The past decade has seen dramatic advances in our knowledge of plant photoreceptors and in our understanding of the signal transduction pathways that they activate (Briggs and Olney, 2001). A major part of these advances has been the identification and characterization of photoreceptors that respond to signals from the blue region of the electromagnetic spectrum. We now know that there are at least two classes of blue light photoreceptors: the cryptochromes and the phototropins. The purpose of this letter is to establish a uniform terminology for the phototropins. Knowledge of their occurrence across the plant kingdom, their structure, and their unique photochemistry is growing at a rapid rate, and it is timely to provide a consistent and simple nomenclature based on their known properties, both structural and functional.
In 1995, Liscum and Briggs identified a genetic locus, designated NPH1 (nonphototropic hypocotyl 1), which encodes a plasma membrane–associated protein known to be essential for most phototropic responses in Arabidopsis. The phototropism mutant JK224, previously described by Khurana and Poff (1989), turned out to be mutated at the NPH1 locus. When plasma membrane preparations from dark-grown seedlings are irradiated before adding ATP, a 120-kD plasma membrane protein becomes heavily phosphorylated. This protein is present at most in trace amounts in the mutant JK224 (Reymond et al., 1992), providing genetic evidence for its role in phototropism. Cloning and sequencing of the NPH1 gene showed that the encoded protein contains all 11 classic motifs expected of a serine/threonine protein kinase (Hanks and Hunter, 1995), indicating that the protein itself was the kinase involved and that the light-activated reaction was an autophosphorylation event (Huala et al., 1997). When the protein was expressed in insect cells via Baculovirus transfection, it still retained light-activated phosphorylation, leading Christie et al. (1998) to conclude that it was itself the photoreceptor for the reaction and therefore a photoreceptor for phototropism. As a result, Christie et al. (1999) assigned the NPH1-encoded protein the trivial name “phototropin” after its functional role in phototropism.
There are now a number of phototropin-like sequences in GenBank. (1) Jarillo et al. (1998) entered a sequence en-coding a slightly smaller Arabidopsis protein with 58% identity and 67% similarity to phototropin. The gene was initially designated NPH2, but this was changed subsequently to NPL1 (NPH1-like 1) because the designation NPH2 was already used to describe another locus involved in phototropism (Liscum and Briggs, 1995). (2) Zacherl et al. (1998) entered sequences for two very similar NPH1 genes from Avena sativa (oat) designated NPH1-1 and NPH1-2. (3) These workers also have entered an NPH1 sequence from Zea mays (maize) (accession number AF033263). (4) Kanegae et al. (2000) reported two Oryza sativa (rice) NPH1-like genes that they designated OsNPH1a and OsNPH1b encoding proteins in rice (cv Nipponbare) that were 61 and 62% identical, respectively, to Arabidopsis phototropin at the protein level. The two genes show different tissue expression and differential regulation by light, suggesting that they might serve different functions. (5) Tyagi et al. (S.B. Tyagi, A.K. Tyagi, and J.P. Khurana, unpublished data) submitted an NPH1 homolog from a different strain of rice (Pusa Basmati 1) to GenBank (accession number CAB65325). (6) The Watson laboratory (R.V. Oakley, S.N. Chary, T. Mitra, and J.C. Watson, unpublished data) recently entered the complete sequence for the pea phototropin homolog PsPK4 (accession number U83281). (7) The Wada laboratory (Nozue et al., 2000) reported a gene with NPH1 homology from the fern Adiantum capillus-veneris. (8) Finally, Nozue et al. (1998) reported a novel chimeric protein from Adiantum. This protein has high homology with phytochrome (including a chromophore binding site and classic phytochrome photoreversibility) at its N-terminal end but with an almost complete phototropin homolog replacing the expected phytochrome sequence at its C-terminal end. These various genes and their GenBank accession numbers and pertinent references are listed in Table 1. (Note that the Arabidopsis NPL1, NPH2, and PK7 entries all report the same sequence.) Also included in Table 1 is a phototropin-like gene from Chlamydomonas reinhardtii, its sequence kindly provided by Dr. Akira Nagatani, and a second phototropin homolog, NPL1, from Adiantum, its sequence kindly provided by Dr. Takatoshi Kagawa.
Table 1.
