Abstract
Production of reactive oxygen intermediates (ROI) and a form of programmed cell death called hypersensitive response (HR) are often associated with disease resistance of plants. We have previously shown that the Rac homolog of rice, OsRac1, is a regulator of ROI production and cell death in rice. Here we show that the constitutively active OsRac1 (i) causes HR-like responses and greatly reduces disease lesions against a virulent race of the rice blast fungus; (ii) causes resistance against a virulent race of bacterial blight; and (iii) causes enhanced production of a phytoalexin and alters expression of defense-related genes. The dominant-negative OsRac1 suppresses elicitor-induced ROI production in transgenic cell cultures, and in plants suppresses the HR induced by the avirulent race of the fungus. Taken together, our findings strongly suggest that OsRac1 has a general role in disease resistance of rice.
Plants develop defense mechanisms to recognize pathogens and protect them from their attack (1). These defense reactions are triggered by the recognition of pathogens by plant disease resistance (R) genes (2). After the recognition of pathogens, a signaling pathway is activated, resulting in resistance to pathogens (3, 4). During the early steps in R gene-mediated disease resistance, reactive oxygen intermediates (ROI) such as O2− and H2O2 are rapidly generated after infection, and hypersensitive response (HR) leading to cell death are observed (5–7). ROI are thought to be mediators of HR cell death (8–10), although unequivocal evidence to support this hypothesis is still lacking. It has been suggested that NADPH oxidase similar to the neutrophil enzyme is involved in ROI production during defense responses of plants (11–13). However, the biochemical nature of plant NADPH oxidase still needs to be studied.
In addition to ROI and HR cell death, involvement of protein phosphorylation cascades in R gene-mediated disease resistance has been suggested by the isolation of the tomato Pto and the rice Xa21 genes (14, 15). More recently, in vivo phosphorylation assays identified mitogen-activated protein kinases (16–18) and calcium-dependent protein kinase (19), the activation of both which is implicated in defense responses. However, whether these protein kinases are indeed involved in R gene-mediated disease resistance still needs to be determined.
One powerful approach to study signaling in plant disease resistance is the identification of mutants which are compromised for the resistance to pathogens (20). Analysis of Arabidopsis mutants which are compromised for the resistance to various pathogens identified a number of mutations involved in disease resistance (20). Among them, two genes, NDR1 (21) and EDS1 (22), were recently cloned, and NDR1 was shown to encode a protein with two predicted transmembrane domains (21). Interestingly, it was shown that these two genes are required by two separate groups of R genes for resistance, suggesting that R genes differ in their requirements for downstream components in Arabidopsis. More recently, a barley gene, Rar 1, which is required for resistance against powdery mildew mediated by several R genes and functions upstream of ROI production, was cloned (23). Rar 1 encodes a novel protein which may be involved in cell death; however, its function in R gene-mediated disease resistance still needs to be studied.
We have previously shown that a constitutively active form of OsRac1, a rice homolog of mammalian Rac GTPase, can induce ROI production as well as cell death in transgenic suspension cultures and plants (24). Because ROI production and cell death are often associated with disease resistance, these results suggest that Rac may be a component of a disease resistance pathway acting downstream of R genes. Here, by the analysis of transgenic rice suspension cultures and plants expressing constitutively active and dominant-negative OsRac1, we show that OsRac1 is a general component of disease resistance in rice.
Materials and Methods
Rice Transformation.
Details of the constitutively active and the dominant-negative OsRac1 constructs and production of suspension cell cultures have been described (24). Agrobacterium-mediated transformation of rice calli was performed according to a published method (25). Plants were regenerated from transformed calli selected by hygromycin resistance.
Infection of Rice Plants with Blast Fungus and Bacterial Blight.
Growth conditions of the blast fungus and methods for the punch infection of the leaf blade with the blast fungus have been described (26). Infection of leaf sheath cells with the rice blast fungus was performed according to a published protocol (27). The rice sheaths were filled with a conidial suspension (1 × 105 conidia/ml) and held at 22°C for 24 h. The sheath cells were microscopically examined for hyphal growth, and photographs were taken 48 h after infection. For bacterial blight infection, leaves were inoculated with the Japanese Xoo race 1, which is compatible with var. Kinmaze, by the clipping method (28), and lengths of lesions were measured 14 days after inoculation.
