off host cell protein synthesis. Thus, the loss of NSs changed BUN from a noninducer to an inducer of IFN.
Figure 7.
Induction of an IFN-β promoter by infection with BUNdelNSs virus. Monolayers of BF cells were transfected in parallel with the IFN-β promoter-containing plasmid (A) or with the herpes simplex virus-thymidine kinase promoter-containing control plasmid (B). At 6 h after transfection, the cells were mock infected or infected with wt BUN or BUNdelNSs virus 9a at 5 pfu per cell. Fifteen hours after infection, luciferase activities in cell lysates were measured. The activity measured in mock-infected cells was taken as 1, and averages (columns) and standard deviations (error bars) of the luciferase activities of three independent experiments are shown.
Discussion
Previous knowledge of the NSs protein encoded by members of the Bunyaviridae is sparse, particularly for viruses in the Bunyavirus genus. There is great variability in both size and amino sequence between the proteins designated NSs in different genera, and it is possible that there may be no functional equivalence between them. Further, members of both the Hantavirus and Nairovirus genera do not encode a nonstructural protein in their S segments. There is also variation in the coding strategies used by viruses in different genera, and for phleboviruses and tospoviruses, the NSs gene is expressed in an ambisense orientation. In addition, these NSs proteins are larger than those of bunyaviruses (32 kDa for Uukuniemi phlebovirus, 52 kDa for tomato-spotted wilt tospovirus, as compared with 11 kDa for Bunyamwera virus; refs. 1–5).
The data presented in this paper provide insights into the possible function(s) of the bunyavirus NSs product. The generation of viruses lacking the NSs gene shows conclusively that the product is not essential for replication in either cultured cells or in mice, and thus NSs can be designated as an accessory protein. The failure to generate a four-segment virus with NSs encoded on a separate segment (all seven isolated plaques were NSs−; data not shown) suggests a selection pressure against viruses in which NSs production is not tightly regulated by being expressed from the same mRNA as N. Alternatively, the rescue of viruses containing a genome diploid for the S-like segment may be less efficient and below the sensitivity of our procedure. An unexpected finding was that the BUN NSs protein may be involved in the inhibition of host cell protein synthesis. Two studies have addressed this mechanism previously. Raju and Kolakofsky (34) reported that La Crosse bunyavirus infection induces mRNA instability in mammalian cells and showed that this instability was independent of the interferon pathway. They suggested that the viral endonuclease activity involved in viral mRNA initiation may mediate this instability; cellular mRNAs would be susceptible to exonuclease degradation if their 5′-capped ends were removed. Frugulhetti and Rebello (35) examined changes in Na+ and K+ ion concentrations in Marituba bunyavirus-infected cells and showed an increase in Na+ and decrease in K+ ion concentration in infected mammalian cells, which correlated temporally with inhibition of host cell protein synthesis. No changes in intracellular ion concentration were noted in infected mosquito cells. The reduction, but not the abrogation, of host cell protein shutoff by BUNdelNSs suggests also that more than one pathway may be involved. This process is clearly highly cell specific, because neither wt BUN (36) nor BUNdelNSs (data not shown) shuts off host cell protein synthesis in mosquito cells.
We also showed that NSs plays an important role in controlling IFN induction after infection by BUN. Therefore, NSs is remarkably similar to the multifunctional NS1 gene product of influenza A virus, which enables the virus to inhibit host cell gene expression and stimulate its own protein synthesis (37–39), but it also is an IFN antagonist (29). Because both influenza A virus and BUN are negative-strand viruses with a segmented genome, they may have similar requirements for growth (shutoff of host cell protein synthesis and IFN antagonism) that have resulted in a convergent evolution of accessory proteins. Influenza A virus NS1 is thought to antagonize the IFN system by binding to and sequestering double-stranded RNA (29), an intermediate product of viral replication that is a potent trigger of IFN induction. Despite the functional similarities, a sequence comparison between BUN NSs and influenza A virus NS1 did not reveal any conserved motifs, and no RNA-binding motif could be identified in the BUN NSs sequence (data not shown). This indicates that BUN NSs might antagonize the IFN system by a mechanism different to influenza A virus NS1.
The different growth properties of mutant and wt viruses appear not to depend on cell type, because we have also observed up to 1 log lower titers of NSs-deleted viruses in CV-1, HeLa, LLCMK2, and VeroE6 cells in addition to BHK cells (data not shown). To determine whether growth of BUNdelNSs is impaired by the induced IFN, multistep growth curve experiments with low multiplicity infection of IFN-competent and IFN-deficient cell lines are in progress.