Phototropin Homologs
| Gene Designation
|
||||
|---|---|---|---|---|
| Species | Old | New | Accession Number | Source |
| Oryza sativa | OsNPH1a | PHOT1 | AB018443 | Kanegae et al. (2000) |
| O. sativa | OsNPH1b | PHOT2 | AB018444 | Kanegae et al. (2000) |
| O. sativa | NPH1 | PHOT1 | CAB65325 | Tyagi et al. (2000) |
| Arabidopsis thaliana | NPH1 | PHOT1 | AF030864 | Huala et al. (1997) |
| A. thaliana | NPL1 | PHOT2 | AF053941 | Jarillo et al. (1998) |
| A. thaliana | NPH2 | PHOT2 | AF053941 | Jarillo et al. (1998) |
| A. thaliana | PK7, partial | PHOT2 | Q05999 | Accession only |
| Avena sativa | NPH1-1 | PHOT1a | AF033096 | Zacherl et al. (1998) |
| A. sativa | NPH1-2 | PHOT1b | AF033097 | Zacherl et al. (1998) |
| Zea mays | NPH1 | PHOT1 | AF033263 | Accession only |
| Pisum sativum | PsPK4 | PHOT1 | U83281 | Accession only |
| Adiantum capillus-veneris | PHY3 | PHY3 | AB012082 | Nozue et al. (1998) |
| A. capillus-veneris | NPH1 | PHOT1? | AB037188 | Nozue et al. (2000) |
| A. capillus-veneris | NPL1 | PHOT2 | Courtesy of T. Kagawa | |
| Chlamydomonas reinhardtii | NPH1 | Phot | Courtesy of A. Nagatani | |
| Pisum sativum | PsPK5 | None | M69034, M92989 | Lin and Watson (1992) |
| Spinacia oleracea | Protein kinase | None | Z30332 | Accession only |
| Mesembryanthemum crystallinum | Protein kinase | None | Z30331, Z30333 | Bauer et al. (1994) |
Table 1 also includes three fairly long partial sequences for apparent phototropin homologs that have been identified in Spinacia oleracea (spinach) (accession number Z30332), Mesembryanthemum crystallinum (ice plant) (Bauer et al., 1994), and Pisum sativum (pea) (Lin and Watson, 1992) that have been submitted to GenBank. Although GenBank contains other partial sequences that may represent phototropins, these are partial sequence fragments and are not included here.
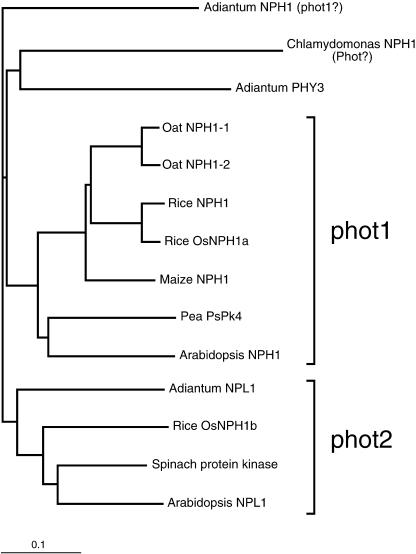
A phylogram showing relationships between the various phototropin proteins is presented in Figure 1. For the angiosperms, the sequences cluster into two branches. The lower branch contains Adiantum NPL1, OsNPH1b, the spinach NPH1 homolog, and Arabidopsis NPL1, with all of the remaining angiosperm sequences clustered in the upper branch. The Adiantum truncated PHY3 (with NPH1 homolog domain) and the Chlamydomonas NPH1 homolog are more distantly related. The second Adiantum NPH1 homolog is the most distant, which is not surprising because Adiantum is not an angiosperm but a fern. Note that the two oat protein sequences, NPH1-1 and NPH1-2, are almost identical, probably reflecting the polyploid origin of cultivated oat. The pea PsPK5 and ice plant phototropin protein sequences are not shown in this analysis because the submitted sequences were considered too short for inclusion. However, the somewhat more complete spinach sequence was included.
Figure 1.
A Phylogram of the Phototropin Family of Blue Light Receptors.
Clustal X (version 1.64b) was used to align the protein sequences. The family tree was obtained by the neighbor-joining method using 1000 bootstrap replicates and was visualized using TreeView (version 1.6.2). The scale represents 0.1 substitutions per site.
