Abstract
Archaeal box C/D sRNAs guide the methylation of specific nucleotides in archaeal ribosomal and tRNAs. Three Methanocaldococcus jannaschii sRNP core proteins (ribosomal protein L7, Nop56/58, and fibrillarin) bind the box C/D sRNAs to assemble the sRNP complex, and these core proteins are essential for nucleotide methylation. A distinguishing feature of the Nop56/58 core protein is the coiled-coil domain, established by α-helices 4 and 5, that facilitates Nop56/58 self-dimerization in vitro. The function of this coiled-coil domain has been assessed for box C/D sRNP assembly, sRNP structure, and sRNP-guided nucleotide methylation by mutating or deleting this protein domain. Protein pull-down experiments demonstrated that Nop56/58 self-dimerization and Nop56/58 dimerization with the core protein fibrillarin are mutually exclusive protein:protein interactions. Disruption of Nop56/58 homodimerization by alteration of specific amino acids or deletion of the entire coiled-coil domain had no obvious effect upon core protein binding and sRNP assembly. Site-directed mutation of the Nop56/58 homodimerization domain also had no apparent effect upon either box C/D RNP- or C′/D′ RNP-guided nucleotide modification. However, deletion of this domain disrupted guided methylation from both RNP complexes. Nuclease probing of the sRNP assembled with Nop56/58 proteins mutated in the coiled-coil domain indicated that while functional complexes were assembled, box C/D and C′/D′ RNPs were altered in structure. Collectively, these experiments revealed that the self-dimerization of the Nop56/58 coiled-coil domain is not required for assembly of a functional sRNP, but the coiled-coil domain is important for the establishment of wild-type box C/D and C′/D′ RNP structure essential for nucleotide methylation.
Keywords: archaeal sRNP, box C/D RNA, nucleotide methylation, sRNP core proteins, Nop56/58
INTRODUCTION
Box C/D RNPs are RNA-guided nucleotide modification complexes that direct the post-transcriptional nucleotide modification of both archaeal and eukaryotic RNAs (Tollervey 1996; Kiss 2001; Bachellerie et al. 2002; Terns and Terns 2002; Decatur and Fournier 2003). The guide RNA of the RNP complex base-pairs with the target RNA to determine the specific nucleotide for modification while the RNA-bound proteins catalyze the nucleotide modification reaction. The occurrence of box C/D guide RNAs in both Eukarya and Archaea suggests that this RNA:protein enzyme arose before the divergence of these two kingdoms more than two billion years ago (Gaspin et al. 2000; Omer et al. 2000; Speckmann et al. 2002; Tang et al. 2002; Tran et al. 2004).
Archaeal box C/D RNAs guide the 2′-O-methylation of target nucleotides contained in both ribosomal and tRNAs (Omer et al. 2003). The box C/D sRNAs characteristically possess terminal box C/D core and internal C′/D′ motifs (Omer et al. 2002; Rashid et al. 2003; Tran et al. 2003). Both motifs possess highly conserved nucleotide box C and D sequences, and each motif, in conjunction with associated guide regions, directs nucleotide methylation. Box D and D′ guide regions base-pair with the target RNA, and the fifth nucleotide of the target RNA within the guide RNA:target RNA duplex is methylated. The box C/D and C′/D′ motifs both fold into three-dimensional RNA configurations designated “kink-turns” or “K-turns” because of their inherent kinked structure (Vidovic et al. 2000; Watkins et al. 2000; Klein et al. 2001; Kuhn et al. 2002; Roshdestvensky et al. 2003; Goody et al. 2004; Moore et al. 2004). In Archaea, the box C/D core and C′/D′ motifs are each bound with three core proteins to assemble symmetric RNP complexes (Omer et al. 2002; Rashid et al. 2003; Tran et al. 2003). These three core proteins bind in an ordered succession with ribosomal protein L7 first recognizing the K-turn. Nop56/58 binds next followed by fibrillarin. Several lines of evidence have demonstrated the ability of Nop56/58 and fibrillarin to dimerize (Aittaleb et al. 2003; Rashid et al. 2003; Tran et al. 2003). The enhanced binding of Nop56/58 in the presence of fibrillarin suggests that these two core proteins may indeed bind the assembling sRNP as a heterodimer (Tran et al. 2003).
The three box C/D sRNP core proteins are essential for sRNA-guided nucleotide methylation. It is clear that ribosomal protein L7 is the first core protein to bind, recognizing the K-turn motifs and initiating sRNP assembly (Kuhn et al. 2002; Omer et al. 2002; Rashid et al. 2003; Tran et al. 2003; Suryadi et al. 2005). Fibrillarin functions as the methylase enzyme. Fibrillarin possesses an S-adenosyl-L-methionine (SAM) binding motif, exhibits homology with other known methylases, and site-specific mutagenesis of putatively critical amino acids for both SAM binding and methylation activity disrupt sRNP-guided methylation (Wang et al. 2000; Omer et al. 2002; Aittaleb et al. 2004). More enigmatic is the third core protein Nop56/58. It is believed that this core protein plays largely structural roles in the sRNP complex, although a supporting role in methylation has not been ruled out. A recent crystal structure of the Archaeoglobus fulgidus Nop56/58-fibrillarin dimer has revealed the potential for Nop56/58 self-dimerization utilizing α-helices 4 and 5, which fold upon each other to establish the coiled-coil domain (Aittaleb et al. 2003). The dimerization of the coiled-coil domains of two individual Nop56/58 proteins to form a heterotetramer complex in the crystal structure led to the suggestion that coiled-coil domain interactions between the Nop56/58 proteins of the box C/D and C′/D′ RNPs could establish inter-RNP protein:protein contacts. Such interactions could explain the previously described cross-talk interactions between these two RNP subcomplexes that are essential for box C/D and C′/D′ RNP-guided nucleotide modification (Tran et al. 2003, 2005).
To explore the function of the Nop56/58 core protein in box C/D sRNP structure and function, we have investigated the importance of the coiled-coil domain for sRNP assembly and sRNP-guided nucleotide methylation. Nop56/58 proteins with mutated or deleted coiled-coil domains revealed that Nop56/58p self-dimerization and interation of Nop56/58 with fibrillarin are mutually exclusive protein:protein interactions. Disruption of Nop56/58 self-dimerization had no effect upon sRNP assembly or nucleotide methylation. However, deletion of the coiled-coil domain, while not affecting sRNP assembly, completely disrupted nucleotide methylation. Probing of box C/D and C′/D′ RNPs revealed structural alterations when the coiled-coil domain was absent. Collectively, these experiments demonstrated that the coiled-coil domain of Nop56/58 is not required for sRNP assembly but is essential for establishment of sRNP structure critical for sRNP-guided nucleotide methylation activity.