Green Fluorescent Protein (GFP) Fusions and Transient Expression in Rice Protoplasts.
The GFP sequence derived from smRS-GFP (29) was fused to the N terminus of OsRac1. Methods for protoplast isolation and electroporation were essentially as described (30). Protoplasts (3 × 107) were isolated from rice Oc suspension cells, mixed with 50 μg of the test plasmid, and electroporated by using the Gene Pulser (Bio-Rad). After an 8-h incubation at 30°C, the protoplasts were examined under a confocal microscope (LSM510, Zeiss) by using ×40 and ×63 (oil immersion) objectives with fluorescein filters.
Momilactone A Measurement.
Methods for the extraction and quantification of momilactone A have been described (26).
Results and Discussion
OsRac1 Is a Component of the N-Acetylchitooligosaccharide Elicitor-Signaling Pathway in Rice Cells.
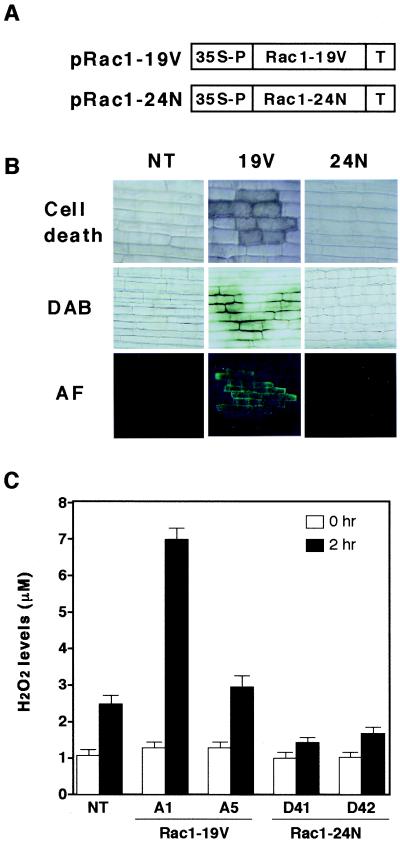
To address the possible role of OsRac1 in disease resistance of rice, we initially examined the effects of the expression of the constitutively active Rac1-G19V and the dominant-negative Rac1-T24N in transgenic rice plants (Fig. 1 A and B). Browning of leaf sheath cells leading to cell death, H2O2 production revealed by brown color developing after staining with diaminobenzidine, and autofluorescence were occasionally detected in the leaf sheath cells of transgenic rice plants expressing the constitutively active Rac1-G19V (Fig. 1B). These three cellular events are typical cellular responses observed during R gene-mediated blast resistance in rice (26, 27, 31). None of these responses were found in untransformed control plants or transgenic plants expressing the dominant-negative OsRac1 (Fig. 1B). Therefore, these results suggest that rice plants transformed with the constitutively active OsRac1 may develop resistance against compatible races of the rice blast fungus.
Figure 1.
Effects of the constitutively active and dominant-negative OsRac1 in transgenic rice plants and cell cultures. (A) Diagrams of vector constructs introduced into rice. pRac1–19V and pRac1–24N are the constitutively active and dominant-negative forms of OsRac1, respectively. pRac1–19V was made by G19V, and pRac1–24N was made by T24N. (B) Cell death, H2O2 production, and autofluorescence (AF) in leaf sheath cells of transgenic rice expressing pRac1–19V and pRac1–24N. Plants ca. 2 months old were used for assays. DAB, diaminobenzidine. (C) Effects of a N-acetylchitooligosaccharide elicitor (3 μg/ml) on H2O2 production in untransformed and transgenic cell cultures expressing pRac1–19V and pRac1–24N. H2O2 levels in media were measured 2 h after addition of the elicitor by a published method (35). NT indicates untransformed control cell cultures. A1 and A5 are transgenic rice cell lines expressing the constitutively active OsRac1, and D41 and D42 are transgenic cell lines expressing the dominant-negative OsRac1. Bars indicate SEM obtained from seven to nine measurements. Student's t test (P < 0.05) was used for statistical analysis of the results.