All of these putative photoreceptor proteins contain two domains, designated LOV domains (PAS domains found in a wide range of signaling proteins activated by light, oxygen, or voltage; Huala et al., 1997, Taylor and Zhulin, 1999). Although it is unlikely, spinach could be an exception, because the incomplete sequence submitted terminates upstream from the second LOV domain and therefore is missing LOV1. The LOV domains from oat and Arabidopsis NPH1 subsequently were shown to bind flavin mononucleotide (FMN; Christie et al., 1999), and those from oat were demonstrated to undergo a unique photocycle involving the light-activated formation of a flavin C(4a)-cysteinyl adduct (Miller et al., 1990) followed by a subsequent dark decay (Salomon et al., 2000). Both LOV domains from Arabidopsis NPL1 and Adiantum NPH1 and PHY3 as well as those from both rice homologs have been shown to bind FMN, and all of these domains (including those from Arabidopsis phototropin) show light-activated spectral changes and subsequent dark recovery, strong evidence that they undergo the same photocycle demonstrated for the oat LOV domains (Sakai et al., 2001; M. Kasahara, T.E. Swartz, M.A. Olney, J.M. Christie, and W.R. Briggs, unpublished data).
Liscum and Briggs (1995) originally reported that the nph1-5 null mutant of phototropin lacked all phototropic responses tested. However, Sakai et al. (2000) recently found that the Arabidopsis nph1-101 mutant, in their hands, showed strong phototropic curvatures in response to continuous light of relatively high fluence rates. They hypothesized that this phototropic response in Arabidopsis involves a second photoreceptor. It was only when they examined an Arabidopsis nph1 npl1 double mutant that this response was almost eliminated (Sakai et al., 2001). Hence, apparently, the NPH1-encoded protein can mediate phototropism in response to light pulses or low levels of continuous light, whereas the NPL1-encoded protein is activated only at higher fluence rates of continuous light.
These photoreceptors are not involved only in phototropism. Kagawa et al. (2001) and Jarillo et al. (2001) have demonstrated that the chloroplast avoidance response observed under strong light (movement of chloroplasts from the periclinal to the anticlinal walls of leaf mesophyll cells, leading to maximal mutual shading) is lacking in an Arabidopsis npl1 mutant. However, chloroplast accumulation on the periclinal walls under dim light, maximizing light interception, appears relatively normal, although the sensitivity to low-fluence-rate blue light is reduced slightly (Kagawa and Wada, 2000). In a recent study, Sakai et al. (2001) found that neither the accumulation nor the avoidance response could be detected in the nph1 npl1 double mutant. Thus, evidently, the NPH1 protein can mediate light-activated chloroplast accumulation over a wide fluence range, whereas NPL1 by itself can mediate accumulation over a low-fluence-rate range and avoidance at higher fluence rates.
It is intriguing that rice NPH1a, most closely related to Arabidopsis NPH1, predominates in rice coleoptiles and is downregulated by light, whereas rice NPH1b, most closely related to Arabidopsis NPL1, predominates in leaves and is upregulated by light. Given the relative importance of these two photoreceptors in phototropism and chloroplast movement, respectively, under different fluence rates, this distribution pattern and the difference in light regulation are not surprising.
The gene terminology as it exists is complex and somewhat confusing (Table 1, second column). Because the angiosperm proteins fall naturally into two phylogenetic groups on the basis of sequence, and also likely fall into two groups based on function, we propose renaming the groups phot1 and phot2 (Figure 1, Table 2). In this scheme, as with the phytochromes (Quail et al., 1994), the wild-type genes are designated PHOT1 and PHOT2, respectively, and the mutants are designated phot1 and phot2. The holoproteins are designated phot1 and phot2, and the apoproteins are designated PHOT1 and PHOT2, in keeping with the current practice for phytochrome (Table 2). This uni-fication of terminology parallels that introduced for other plant photoreceptors: hy4 mutant alleles are now designated cry1 (Lin et al., 1995), hy3 alleles are now designated phyB (Reed et al., 1993), and hy8 (Dehesh et al., 1993), fre1 (Nagatani et al., 1993), and fhy2 (Whitelam et al., 1993) alleles are now all designated phyA (Quail et al., 1994). Where two proteins from the same species fall within one cluster, as in the case of the oat phototropins, their genes are designated PHOT1a and PHOT1b, as shown in Table 1. In the case of Chlamydomonas NPH1 and Adiantum PHY3, we have simply designated the former Phot (for consistency with common gene nomenclature in Chlamydomonas, the first letter is capitalized) and have retained PHY3 for the latter.