RESULTS
The Nop56/58-fibrillarin core protein heterodimer and sR8 box C/D sRNA structure
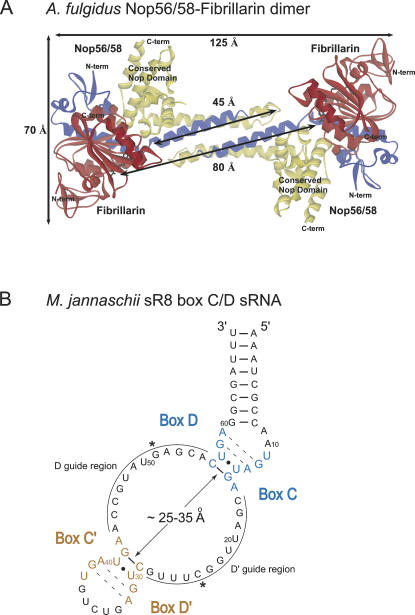
Our previous analysis of Methanocaldococcus jannaschii sR8 box C/D sRNP assembly demonstrated that core proteins Nop56/58 and fibrillarin can dimerize and that heterodimer formation likely facilitates sRNP assembly (Tran et al. 2003). The cocrystallization of the A. fulgidus Nop56/58 and fibrillarin core proteins has provided a more detailed look at this dimer's structure (Aittaleb et al. 2003; Fig. 1A). Interestingly, the crystal structure revealed self-dimerization of the Nop56/58-fibrillarin dimer using Nop56/58 α-helices 4 and 5, which have been designated the coiled-coil domain. This self-dimerization suggested the possibility that the individual Nop56/58 core proteins bound to the box C/D core and C′/D′ RNPs could interact via their coiled-coil domains when in the complete sRNP complex (Rashid et al. 2003). Such inter-RNP contacts could explain box C/D and C′/D′ RNP cross-talk interactions that we demonstrated are critical for box C/D sRNP-guided 2′-O-methylation activity (Tran et al. 2003). However, preliminary modeling of this core protein heterodimer against the M. jannaschii sR8 box C/D sRNA (Fig. 1B) suggested that this inter-RNP interaction is inconsistent with the typically small size of the archaeal box C/D sRNAs and limited inter-RNP spatial constraints (Gaspin et al. 2000; Tran et al. 2005). Specifically, we have estimated an inter-RNP distance between the box C/D and C′/D′ motifs of between 25 and 35 Å (Fig. 1B). The smaller distance was estimated if the two guide regions could base-pair with each other (lower limit) or if both guide sequences remained extended and single-stranded (upper limit). This range of intermotif distance could not easily accommodate the ∼45 Å distance required for the self-dimerized coiled-coil domains, while coincidently positioning the guide: target duplex in the vicinity of the fibrillarin active site. Also noteworthy are the active sites of the two respective fibrillarin proteins are ∼80 Å apart. This is a considerably greater distance than that which separates the guide nucleotides of the D and D′ guide sequences (Fig. 1B) where the nucleotide modification reaction must occur.
FIGURE 1.
Crystal structure of archaeal A. fulgidus Nop56/58-fibrillarin core protein heterotetramer and M. jannaschii sR8 box C/D sRNA. (A) Crystal structure of the A. fulgidus Nop56/58-fibrillarin core protein heterodimer determined by Aittaleb and coworkers (Aittaleb et al. 2003). Fibrillarin is presented in red, and Nop56/58 is presented in blue, with the Nop domain indicated in yellow. Extended α-helices 4 and 5 of Nop56/58 (∼45 Å) establish the coiled-coil domain, which self-associates to form a heterotetramer complex. The distance between the catalytic sites of fibrillarin (∼80 Å) and overall dimensions of the Nop56/58-fibrillarin heterodimer are indicated. (B) Folded secondary structure of M. jannaschii sR8 box C/D sRNA, a model box C/D sRNA. Nucleotide boxes C, D, C′, and D′ are indicated, as well as the terminal box C/D core and internal C′/D′ motifs separated by the D and D′ sRNA guide sequences. Asterisks indicate guide nucleotides of each guide region that base-pair with the nucleotide targeted for 2′-O-methylation.
Nop56/58p self-dimerization and interaction of Nop56/58p with fibrillarin are mutually exclusive protein:protein interactions
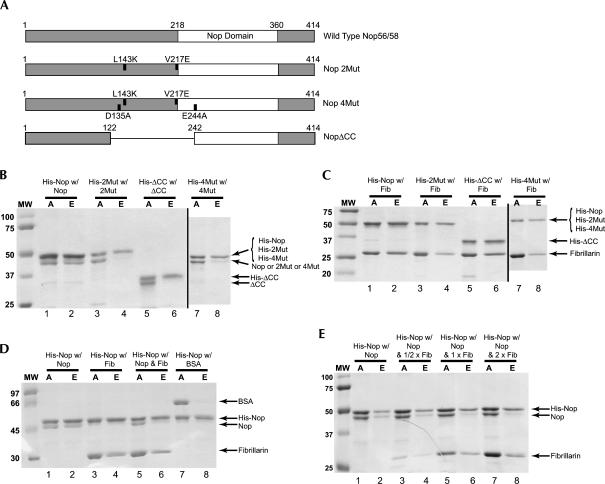
To further explore the potential role of Nop56/58's coiled-coil domain in box C/D sRNP assembly, recombinant M. jannaschii Nop56/58 proteins mutated in the coiled-coil domain (Fig. 2A) were assessed for their ability to self-dimerize as well as interact with fibrillarin. Mutant 2Mut (L143K; V217E) was patterned after a previous A. fulgidus Nop56/58 construct that was shown to disrupt Nop56/58 self-dimerization (Aittaleb et al. 2003). The Nop56/58 4Mut protein contained an additional two altered amino acids in the coiled-coil domain (D135A and E244A), whereas the ΔCC mutant deleted α-helices 4 and 5 of the coiled-coil domain (amino acid residues D123–K241). These Nop56/58 mutants were constructed as both His-tagged and untagged versions to facilitate self-dimerization analysis in protein pull-down experiments. As previously observed (Rashid et al. 2003), mutation of specific coiled-coil domain amino acids (2Mut) greatly reduces Nop56/58 self-dimerization by disrupting coiled-coil interactions (Fig. 2B, lanes 3–4). Some residual self-dimerization is observed with the 4Mut mutant (Fig. 2B, lanes 7–8), but this interaction is weak. As expected, complete loss of self-dimerization was observed when the coiled-coil domain (Fig. 2B, lanes 5–6) was completely deleted (ΔCC mutant). Notably, all the mutant Nop56/58 proteins still retained their ability to interact with fibrillarin and form a heterodimer (Fig. 2C, lanes 4,6,8). Strikingly, however, when His-tagged wild-type Nop56/58 was incubated with nontagged Nop56/58 and fibrillarin, the Nop56/58-fibrillarin heterodimer predominated with a severe reduction of Nop56/58 self-dimerization (Fig. 2D, lanes 5–6). This result indicated that Nop56/58 interaction with fibrillarin is mutually exclusive with and dominant over Nop56/58 self-dimerization. Titration of the Nop56/58 homodimer with increasing amounts of fibrillarin confirmed the dominance of fibrillarin interaction with Nop56/58 over self-dimerization (Fig. 2E). Notably, these experiments were carried out at room temperature, but similar results were obtained at elevated temperature (data not shown). Similar pull-down experiments were also attempted using A. fulgidus Nop56/58 and fibrillarin. Unfortunately, solubility of Nop56/58 as an isolated protein was a problem, as previously noted (Aittaleb et al. 2003; Rashid et al. 2003), and we were unable to carry out these experiments.
FIGURE 2.