N-acetylchitooligosaccharide elicitors have been shown to induce a cascade of cellular responses in cultured rice cells. They include induction of momilactone A, a phytoalexin of rice (32), ROI production (33), and expression of several defense-related genes (34). Because a series of events caused by N-acetylchitooligosaccharide elicitors in cultured rice cells mimics early signaling events induced by avirulent races of blast fungus in leaf cells, we examined whether OsRac1 is involved in the elicitor signaling pathway by treating transgenic cell cultures expressing the constitutively active and the dominant-negative forms of OsRac1 with the elicitor and measured H2O2 levels (Fig. 1C). In the untransformed cells, increased levels of H2O2 were detected 2 h after elicitor treatment. In contrast, in transgenic cell cultures expressing the constitutively active OsRac1, levels of H2O2 production in two independent transgenic cultures were significantly higher than controls both at 0 and 2 h. In contrast, in each of the two transgenic cell cultures expressing the dominant-negative OsRac1, H2O2 production was suppressed. These results indicate that OsRac1 is an intermediate in the elicitor signaling pathway leading to ROI production in cultured rice cells.
OsRac1 Is Essential for R Gene-Mediated Blast Resistance in Rice.
To test whether OsRac1 is involved in disease resistance in rice, we chose the rice blast fungus Magnaporthe grisea, because a series of R genes corresponding to a number of races has been identified (36) and a gene-for-gene relationship has been established in this plant–pathogen interaction (37). More recently, two R genes for blast resistance, Pi-b and Pi-ta, were isolated and shown to be members of the NBS-LRR class of plant R genes (37, 38).
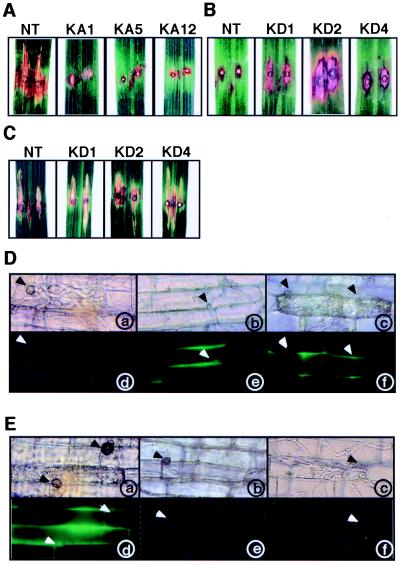
The japonica rice var. Kinmaze, which was used for the production of transgenic plants, carries the Pi-a resistance gene that is incompatible with race 031 of the blast fungus (36). First, we infected the leaf blade of transgenic rice plants expressing the constitutively active OsRac1 with race 007 of the blast fungus by the punch infection method (26). Because race 007 is compatible with var. Kinmaze, infected leaves of untransformed control plants developed typical disease symptoms (Fig. 2A, NT). Of eight independent transgenic lines which were repeatedly tested, six showed HR-like responses (Fig. 2A, KA5 and KA12) or a greatly reduced level of symptoms (Fig. 2A, KA1), and two lines showed an intermediate level of resistance (data not shown). We did not find a clear correlation between the response and expression level of the transgene (data not shown). These results indicate that rice plants transformed with the constitutively active OsRac1 acquire resistance to a compatible race of the rice blast fungus and exhibit HR-like responses which are normally induced by avirulent races.
Figure 2.
Responses of transgenic rice plants expressing the constitutively active and the dominant-negative OsRac1 to infection by the rice blast fungus. (A) Responses of the leaf blade of transgenic rice plants expressing the constitutively active OsRac1 to infection by the compatible race 007. NT indicates untransformed control plants, and KA1, KA5, and KA12 are independent transgenic rice plants. A total of eight lines were tested, and results of three lines are shown. Lesions were observed three weeks after infection. (B) Responses of transgenic rice plants expressing the dominant-negative OsRac1 to infection by the incompatible race 031. NT indicates untransformed control plants, and KD1, KD2, and KD4 are independent transgenic rice plants. Lesions were observed 2 weeks after infection. (C) Responses of transgenic rice plants expressing the dominant-negative OsRac1 to infection by the compatible race 007. NT indicates untransformed control plants, and KD1, KD2, and KD4 are independent transgenic rice plants. Lesions were observed 2 weeks after infection. (D) Microscopic observation of the leaf sheath cells in transgenic plants expressing the constitutively active OsRac1 after infection with the compatible race 007. Representative photographs of KA7 (b and e) and KA12 (c and f) are shown. Excised leaf sheaths were infected with the fungus, and photographs were taken 48 h after infection. Arrows indicate appressoria formed from fungal spores. (a–c) Leaf sheath cells under normal light. (d–f) Leaf sheath cells under fluorescence light. (E) Microscopic observation of the leaf sheath cells in transgenic plants expressing the dominant-negative OsRac1 after infection with the incompatible race 031. Infection of the fungus and microscopic observations were performed as in D. Representative photographs of KD4 taken at 48 h (b and e) and 72 h (c and f) after infection are shown. (a–c) Leaf sheath cells under normal light. (d–f) Leaf sheath cells under fluorescence light.