After inoculation of 10,000 pfu virus i.p., neither wt nor the NSs-deleted viruses appeared to be efficiently neuroinvasive in mice. Studies on La Crosse and Tahyna bunyaviruses indicate that neuroinvasion is highly variable and requires a threshold dose of inoculum (40). In contrast, the i.c. inoculations were lethal and highlighted a clear phenotypic difference between wt and NSs-deleted viruses. The absence of NSs did not affect the tropism of the virus in the mouse brain but did affect the ability of the virus to replicate in or to spread between central nervous system cells. Furthermore, mice inoculated i.c. with BUNdelNSs survived at least 3 days longer than mice inoculated with wt BUN. We suggest that the failure to counteract the induction of IFN is also responsible for the impaired ability of the NSs-deleted virus to replicate and spread in the adult mouse brain. In addition, the reduced shutoff observed in cultured cells may also contribute to the attenuated phenotype. To distinguish between these possibilities, infection experiments with mice that are deficient in IFN responses are currently under way.
In summary, our results suggest that BUN NSs is an accessory protein that acts as a virulence factor by helping the virus to shut off host cell protein synthesis and to inhibit IFN induction.
Acknowledgments
We thank Stephen Goodbourn for providing the reporter plasmids, Rick Randall and members of our laboratory for helpful discussions and suggestions, and Susan Jacobs for critically reviewing the manuscript. This work was supported by Medical Research Council Grant G9535974 and Wellcome Trust Grants 048387 and 046745 (to R.M.E.). F.W. received a Long Term Fellowship ALTF 36-1998 from the European Molecular Biology Organization.
Abbreviations
- wt
wild type
- BUN
Bunyamwera virus
- RT
reverse transcriptase
- i.c.
intracerebrally
- RT-PCR
reverse transcriptase–PCR
- pfu
plaque-forming unit
References
- 1.Elliott R M. Mol Med. 1997;3:572–577. [PMC free article] [PubMed] [Google Scholar]
- 2.Bishop D H L. In: The Bunyaviridae. Elliott R M, editor. New York: Plenum; 1996. pp. 19–61. [Google Scholar]
- 3.Bouloy M. Adv Virus Res. 1991;40:235–275. doi: 10.1016/s0065-3527(08)60281-x. [DOI] [PubMed] [Google Scholar]
- 4.Elliott R M. J Gen Virol. 1990;71:501–522. doi: 10.1099/0022-1317-71-3-501. [DOI] [PubMed] [Google Scholar]
- 5.Schmaljohn C S. In: Virology. 2nd Ed. Fields B N, Knipe D M, editors. New York: Raven; 1996. pp. 1447–1471. [Google Scholar]
- 6.Dunn E F, Pritlove D C, Elliott R M. J Gen Virol. 1994;75:597–608. doi: 10.1099/0022-1317-75-3-597. [DOI] [PubMed] [Google Scholar]
- 7.Bowen M D, Jackson A O, Bruns T D, Hacker D L, Hardy J L. J Gen Virol. 1995;76:559–572. doi: 10.1099/0022-1317-76-3-559. [DOI] [PubMed] [Google Scholar]
- 8.Saeed M F, Wang H, Nines M, Vasconcelos P F C, Weaver S C, Shope R E, Watts D M, Tesh R B, Barrett A D T. J Gen Virol. 2000;81:743–745. doi: 10.1099/0022-1317-81-3-743. [DOI] [PubMed] [Google Scholar]
- 9.Bellocq C, Kolakofsky D. J Virol. 1987;61:3960–3967. doi: 10.1128/jvi.61.12.3960-3967.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bellocq C, Raju R, Patterson J L, Kolakofsky D. J Virol. 1987;61:87–95. doi: 10.1128/jvi.61.1.87-95.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bouloy M, Hannoun C. Virology. 1976;69:258–264. doi: 10.1016/0042-6822(76)90212-9. [DOI] [PubMed] [Google Scholar]
- 12.Gerbaud S, Pardigon N, Vialat P, Bouloy M. In: The Biology of Negative-Strand Viruses. Mahy B W J, Kolakofsky D, editors. Amsterdam: Elsevier; 1987. pp. 191–198. [Google Scholar]