Nop56/58 Self-dimerization and Nop56/58 dimerization with fibrillarin are mutually exclusive interactions. (A) Schematic presentation of the M. jannaschii Nop56/58 wild-type and mutant proteins. Mutated amino acids in the coiled-coil domain of the 2Mut and 4Mut Nop56/58 mutant proteins and the deleted domain of the ΔCC mutant protein are indicated. (B) Protein–protein pull-down analysis of wild-type and mutant Nop56/58 self-dimerization. Wild-type, 2Mut, 4Mut, or ΔCC Nop56/58 proteins were mixed with His-tagged protein and then self-dimerized protein pairs selected by nickel column chromatography. Total or applied protein (A) and His-tagged and nickel-selected or eluted (E) proteins were resolved by SDS gel electrophoresis and revealed by staining. Proteins used in individual incubations and selected in the eluted fractions are indicated above and at the side, respectively. (C) Mutation or deletion of the Nop56/58 coiled-coil domain does not affect dimerization with fibrillarin. Proteins used in the individual incubations (A) and observed in the selected fractions (E) are indicated above and at the side, respectively. (D) Nop56/58p self-dimerization and Nop56/58 dimerization with fibrillarin are mutually exclusive protein:protein interactions. Proteins used in the individual incubations (A) and observed in the selected fractions (E) are indicated above and at the side, respectively. (E) Titration of the Nop56/58 homodimer with increasing amounts of fibrillarin disrupts Nop56/58 self-dimerization. Constant amounts of His-tagged and untagged Nop56/58 were incubated with increasing concentrations of fibrillarin protein (lanes 3,5,7). Interacting proteins were selected by nickel chromatography of the His-tagged Nop56/58 and resolved by SDS gel electrophoresis (lanes 4,6,8).
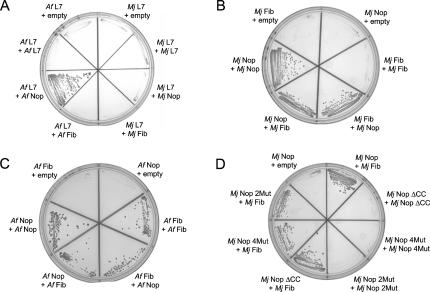
Archaeal box C/D core protein:protein interactions were also tested in vivo using the yeast two-hybrid system. Consistent with the in vitro pull-down analysis, M. jannaschii L7 does not interact with Nop56/58 or fibrillarin, whereas Nop56/58 is able to self-dimerize with itself and interact with fibrillarin (Figs. 3 A,B). The effect of mutations in the coiled-coil region (2Mut and 4Mut) as well as deletion of this region (ΔCC) clearly revealed a loss of Nop56/58 self-dimerization (Fig. 3D). However, disruption of the coiled-coil interaction had no effect upon Nop56/58's ability to interact with fibrillarin. Parallel in vivo analysis of the A. fulgidus box C/D core protein interactions was also carried out. Strikingly, novel core protein interactions were observed, distinct from the M. jannaschii core protein interactions. Notably, A. fulgidus L7 interacts with Nop56/58 (Fig. 3A). Again, Nop56/58 self-dimerizes and interacts with fibrillarin as expected. However, A. fulgidus fibrillarin is also able to interact with itself (Fig. 3C). These unique core protein interactions displayed by these two archaeal organisms may have implications for the final assembled structure of their respective box C/D sRNP complexes (see Discussion).
FIGURE 3.
In vivo analysis of M. jannaschii and A. fulgidus box C/D sRNP core proteins reveal organism-specific protein:protein interactions. M. jannaschii and A. fulgidus core protein interactions were assessed in vivo using the yeast two-hybrid system. Yeast carrying a Gal4:HIS3 reporter gene were transformed sequentially with a bait vector (indicated on top) and a prey vector (indicated on bottom), and the resulting strains were tested for growth. (A) M. jannaschii (Mj) and A. fulgidus (Af) L7 core protein interactions with L7, Nop56/58, and fibrillarin. (B) M. jannaschii (Mj) core protein interactions between Nop56/58 and fibrillarin. (C) A. fulgidus (Af) core protein interactions between Nop56/58 and fibrillarin. (D) M. jannaschii (Mj) Nop56/58 coiled-coil mutant core protein interactions between mutant Nop56/58 and fibrillarin proteins.
The Nop56/58 coiled-coil domain is dispensable for box C/D sRNP assembly but essential for sRNP-guided methylation
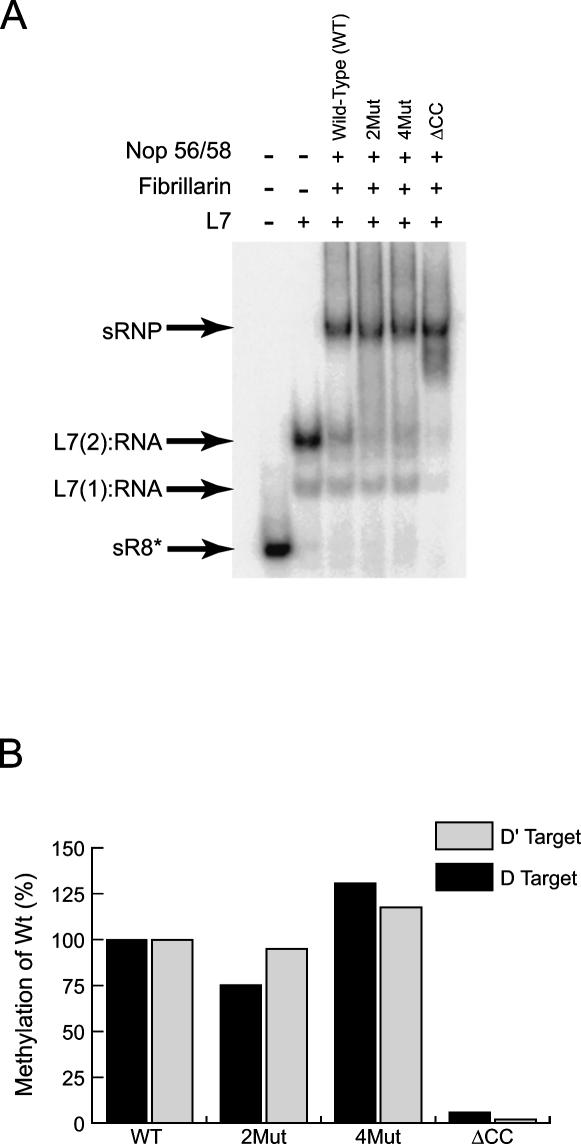
The ability of the mutant Nop56/58 proteins to be assembled into sRNP complexes was assessed in electrophoretic mobility gel-shift analysis. Mutations in the coiled-coil region of Nop56/58 that disrupt or reduce self-dimerization via the coiled-coil domain had no effect upon sRNP assembly (Fig. 4A). Similarly, when the coiled-coil domain was deleted from Nop56/58 (ΔCC mutant) the sR8 sRNP complex was fully assembled. sRNP assembly with the ΔCC mutant yields a minor and slightly faster migrating RNP. At this time we do not know the structure of this complex but suspect that deletion of the coiled-coil domain may allow alternative conformations of the fully assembled sRNP. The Nop56/58 coiled-coil mutant proteins were also tested for assembly on an sR8 halfmer sRNAs that contained only the box C/D core motif or the C′/D′ motif. While binding of L7, Nop56/58, and fibrillarin was less efficient upon these individual halfmer RNAs as previously observed (Tran et al. 2003), complete RNP complexes could be formed with the mutant Nop56/58 proteins (data not shown).