To examine the cellular events leading to the resistance, leaf sheath cells were infected with the same compatible race of the fungus and examined under the microscope. At 48 h after infection, invading hyphae with branches were visible in leaf epidermal cells of untransformed control plants and had spread to neighboring cells (Fig. 2D, a). No autofluorescence in infected cells was observed (Fig. 2D, d). In contrast, in leaf sheath cells of transgenic plants expressing the constitutively active OsRac1, hyphal development was greatly suppressed although attachment of appressorium to leaf sheath cells was observed (Fig. 2D, b and c). Furthermore, cell death (Fig. 2D, c) and autofluorescence (Fig. 2D, e and f) were detected in the leaf sheath cells of transgenic plants. In conjunction with the results of the leaf blade infection assays, these results indicate that constitutively active OsRac1 confers resistance to the compatible race of blast fungus in leaf cells and that cellular events normally observed in the resistance reactions are found in the leaves of transgenic plants.
Next, we tried to ascertain whether HR caused by an avirulent race of the rice blast fungus is suppressed by expression of the dominant-negative OsRac1. The leaf blades of three independent transgenic lines expressing the dominant-negative OsRac1 were infected with an avirulent race 031 of the blast fungus, and lesions developed after infection were analyzed. Typical HR accompanied by cell death was observed in nontransformed control plants (Fig. 2B, NT). In contrast, in all three transgenic lines examined, HR was clearly suppressed in the leaf blade, and suppression was observed in repeated experiments (Fig. 2B, KD1, KD2, and KD4), indicating that the dominant-negative OsRac1 suppresses R gene-specific resistance against rice blast fungus. To see whether OsRac1 has any role in the basal resistance against rice blast, leaf blades expressing the dominant-negative OsRac1 were infected with a compatible race 007 of the blast fungus. The results showed that the dominant-negative OsRac1 caused no severer symptoms than untransformed control plants, suggesting that OsRac1 may not play a role in the basal resistance against rice blast.
In leaf sheath cells of untransformed control plants, HR cell death and autofluorescence were clearly visible after infection with an avirulent race of the rice blast fungus (Fig. 2E, a and d). In contrast, in three independent transgenic plants expressing the dominant-negative OsRac1, HR cell death and autofluorescence, two primary cell markers for the resistance response, were simultaneously abolished or greatly reduced (Fig. 2E, b, c, e, and f). Microscopic examination of infected cells revealed that at 48 h after infection, hyphae started to develop (Fig. 2E, b), and at 72 h after infection, invasion of branched hyphae in neighboring cells was observed (Fig. 2E, c). These results indicate that the dominant-negative OsRac1 delays the resistance reactions and that suppression of the resistance was accompanied by loss of HR cell death and autofluorescence in infected cells. Taken together, our results strongly suggest that OsRac1 is an essential component of a resistance-signaling pathway for the blast fungus acting downstream of R genes.
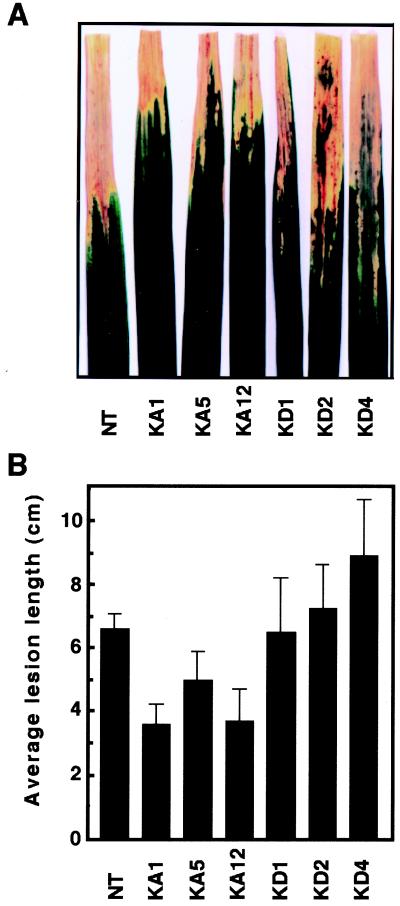
Constitutively Active OsRac1 Confers Resistance to a Compatible Race of Rice Bacterial Blight.