- 13.Patterson J L, Holloway B, Kolakofsky D. J Virol. 1984;52:215–222. doi: 10.1128/jvi.52.1.215-222.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vialat P, Bouloy M. J Virol. 1992;66:685–693. doi: 10.1128/jvi.66.2.685-693.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunn E F, Pritlove D C, Jin H, Elliott R M. Virology. 1995;211:133–143. doi: 10.1006/viro.1995.1386. [DOI] [PubMed] [Google Scholar]
- 16.Bridgen A, Elliott R M. Proc Natl Acad Sci USA. 1996;93:15400–15404. doi: 10.1073/pnas.93.26.15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Didcock L, Young D F, Goodbourn S, Randall R E. J Virol. 1999;73:3125–3133. doi: 10.1128/jvi.73.4.3125-3133.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watret G E, Pringle C R, Elliott R M. J Gen Virol. 1985;66:473–482. doi: 10.1099/0022-1317-66-3-473. [DOI] [PubMed] [Google Scholar]
- 19.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current Protocols in Molecular Biology. New York: Wiley; 1993. [Google Scholar]
- 20.King P, Goodbourn S. J Biol Chem. 1994;269:30609–30615. [PubMed] [Google Scholar]
- 21.Rose J K, Buonocore L, Whitt M A. BioTechniques. 1991;4:520–525. [PubMed] [Google Scholar]
- 22.Fazakerley J K, Pathak S, Scallan M, Amor S, Dyson H. Virology. 1993;195:627–637. doi: 10.1006/viro.1993.1414. [DOI] [PubMed] [Google Scholar]
- 23.Pringle C R. In: The Bunyaviridae. Elliott R M, editor. New York: Plenum; 1996. pp. 189–226. [Google Scholar]
- 24.Iroegbu C U, Pringle C R. J Virol. 1981;37:383–394. doi: 10.1128/jvi.37.1.383-394.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rozhon E J, Gensemer P, Shope R E, Bishop D H L. Virology. 1981;111:125–138. doi: 10.1016/0042-6822(81)90659-0. [DOI] [PubMed] [Google Scholar]
- 26.Urquidi V, Bishop D H L. J Gen Virol. 1992;73:2255–2265. doi: 10.1099/0022-1317-73-9-2255. [DOI] [PubMed] [Google Scholar]
- 27.Enami M, Sharma G, Benham C, Palese P. Virology. 1991;185:291–298. doi: 10.1016/0042-6822(91)90776-8. [DOI] [PubMed] [Google Scholar]
- 28.Garcin D, Pelet T, Calain P, Roux L, Curran J, Kolakofsky D. EMBO J. 1995;14:6087–6094. doi: 10.1002/j.1460-2075.1995.tb00299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy D E, Durbin J E, Palese P, Muster T. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 30.Garcin D, Latorre P, Kolakofsky D. J Virol. 1999;73:6559–6565. doi: 10.1128/jvi.73.8.6559-6565.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu Y, Wambach M, Katze M G, Krug R M. Virology. 1995;214:222–228. doi: 10.1006/viro.1995.9937. [DOI] [PubMed] [Google Scholar]
- 32.Goodbourn S, Didcock L, Randall R E. J Gen Virol. 2000;81:2341–2364. doi: 10.1099/0022-1317-81-10-2341. [DOI] [PubMed] [Google Scholar]
- 33.Talon J, Salvatore M, O'Neill R E, Nakaya Y, Zheng H, Muster T, Garcia-Sastre A, Palese P. Proc Natl Acad Sci USA. 2000;97:4309–4314. doi: 10.1073/pnas.070525997. . (First Published March 21, 2000; 10.1073/pnas.070525997) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raju R, Kolakofsky D. J Virol. 1988;62:27–32. doi: 10.1128/jvi.62.1.27-32.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frugulhetti I C P P, Rebello M A. J Gen Virol. 1989;70:3493–3499. doi: 10.1099/0022-1317-70-12-3493. [DOI] [PubMed] [Google Scholar]
- 36.Elliott R M, Wilkie M L. Virology. 1986;150:21–32. doi: 10.1016/0042-6822(86)90262-x. [DOI] [PubMed] [Google Scholar]
- 37.Enami K, Sato T A, Nakada S, Enami M. J Virol. 1994;68:1432–1437. doi: 10.1128/jvi.68.3.1432-1437.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fortes P, Beloso A, Ortin J. EMBO J. 1994;13:704–712. doi: 10.1002/j.1460-2075.1994.tb06310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu Y, Quian X Y, Krug R M. Genes Dev. 1994;8:1817–1828. doi: 10.1101/gad.8.15.1817. [DOI] [PubMed] [Google Scholar]
- 40.Janssen R, Gonzalez-Scarano F, Nathanson N. Lab Invest. 1984;50:447–455. [PubMed] [Google Scholar]