FIGURE 4.
The coiled-coil domain of Nop56/58 is dispensable for box C/D sRNP assembly but essential for sRNP-guided nucleotide methylation. (A) Wild-type or mutant Nop56/56 proteins along with wild-type L7 and fibrillarin were incubated with radiolabeled M. jannaschii sR8 sRNA to assemble the sRNP. Assembled complexes were resolved on a native polyacrylamide gel and RNPs revealed by autoradiography. Combinations of core proteins in the various RNP assembly reactions are indicated above each lane and assembled RNP complexes designated at the side. (B) sRNP complexes were assembled with wild-type and mutant Nop56/58 proteins and incubated with 3H-S-adenosyl-L-methionine and target RNAs complementary to D or D′ guide regions. Nucleotide methylation guided by the box C/D core and C′/D′ RNPs was assessed by measuring incorporation of TCA precipitable 3H-CH3 into the individual target RNAs. Target nucleotide methylation of the mutant Nop56/58 sRNPs was compared with methylation activities of the box C/D and C′/D′ RNPs (100%) assembled with wild-type core proteins.
Given that the Nop56/58 mutant proteins could assemble sRNP with equal efficiency to the wild-type core protein, the ability of these sRNPs to function in nucleotide methylation was assessed. The methylation capabilities of the mutant sRNPs were determined by measuring incorporation of 3H-CH3 donated from SAM into target RNAs complementary to both D and D′ guide sequences. Amino acid mutations within the coiled-coil domain that disrupted Nop56/58 self-dimerization had little effect upon nucleotide methylation guided by either the box C/D or C′/D′ RNPs (Fig. 4B). However, deletion of the coiled-coil domain completely disrupted nucleotide methylation from both RNPs. Collectively, these results demonstrated that the coiled-coil domain, while not required for sRNP assembly, is necessary for both box C/D and C′/D′ RNP-guided nucleotide methylation activity.
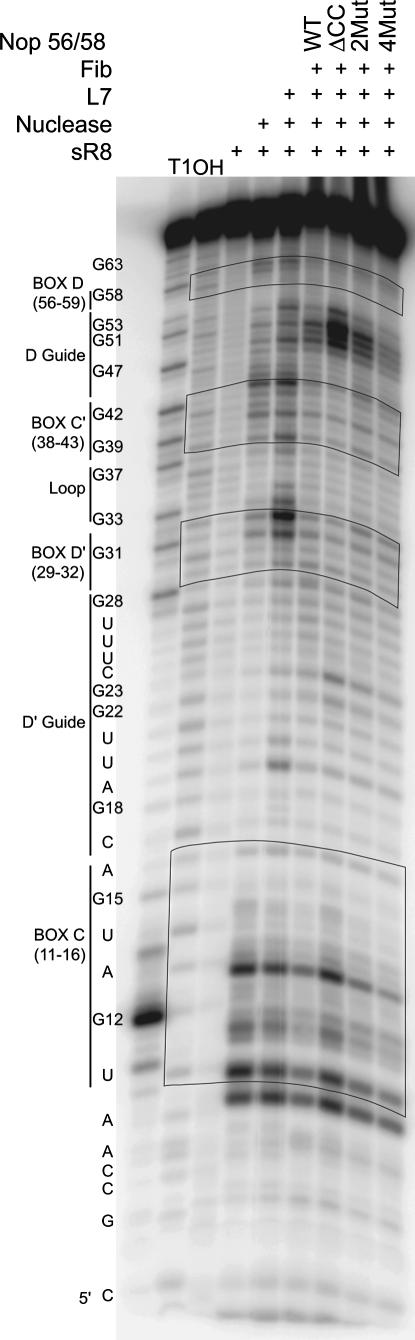
sR8 sRNP complexes assembled with mutant Nop56/58 core protein exhibited a perturbed box C/D and C′/D′ RNP structure
To explore the possibility that Nop56/58's coiled-coil domain was important for establishment of sRNP structure, box C/D core and C′/D′ RNPs were probed using nuclease cleavage. Wild-type and Nop56/58 mutant sRNPs were assembled on 5′-terminally radiolabeled sR8 sRNA and incubated in buffer containing mung bean nuclease to cleave sRNA-accessible regions. Resolution of the resulting radiolabeled sR8 fragments on polyacrylamide gels indicated distinctive nuclease cleavage patterns for wild-type and mutant sRNPs (Fig. 5). These differences were most pronounced for the ΔCC coiled-coil deletion mutant in the D guide region, revealing an enhanced cleavage around the guide nucleotide and extending away from box D. A similar but less pronounced increase in nuclease digestion was seen for the D′ guide region. Small changes, both increases and decreases in digestion patterns, were also noted for individual nucleotides within the box C/D and C′/D′ motifs. From these results, we concluded that assembly of the sRNP complex with mutant Nop56/58 core proteins, most notably Nop56/58 missing the coiled-coil region, altered the overall structure of both box C/D and C′/D′ RNPs within the assembled sR8 sRNP complex. Strikingly, the most profound effects upon sRNP structure were noted in the conformation of the box C/D- and C′/D′-associated guide regions.
FIGURE 5.
sR8 sRNP assembly with mutant Nop56/58 core protein alters box C/D and C′/D′ RNP structure. sR8 sRNA was radiolabeled at the 5′ terminus, assembled into sRNP complexes using wild-type or mutant Nop56/58 core proteins, and then digested with limiting amounts of mung bean nuclease. After digestion, sRNA fragments were precipitated with LiClO4, resolved on 14% polyacrylamide-urea denaturing gels, and revealed by autoradiography. Core proteins used to assemble sRNP complexes are indicated at the top. RNase T1 digestion (T1) and alkaline hydrolysis (OH) of 5′-radiolabeled sR8 provided nucleotide sequence ladders. The sR8 sequence is indicated at the side along with designated C, D, C′, and D′ boxes and associated D and D′ guide sequences. Corresponding RNA cleavage patterns within boxes C, D, C′, and D′ are enclosed in blocks.
DISCUSSION
Nop56/58 is one of three core proteins that bind the archaeal box C/D sRNAs to assemble a methylation-competent, RNA-guided nucleotide modification complex. Emerging crystal structures of the archaeal core proteins are now facilitating a more detailed examination of core protein structure–function relationships and the potential role(s) of individual core proteins in sRNP assembly and nucleotide methylation. In this work, we have examined the importance of the coiled-coil domain of Nop56/58 for sRNP assembly and nucleotide methylation. sRNP assembly studies revealed that this domain is not essential for Nop56/58 binding to the sRNA or assembly of the complete sRNP. However, methylation activities of both the box C/D and C′/D′ RNPs are disrupted when the coiled-coil domain is absent. The observation that loss of this domain perturbs sRNP structure suggests that the coiled-coil domain contributes to overall sRNP organization, which, in turn, is critical for nucleotide methylation activity.