Table 2.
Proposed Phototropin Nomenclature
| Wild-type gene | PHOT1, PHOT2 |
|---|---|
| Mutated gene | phot1, phot2 |
| Mutant alleles (from different laboratories) | phot1-1, phot1-101, etc. |
| Holoprotein | phot1, phot2 |
| Apoprotein | PHOT1, PHOT2 |
The scientific community performing research on phototropins has agreed to redesignate the photoreceptors NPH1 and NPL1 as phot1 and phot2. The phot designation is restricted to photoreceptor proteins with the following properties: two LOV domains in the N-terminal half, each of which binds FMN as a chromophore; a serine/threonine protein kinase in the C-terminal half; light-activated autophosphorylation; and light-activated formation of a flavin C(4a)-cysteinyl adduct and its subsequent dark decay. We further suggest that mutant alleles be designated according to the laboratory of origin, for example, phot1-1 instead of nph1-1 for the Briggs laboratory, phot1-101 instead of nph1-101 for the Okada laboratory, etc. (Table 2). A similar program was established previously to designate phytochrome mutants isolated in different laboratories (see Table 2 in Quail et al., 1994) and has been adopted universally.
References
- Bauer, B., Fischer, K., and Dietz, K.-J. (1994). cDNA sequence of a protein kinase from the inducible Crassulacean acid metabolism plant Mesembryanthemum crystallinum L., encoding a SNF-1 homolog. Plant Physiol. 106 1225–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs, W.R., and Olney, M.A. (2001). Photoreceptors in plant photomorphogenesis to date: Five phytochromes, two cryptochromes, one phototropin, and one superchrome. Plant Physiol. 125 85–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie, J.M., Reymond, P., Powell, G.K., Bernasconi, P., Raibekas, A., Liscum, E., and Briggs, W.R. (1998). Arabidopsis NPH1: A flavoprotein with the properties of a photoreceptor for phototropism. Science 282 1698–1701. [DOI] [PubMed] [Google Scholar]
- Christie, J.M., Salomon, M., Nozue, K., Wada, M., and Briggs, W.R. (1999). LOV (light, oxygen, or voltage) domains of the blue light photoreceptor phototropin (nph1): Binding sites for the chromophore flavin mononucleotide. Proc. Natl. Acad. Sci. USA 96 8779–8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehesh, K., Franci, C., Parks, B.M., Seeley, K.A., Short, T.W., Tepperman, J.M., and Quail, P.H. (1993). Arabidopsis HY8 en-codes phytochrome A. Plant Cell 5 1081–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks, S.K., and Hunter, T. (1995). The eukaryotic protein kinase superfamily: Kinase (catalytic) domain structure and classification. FASEB J. 9 576–596. [PubMed] [Google Scholar]
- Huala, E., Oeller, P.W., Liscum, E., Han, I.-S., Larsen, E., and Briggs, W.R. (1997). Arabidopsis NPH1: A protein kinase with a putative redox-sensing domain. Science 278 2121–2123. [DOI] [PubMed] [Google Scholar]
- Jarillo, J.A., Ahmad, M., and Cashmore, A.R. (1998). NPL1 (accession No. AF053941): A second member of the NPH serine/threonine kinase family of Arabidopsis (PGR98–100). Plant Physiol. 117 719. [Google Scholar]
- Jarillo, J.A., Gabrys, H., Capel, J., Alonso, J.M., Ecker, J.R., and Cashmore, A.R. (2001). Phototropin-related NPL1 controls blue light-induced chloroplast relocation. Nature (in press). [DOI] [PubMed]
- Kagawa, T., and Wada, M. (2000). Blue light-induced chloroplast relocation in Arabidopsis thaliana as analyzed by microbeam irradiation. Plant Cell Physiol. 41 84–93. [DOI] [PubMed] [Google Scholar]
- Kagawa, T., Sakai, T., Suetsugu, N., Oikawa, K., Ishiguro, S., Kato, T., Tabata, S., Okada, K., and Wada, M. (2001). Arabidopsis NPL1: A phototropin homologue controlling the chloroplast high-light avoidance response. Science 291 2138–2141. [DOI] [PubMed] [Google Scholar]
- Kanegae, H., Tahir, M., Savazzini, F., Yamamoto, K., Yano, M., Sasaki, T., Kanegae, T., Wada, M., and Takano, M. (2000). Rice homologues OsNPH1a and OsNPH1b are differently photoregulated. Plant Cell Physiol. 41 415–423. [DOI] [PubMed] [Google Scholar]