Nop56/58 forms a stable complex with fibrillarin in solution, and this protein dimer “platform” interacts with the L7–sRNA complex to promote target RNA binding upon the sRNA guide sequence for nucleotide methylation. The conserved Nop domain (residues 109–258 in A. fulgidus and 218–360 in M. jannaschii Nop56/58) is ostensibly important, since it has been highly conserved throughout evolution. Recent studies have shown that the looped region of U4 snRNA (similar to box C′/D′) interacts specifically with the conserved Nop domain of the 61-kDa snRNP protein, which has high sequence similarity with Nop56/58 (Nottrott et al. 2002). These results suggest that the conserved Nop domain is essential for establishing interactions with L7–sRNA K-turn complexes by interacting with a region of the K-turn motif as well as through probable L7 interactions. In turn, Nop56/58, in binding with fibrillarin to establish a platform for catalytic activity, likely positions and orients fibrillarin for substrate recognition and catalysis. Thus, Nop56/58 might be considered a bridge between L7 in its recognition of the K-turn and the methylase fibrillarin. The observation that A. fulgidus L7 interacts with Nop56/58 as seen in our yeast two-hybrid experiments is consistent with this suggestion.
The cocrystal structure of the A. fulgidus Nop56/58-fibrillarin dimer revealed the potential for Nop56/58 self-dimerization via its coiled-coil domain (Aittaleb et al. 2003). This coiled-coil domain is formed by α-helices 4 and 5 of Nop56/58 that prominently extend outward from the globular conserved Nop domain which is responsible for interactions with fibrillarin. Both our in vitro protein pull-down and in vivo yeast two-hybrid experiments have experimentally verified this protein:protein interaction. Nop56/58 homodimerization could be a particularly attractive way to explain the inter-RNP cross-talk interactions that are vital for box C/D and C′/D′ RNP methylation activities and conservation of distances between the two motifs in Archaea. Our previous work has revealed the importance of juxtaposed RNP complexes within the complete sRNP for nucleotide modification (Tran et al. 2003). The fact that inter-RNP spacing is also critical could be consistent with the need to establish self-dimerized coiled-coil domains between the individual Nop56/58 proteins located in the box C/D and C′/D′ RNPs (Gaspin et al. 2000; Tran et al. 2005). However, our results presented here demonstrate that such Nop56/58 coiled-coil interactions between the box C/D and C′/D′ RNPs are not critical for either sRNP assembly or nucleotide methylation.
Analysis of Nop56/58 domain structure in Archaea and Eukarya indicates that self-dimerization is unlikely to be a common feature in box C/D RNP complexes. Comparison of the sequences of archaeal Nop56/58 homologs has shown that the A. fulgidus Nop56/58 protein is considerably smaller than other homologs (Fig. 6; Table 1). Notably, typical Nop56/58 proteins have an elongated N-terminal region, an expanse of ∼60 amino acids between α-helices 4 and 5, and a poly-lysine/arginine basic C terminus. Interestingly, the region between α-helices 4 and 5 is not highly conserved in sequence, but is in length, suggesting that spatial distancing is more important than the structure. Previous work with Nop56/58 from A. fulgidus has indicated that self-dimerization is important for methylation activity (Rashid et al. 2003); these results directly contrast our results. The reason for this discrepancy is unclear, but the fact that A. fulgidus Nop56/58 is an uncharacteristically small protein (Fig. 6; Table 1) may account for the contradictory results. Strikingly, yeast two-hybrid analysis has demonstrated distinct differences in core protein:protein interactions between M. jannaschii and A. fulgidus. Perhaps A. fulgidus has unique or species-specific core protein interactions that account for the observed differences. Table 1 shows the distribution of Nop56/58 homolog sizes in both archaeal and eukaryotic organisms. Of the Nop56/58 protein sequences analyzed, only five are shorter than 300 amino acids: A. fulgidus (261), Haloarcula marismortui (280), Halobacterium salinarum (277), Thermoplasma acidophilum (260), and Thermoplasma volcanium (264). The average size of archaeal Nop56/58 homologs is 370 amino acids. The average size of eukaryotic Nop56 is 532 amino acids, and for Nop58, 521 amino acids. Thus, the A. fulgidus Nop56/58 homolog is not representative of typical Nop56/58 proteins.
FIGURE 6.
Archaeal and eukaryotic Nop56/58 core proteins sequence comparison of archaeal and eukaryotic Nop56/58 core proteins. Sequence alignment of archaeal and eukaryotic Nop56/58 homologs annotated with the secondary structure of A. fulgidus Nop56/58. Note that the N terminus of A. fulgidus Nop56/58 is shortened, there is a large gap of ∼60 amino acids that is missing between α-helix 4 and α-helix 5, and the virtual absence of the poly-lysine/arginine-rich C terminus. In the secondary structure annotation, cylinders indicate α-helices, arrows indicate β-sheets, and continuous lines indicate coiled or unstructured regions. Dashed lines indicate regions of the protein that were not discernable in the crystal structure, presumably due to disorder or flexibility; amino acid residues in this region (A. fulgidus Nop56/58) are italicized in the alignment. ClustalW sequence alignment conservation is indicated as follows: (*), identical residues; (:), strong conservation; (.), semiconservation. Amino acids are colored as follows: nonpolar (A, G, I, L, M, P, V), dark gray; uncharged polar (C, Q, N, S, T), teal; basic (H, K, R), red; acidic (D, E), blue; aromatic (F Y W), green.
TABLE 1.
Size of archaeal and eukaryotic Nop56 and Nop58 homologs

In eukaryotes, the single archaeal Nop56/58 core protein is replaced with the two distinct but highly homologous Nop56 and Nop58 homologs (Gauthier et al. 1997; Lafontaine and Tollervey 1999; Newman et al. 2000). The gene encoding this box C/D core protein has presumably undergone duplication during evolution, after the split between Archaea and Eukarya some two billion years ago. Strikingly, the RNA-binding capabilities of the eukaryotic homologs have also evolved with Nop56 and Nop58 specifically cross-linking to the C′/D′ and box C/D core motifs, respectively (Cahill et al. 2002). This differential distribution of Nop56 and Nop58 along with the differential binding of the 15.5-kDa core protein to the box C/D core motif results in the formation of an “asymmetric” snoRNP as opposed to the symmetric archaeal complex (Weinstein Szewczak et al. 2002). It is not yet clear if eukaryotic Nop56 and Nop58 proteins have retained the ability to self-dimerize via their coiled-coil domains. Evaluation of the coiled-coil domain in the eukaryotic Nop56 and Nop58 homologs reveals that α-helices 4 and 5 are similar in size to the typical archaeal homologs and are interspersed by a 50– to 60–amino acid region between the two helices (Fig. 6). This would suggest that even if these two proteins have retained the ability to dimerize, steric hindrance due to the α-helix 4–5 linker domain makes it improbable that they would do so in an assembled box C/D snoRNP. Additionally, spacing between the box C/D and C′/D′ motifs in eukaryotic box C/D snoRNAs is not highly conserved as it is for the archaeal sRNAs (Gaspin et al. 2000; Tran et al. 2005). Therefore, the dimerization of Nop56-Nop58 via their coiled-coil domains as an important and shared feature of eukaryotic snoRNP complex is even more improbable.
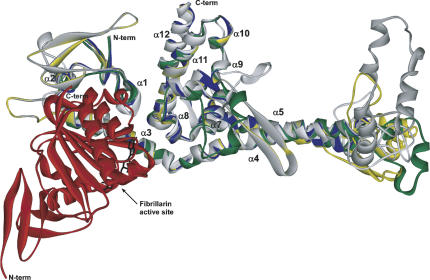
Molecular modeling experiments have been conducted with the M. jannaschii Nop56/58 protein, using the A. fulgidus protein as a starting structure (Fig. 7). These studies have provided some insight into the structure of a typical Nop56/58 homolog. The extended N terminus likely forms a β-sheet region near the fibrillarin-interaction domain. The region between α-helices 9 and 10, which is disordered in the A. fulgidus structure, can be folded into several possible structures and is probably reflective of the flexibility of this region. Lastly, a large, potentially globular domain can be folded from the ∼65 amino acids between α-helices 4 and 5 (α-helix 4–5 linker domain). Several different structures were obtained by modeling programs, and four superimposed models are shown in Figure 7. Although the actual structure of the α-helix 4–5 linker domain is nebulous, it provides a rational explanation for the pull-down and yeast two-hybrid results. In the absence of fibrillarin, Nop56/58 proteins are capable of homodimerization via the coiled-coil domains. However, when fibrillarin is complexed to Nop56/58, steric hindrance between the α-helix 4–5 linker domain and fibrillarin bound to the Nop56/58 dimerization partner likely prevents Nop56/58-Nop56/58 homodimerization. Consequently, it is unlikely that coiled-coil dimerization of the Nop56/58 homologs is a general phenomenon in box C/D sRNP assembly, but possibly an idiosyncrasy of the small, atypical A. fulgidus Nop56/58 protein.
FIGURE 7.
Modeled structure of the M. jannaschii Nop56/58-fibrillarin heterodimer. Structural model of M. jannaschii Nop56/58 and fibrillarin based on the A. fulgidus complex. Four independent Nop56/58p models are shown, superimposed on the A. fulgidus Nop56/58 crystal structure (blue). The four models indicate several potential ways of folding the extended N terminus, the region interspersed between α-helix 4 and α-helix 5, and the region between α-helix 9 and α-helix 10, which was disordered in the A. fulgidus Nop56/58 structure. The basic C-terminal region (poly lysine/arginine) was not modeled. Fibrillarin is shown in red with bound S-adenosyl-L-methionine in black. Models 1 (yellow) and 2 (dark gray) were initially generated using Swiss-Model. Model 3 (green) was generated with 3DJIGSAW, and Model 4 (light gray) was generated with CPH models. Note the predicted high structural conservation of the consensus Nop domain (residues 218–360 for M. jannaschii Nop56/58). The potentially globular domain at the end of the α-helix 4 and 5 coiled-coil domain may be responsible for preventing Nop56/58 homodimerization in the presence of fibrillarin due to steric hindrance.
Finally, emerging crystal structures and molecular modeling are providing important insight into archaeal box C/D sRNP structure. A firm understanding of sRNP structure will provide a “simplest model complex” for subsequent analysis of the larger, more structurally complex and more functionally diverse eukaryotic box C/D snoRNP. For the archaeal sRNP, it is important that the box C/D and C′/D′ RNPs be juxtaposed for efficient nucleotide modification activity. While Nop56/58 self-dimerization linking the box C/D and C′/D′ RNPs is apparently not a common and conserved feature of the archaeal complexes, it is an attractive idea that the three core proteins contained in each RNP do indeed interact in some fashion via inter-RNP protein:protein contacts. Such cross-talk interactions could be essential for nucleotide modification. In vivo yeast two-hybrid studies have revealed additional, species-specific core protein interactions in A. fulgidus. These additional core protein interactions may well suggest that individual organisms utilize unique core protein contacts, not only for sRNP assembly, but also for inter-RNP cross-talk interactions that are potentially important for RNA-guided methylation activity. Clearly, additional biochemical and structural studies are needed to define such inter-RNP protein:protein interactions and help elucidate the enzymatic functions of this evolutionarily ancient RNA-guided nucleotide modification complex.
MATERIALS AND METHODS
Core protein production and purification
Recombinant M. jannaschii ribosomal protein L7, Nop56/58, and fibrillarin were expressed and purified as previously described (Tran et al. 2003). Mutant Nop56/58 proteins were created using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) and the following DNA oligonucleotide primers:
GATTTGGATAAAACTCTAAACTTGAAATCTGAGAGATTGAGAGAGTGG
CCACTCTCTCAATCTCTCAGATAACAAGTTTAGAGTTTTATCCAAATC
GGAGAATTGGAAGATTATGATTTAGATGAAATTGTTAAATTTGCTGAGG
CCTCAGCAAATTTAACAATTTCATCTAAATCATAATCTTCCAATTCTCC
GTTGCAGAGGCAATATCTGCTTTGGATAAAACTCTAAACTTG
CAAGTTTAGAGTTTTATCCAAAGCAGATATTGCCTCTGCAAC
GAAATTGTTAAATTTGCTGAGGCAATAAATCACCTATATG
CATATAGGTGATTTATTGCCTCAGCAAATTTAACAATTTC
CTTTTGTTGAGCATAGCTCTTTATAACTTTCTTTGTTAGCTC
TTGATGAACGAAGAAGCTCCAAATATTACAAAACTTGCTGG
CTATGCTCAACAAAAGTTGATGAACGAAGAAGCTCC
GGAGCTTCTTCGTTCATCAACTTTTGTTGAGCATAG
Nop56/58 mutant proteins 2Mut (L143K; V217E), 4Mut (L143K; V217E; D135A; E244A) and ΔCC (D123–K241) were constructed using the following PCR primer pairs: 2Mut (primers1 and 2, then 3 and 4), 4 Mut (primers 5 and 6, then 7 and 8), ΔCC (primers 9 and 10, then 11 and 12). N-terminal hexa-histidine tagged Nop56/58p wild-type and mutant protein expression constructs were created by subcloning the gene inserts into the NdeI and BamHI sites of the pET28a vector (Novagen). Upstream NdeI and downstream BamHI sites were created in each insert during PCR amplification. Wild-type and mutant Nop56/58 constructs were expressed in Rosetta DE3 cells (Novagen) and purified using cation exchange SP Sepharose Fast Flow (Sigma-Aldrich) as previously detailed (Tran et al. 2003).
Determination of RNP assembly and protein:protein interactions via pull-down analysis
RNP assembly was assessed by electrophoretic mobility-shift analysis as previously described (Tran et al. 2003). Protein:protein interactions were assessed in pull-down analysis using His-tagged wild-type or mutant Nop56/58 proteins. Tagged Nop56/58 protein(s) was incubated with untagged wild-type Nop56/58, mutant Nop56/58, or fibrillarin at equimolar concentrations (5 μM each) in buffer A (20 mM HEPES at pH 7.0, 100 mM NaCl, 1 mM MgCl2) for 10 min at room temperature. (Note: Similar pull-down experiments carried out at 70°C gave the same results.) Approximately 20% of the mixture was removed and designated the “applied” sample. Approximately 30 μL of 50% His-bind slurry (Novagen) was added to the remaining mixture and incubated an additional 20 min. Protein-bound resin was washed four times with buffer A supplemented with 25 mM imidazole, 0.007% SDS, and 0.1% Triton X-100. Bound proteins were eluted with buffer A containing 1 M imidazole. Applied and eluted proteins were resolved by SDS-polyacrylamide gel electrophoresis and visualized by Coomassie Blue staining.
sRNP-guided in vitro methylation
sRNP complexes were assembled at 70°C for 10 min in 80-μL volumes containing 0.65 μM guide RNA, 0.8 μM L7, 3.1 μM Nop56/58, and 1.1 μM fibrillarin (Tran et al. 2003). Assembled RNPs were then mixed with 6.5 μM target RNA (Dharmacon), 3.3 μM SAM (Calbiochem), and 3 μL of H3-SAM (55 mCi/mL; MP Biomedicals) to a final volume of 110 μL. After incubation at 68°C for 1 h, 20-μL aliquots were removed and TCA precipitated onto filter paper. Methyl incorporation into target RNAs was measured by scintillation counting. Methylation assays were performed in triplicate and are reported as the average of three experiments.