- Khurana, J.P., and Poff, K.L. (1989). Mutants of Arabidopsis thaliana with altered phototropism. Planta 178 400–406. [PubMed] [Google Scholar]
- Lin, C., Robertson, D.E., Ahmad, M., Raibekas, A.A., Schuman, M.S., Dutton, P.L., and Cashmore, A.R. (1995). Association of flavin adenine dinucleotide with the Arabidopsis blue light receptor CRY1. Science 269 968–970. [DOI] [PubMed] [Google Scholar]
- Lin, X., and Watson, J.C. (1992). cDNA sequence of PsPK5, a protein kinase homolog from Pisum sativum L. Plant Physiol. 100 1072–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum, E., and Briggs, W.R. (1995). Mutations in the NPH1 locus of Arabidopsis disrupt the perception of phototropic stimuli. Plant Cell 7 473–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, S.M., Massey, V., Ballou, D., Williams, C.H., Jr., Distefano, M.D., Moore, M.J., and Walsh, C.T. (1990). Use of a site-directed triple mutant to trap intermediates: Demonstration that the flavin C(4a)-thiol adduct and reduced flavin are kinetically competent intermediates in mercuric ion reductase. Biochemistry 29 2831–2841. [DOI] [PubMed] [Google Scholar]
- Nagatani, A., Reed, J.W., and Chory, J. (1993). Isolation and initial characterization of Arabidopsis mutants that are deficient in phytochrome A. Plant Physiol. 102 269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozue, K., Kanegae, T., Imaizumi, T., Fukuda, S., Okamoto, H., Yeh, K.-C., Lagarias, J.C., and Wada, M. (1998). A phytochrome from the fern Adiantum with features of the putative photoreceptor NPH1. Proc. Natl. Acad. Sci. USA 95 15826–15830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozue, K., Christie, J.M., Kiyosue, T., Briggs, W.R., and Wada, M. (2000). Isolation and characterization of a fern phototropin (accession No. AB037188), a putative blue-light photoreceptor for phototropism (PGR00–039). Plant Physiol. 122 1457.10759541 [Google Scholar]
- Quail, P.H., Briggs, W.R., Chory, J., Hangarter, R., Harberd, N.P., Kendrick, R.E., Koorneef, M., Parks, B., Sharrock, R.A., Schäfer, E., Thompson, W.F., and Whitelam, G.C. (1994). Spotlight on phytochrome nomenclature. Plant Cell 6 468–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, J.W., Nagpal, P., Poole, D.S., Furuya, M., and Chory, J. (1993). Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond, P., Short, T.W., Briggs, W.R., and Poff, K.L. (1992). Light-induced phosphorylation of a membrane protein plays an early role in signal transduction for phototropism. Proc. Natl. Acad. Sci. USA 89 4718–4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai, T., Wada, T., Ishiguro, S., and Okada, K. (2000). RPT2: A signal transducer of the phototropic response in Arabidopsis. Plant Cell 12 225–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai, T., Kagawa, T., Kasahara, M., Swartz, T.E., Christie, J.M., Briggs, W.R., Wada, M., and Okada, K. (2001). Nph1 and npl1: Blue-light receptors that mediate both phototropism and chloroplast relocation in Arabidopsis. Proc. Natl. Acad. Sci. USA (in press). [DOI] [PMC free article] [PubMed]
- Salomon, M., Christie, J.M., Knib, E., Lempert, U., and Briggs, W.R. (2000). Photochemical and mutational analysis of the FMN-binding domains of the plant blue light receptor, phototropin. Biochemistry 39 9401–9410. [DOI] [PubMed] [Google Scholar]
- Taylor, B.L., and Zhulin, I.B. (1999). PAS domains: Internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 22 479–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelam, G.C., Johnson, E., Peng, J., Carol, P., Anderson, M.L., Cowl, J.S., and Barberd, N.P. (1993). Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5 757–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacherl, M., Huala, E., Rüdiger, W., Briggs, W.R., and Salomon, M. (1998). Isolation and characterization of cDNAs from oat encoding a serine/threonine kinase: An early component in signal transduction for phototropism (Accession Nos. AF033096 and AF033097) (PGR98–028). Plant Physiol. 116 869. [Google Scholar]