Yeast two-hybrid analysis of in vivo protein interactions
Wild-type and mutant core proteins were tested for interaction in vivo using the yeast two-hybrid system (James et al. 1996). L7, fibrillarin, Nop56/58, and Nop56/58 mutant (2Mut, 4Mut, and ΔCC) genes from M. jannaschii were subcloned from the pET28a expression vectors into the pAS2–1 (bait, GAL4BD fusion, Trp marker) and pACT2 (prey, GAL4AD fusion, Leu marker) vectors using the NcoI and BamHI restriction sites. M. jannaschii fibrillarin retains an N-terminal His-tag encoded by the pET28a vector. A. fulgidus L7, fibrillarin and Nop56/58 genes were also subcloned into the two-hybrid vectors using NcoI and XhoI/SalI restriction sites. Each A. fulgidus fusion protein retained the pET28a-encoded His-tag. Bait vectors were transformed into the pJ69–4a host strain, which contains the GAL-HIS3 reporter. Prey vectors were transformed into strains carrying each bait, and double transformants were tested for growth on Dex-His media for 3–5 d. Vectors containing no insert were used as negative controls.
Nuclease mapping of protein binding and induced RNA conformation changes
sRNP complexes were assembled with 26 nM 5′-32P-radiolabeled sR8 sRNA and 1–3 μM L7, wild-type or mutant Nop56/58, and fibrillarin in 9.5 μL reactions of mapping assembly buffer (20 mM HEPES at pH 7, 100 mM NaCl, 3 mM MgCl2, and 0.2 mM EDTA) supplemented with 0.5 μg/μL tRNA for 5 min at 70°C. Samples were cooled to 50°C for 5 min before cooling to room temperature. Mung bean nuclease digestion was accomplished by incubating assembled sRNP for 10 min at room temperature with ∼6 units of mung bean nuclease (Epicentre) in a final volume of 10 μL. RNA and proteins were precipitated with 10 vol of 2% LiClO4 in acetone, and pellets were washed with 20 vol of acetone. Alkaline hydrolysis was accomplished by incubating 1.9 nM 5′-32P-radiolabeled sR8 sRNA in 10 mM sodium bicarbonate (pH 10) and 1 mM EDTA supplemented with 0.5 μg/μL tRNA for 10 min at 90°C. RNase T1 digestion was accomplished by incubating 2.5 nM 5′-32P-radiolabeled sR8 sRNA in mapping assembly buffer supplemented with 0.3 μg/μL tRNA and 0.1 U RNase T1(Ambion) for 10 min at room temperature. RNA samples were heated for 1 min at 90°C in 4 M urea, and RNA fragments then resolved on 14% polyacrylamide-7M urea gels and visualized by autoradiography.
M. jannaschii Nop56/58 analysis and modeling
Protein sequence data used in generating Figure 6 were based on NCBI gene bank entries: M. jannaschii Nop56/58 (15,668,875); A. fulgidus Nop56/58 (11,499,670); Sulfolobus solfataricus Nop56/58 (15,897,820); Aeropyrum pernix Nop56/58 (14,601,908); Pyrococcus furiosus Nop56/58 (18,976,432); Saccharomyces cerevisiae Nop56 (6,323,226); S. cerevisiae Nop58 (6,324,886); Homo sapiens Nop56 (20,141,472); and H. sapiens Nop58 (33,872,137). Sequences were aligned using ClustalW at the EMBI server (Thompson et al. 1994). Molecular models of the M. jannaschii Nop56/58 protein were initially constructed using three online servers (Swiss-Model, 3D-JIGSAW, CPHmodels) based on the structure of A. fulgidus Nop56/58 (Bates et al. 2001; Lund et al. 2002; Aittaleb et al. 2003; Schwede et al. 2003). The resulting initial models were optimized with DS Modeling 1.1 (Accelrys). To generate the M. jannaschii Nop56/58-fibrillain complex model (Fig. 7), the crystal structure of M. jannaschii fibrillarin (Wang et al. 2000) was superimposed on fibrillin in the A. fulgidus complex (1.17 Å RMSD); likewise, the Nop56/58 models were superimposed on A. fulgidus Nop56/58. Single rounds of rigid body and conjugate gradient minimization (with no experimental energy terms) were performed using CNS (Brünger et al. 1998).
ACKNOWLEDGMENTS
We thank Kevin Weeks, Daniel Brandt, and Paul Wollenzien for their assistance and suggestions in probing RNP structure and Nick Watkins for insightful discussion. We thank Jimmy Suryadi for attempting to prepare A. fulgidus core proteins for analysis. This work was supported by NSF grant MCB 0215545 (ESM), NIH grant GM52581 (SB), and NIH grant GM069699 (BAB).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2230106.
REFERENCES
- Aittaleb M., Rashid R., Chen Q., Palmer J., Daniels C., Li H. Structure and function of archael box C/D sRNP core proteins. Nat. Struct. Biol. 2003;10:256–263. doi: 10.1038/nsb905. [DOI] [PubMed] [Google Scholar]
- Aittaleb M., Visone T., Fenley M., Li H. Structural and thermodynamic evidence for a stabilizing role of Nop5p in S-adenosyl-L-methionine binding to fibrillarin. J. Biol. Chem. 2004;279:41822–41829. doi: 10.1074/jbc.M406209200. [DOI] [PubMed] [Google Scholar]
- Bachellerie J.P., Cavaille J., Huttenhofer A. The expanding snoRNA world. Biochimie. 2002;84:775–790. doi: 10.1016/s0300-9084(02)01402-5. [DOI] [PubMed] [Google Scholar]
- Bates P., Kelly L., MacCallum R., Sternberg M. Enhancement of protein modeling by human intervention in applying the automatic programs 3D-JIGSAW and 3D-PSSM. Proteins . Suppl. 2001;5:39–46. doi: 10.1002/prot.1168. [DOI] [PubMed] [Google Scholar]
- Brünger A., Adams P., Clore G., DeLano W., Gros P., Grosse-Kunstleve R., Jiang J., Kuszewski J., Nilges M., Pannu N., et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Cahill N., Friend K., Speckman W., Li Z., Terns R., Terns M., Steitz J. Site-specific cross-linking analyses reveal an asymmetric distribution for a box C/D snoRNP. EMBO J. 2002;21:3816–3828. doi: 10.1093/emboj/cdf376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decatur W., Fournier M.J. RNA-guided nucleotide modification of ribosomal and other RNAs. J. Biol. Chem. 2003;278:695–698. doi: 10.1074/jbc.R200023200. [DOI] [PubMed] [Google Scholar]
- Gaspin C., Cavaille J., Erauso G., Bachellerie J.P. Archaeal homologs of eukaryotic methylation guide small nucleolar RNAs: Lessons from the Pyrococcus genomes. J. Mol. Biol. 2000;297:895–906. doi: 10.1006/jmbi.2000.3593. [DOI] [PubMed] [Google Scholar]
- Gauthier T., Berges T., Tollervey D., Hurt E. Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol. Cell. Biol. 1997;17:7088–7098. doi: 10.1128/mcb.17.12.7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goody T., Melcher S., Norman D., Lilley D. The kink-turn motif in RNA is dimorphic, and metal ion dependent. RNA. 2004;10:254–264. doi: 10.1261/rna.5176604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P., Halladay J., Craig E. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 2001;20:3617–3622. doi: 10.1093/emboj/20.14.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein D., Schmeing T., Moore P., Steitz T. The kink-turn: A new RNA secondary structure motif. EMBO J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn J., Tran E., Maxwell E.S. Archaeal ribosomal protein L7 is a functional homolog of the eukaryotic 15.5kD/Snu13p snoRNP core protein. Nucleic Acids Res. 2002;30:931–941. doi: 10.1093/nar/30.4.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine D., Tollervey D. Nop58p is a common component of the box C+D snoRNPs that is required for snoRNA stability. RNA. 1999;5:455–467. doi: 10.1017/s135583829998192x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund O., Nielson C., Lundegaard P., Worning P. CASP5 Conference; Asilomar, CA: 2002. X3M: A computer program to extract 3D models. CASP5 methods abstracts, p. 102. [Google Scholar]
- Moore T., Zhang Y., Fenley M., Li H. Molecular basis of box C/D RNA-protein interactions: Cocrystal structure of archaeal L7Ae and a box C/D RNA. Structure. 2004;12:807–818. doi: 10.1016/j.str.2004.02.033. [DOI] [PubMed] [Google Scholar]
- Newman D., Kuhn J., Shanab G., Maxwell E.S. Box C/D snoRNA-associated proteins: Two pairs of evolutionarily ancient proteins and possible links to replication and transcription. RNA. 2000;6:861–879. doi: 10.1017/s1355838200992446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nottrott S., Urlaub S., Luhrmann R. Hierarchical, clustered protein interactions with U4/U6 snRNA: A biochemical role for U4/U6 proteins. EMBO J. 2002;21:5527–5538. doi: 10.1093/emboj/cdf544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omer A., Lowe T., Russell A., Ebhardt E., Eddy S., Dennis P. Homologs of small nucleolar RNAs in Archaea. Science. 2000;288:517–522. doi: 10.1126/science.288.5465.517. [DOI] [PubMed] [Google Scholar]
- Omer A., Ziesche S., Ebhardt H., Dennis P. In vitro reconstitution and activity of a C/D box methylation guide ribonucleoprotein complex. Proc. Natl. Acad. Sci. 2002;99:5289–5294. doi: 10.1073/pnas.082101999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omer A., Ziesche S., Decatur W., Fournier M.J., Dennis P. RNA-modifying machines in archaea. Mol. Microbiol. 2003;48:617–629. doi: 10.1046/j.1365-2958.2003.03483.x. [DOI] [PubMed] [Google Scholar]
- Rashid R., Aittaleb M., Chen Q., Spiegel K., Demeler B., Li H. Functional requirement for symmetric assembly of archaeal box C/D small ribonucleoprotein particles. J. Mol. Biol. 2003;333:295–306. doi: 10.1016/j.jmb.2003.08.012. [DOI] [PubMed] [Google Scholar]
- Roshdestvensky T., Tang T., Tchirkova I., Brosius J., Bachellerie J.P., Huttenhofer A. Binding of L7Ae protein to the K-turn of archaeal snoRNAs: A shared RNA binding motif for C/D and H/ACA box snoRNAs in Archaea. Nucleic Acids Res. 2003;31:869–877. doi: 10.1093/nar/gkg175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede T., Kopp J., Guex N., Peitsch M. Swiss-Model: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speckmann W., Li Z., Lowe T., Eddy S., Terns R., Terns M. Archaeal guide RNAs function in rRNA modification in the eukaryotic nucleus. Curr. Biol. 2002;12:1–5. doi: 10.1016/s0960-9822(02)00655-3. [DOI] [PubMed] [Google Scholar]
- Suryadi J., Tran E., Maxwell E.S., Brown B. The crystal structure of the Methanocaldococcus jannaschii multi-functional L7Ae RNA-binding protein reveals an induced-fit interaction with the box C/D RNAs. Biochemistry. 2005;44:9657–9672. doi: 10.1021/bi050568q. [DOI] [PubMed] [Google Scholar]
- Tang T., Bachellerie J.P., Rozhdestvensky T., Bortolin M., Huber H., Drungowski M., Elge T., Brosius J., Huttenhofer A. Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fuldidus . Proc. Natl. Acad. Sci. 2002;99:7536–7541. doi: 10.1073/pnas.112047299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terns M., Terns R. Small nucleolar RNAs: Versatile trans-acting molecules of ancient evolutionary origin. Gene Expr. 2002;10:17–39. [PMC free article] [PubMed] [Google Scholar]
- Thompson J., Higgins D., Gibson T. Clustal W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D. Small nucleolar RNAs guide ribosomal RNA methylation. Science. 1996;273:1056–1057. doi: 10.1126/science.273.5278.1056. [DOI] [PubMed] [Google Scholar]
- Tran E., Zhang X., Maxwell E.S. Efficient RNA 2′-O-methylation requires juxtaposed and symmetrically assembled archaeal box C/D and C′/D′ RNPs. EMBO J. 2003;22:3930–3940. doi: 10.1093/emboj/cdg368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran E., Brown J., Maxwell E.S. Evolutionary origins of the RNA-guided nucleotide-modification complexes: From the primitive translation apparatus? Trends Biochem. Sci. 2004;29:343–350. doi: 10.1016/j.tibs.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Tran E., Zhang X., Lackey L., Maxwell E.S. Conserved spacing between the box C/D and C′/D′ RNPs of the archaeal box C/D sRNP complex is required for efficient 2′-O-methylation of target RNAs. RNA. 2005;11:285–293. doi: 10.1261/rna.7223405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidovic I., Nottrott S., Hartmuth K., Luhrmann R., Ficner R. Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment. Mol. Cell. 2000;6:1331–1342. doi: 10.1016/s1097-2765(00)00131-3. [DOI] [PubMed] [Google Scholar]
- Wang H., Boisvert D., Kim K., Kim R., Kim S.H. Crystal structure of a fibrillarin homologue from Methanococcus jannaschii, a hyperthermophile, at 1.6 Å resolution. EMBO J. 2000;19:7–17. doi: 10.1093/emboj/19.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins N., Segault V., Charpentier B., Nottrott S., Fabrizio P., Bachi A., Wilm M., Roshbash M., Branlant C., Luhrmann R. A common core RNP structure shared between the small nucleolar box C/D RNPs and the spliceosomal U4 snRNP. Cell. 2000;103:457–466. doi: 10.1016/s0092-8674(00)00137-9. [DOI] [PubMed] [Google Scholar]
- Weinstein Szewczak L., DeGregorio S., Strobel S., Steitz J. Exclusive interaction of the 15.5kD protein with the terminal box C/D motif of a methylation guide snoRNP. Chem. Biol. 2002;9:1095–1107. doi: 10.1016/s1074-5521(02)00239-9. [DOI] [PubMed] [Google Scholar]