To examine whether OsRac1 has a general role in rice disease resistance, we inoculated leaves of transgenic plants expressing constitutively active and dominant-negative OsRac1 with a compatible race of bacterial blight, Xanthomonas oryzae pv. oryzae. Each of the three independent transgenic rice plants expressing constitutively active OsRac1 was more resistant to the pathogen than untransformed control plants (Fig. 3). In contrast, leaves of transgenic plants expressing dominant-negative OsRac1 showed lesions similar to (KD1 and KD2) or longer (KD4) than those of the wild type. These results suggest that OsRac1 might also be involved in basal resistance against a virulent bacterial pathogen.
Figure 3.
Resistance of transgenic rice expressing constitutively active OsRac1 to a compatible race of rice bacterial blight. (A) Lesion phenotypes of transgenic plants after infection with a compatible race of Xanthomonas oryzae pv. oryzae. Photographs were taken 12 days after inoculation. NT, untransformed control plants; KA1, KA5, and KA12, independent transgenic rice plants expressing the constitutively active OsRac1; and KD1, KD2, and KD4, independent transgenic rice plants expressing the dominant-negative OsRac1. (B) Average lesion length of bacterial blight disease. Leaves were inoculated with the Japanese Xoo race 1. (Bars indicate SEM obtained from four to eight measurements.) Student's t test (P < 0.05) was used for statistical analysis of the results.
Activation of Phytoalexin Production and Alteration of Defense-Related Gene Expression in Transgenic Rice Expressing the Constitutively Active OsRac1.
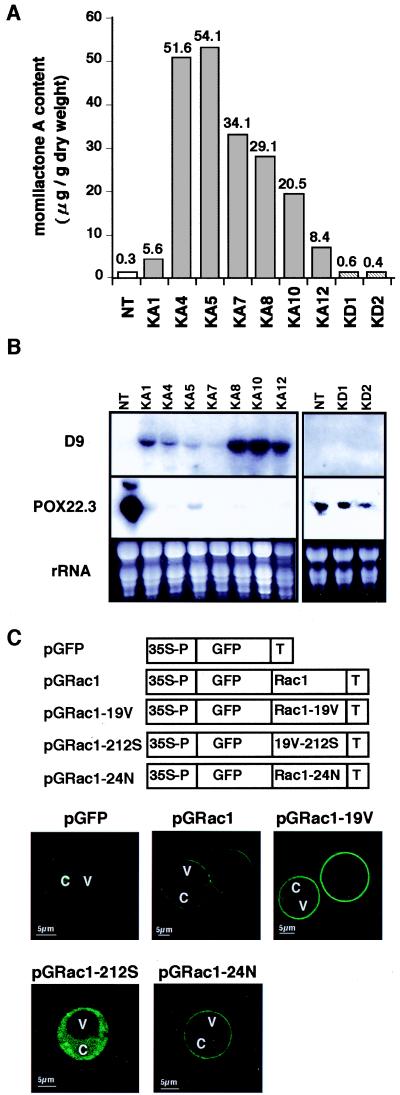
To elucidate the basis for the blast resistance observed in transgenic rice expressing the constitutively active OsRac1, we examined the amounts of momilactone A, a major phytoalexin of rice, in the leaves of transgenic rice (Fig. 4A). In all seven transgenic lines examined, the levels of momilactone A were highly elevated, and the increases were 19- to 180-fold higher than the levels of the untransformed control plants. Transgenic lines expressing the dominant-negative OsRac1 showed momilactone A levels similar to control plants (Fig. 4A).
Figure 4.
Phytoalexin production, alteration of gene expression in transgenic plants, and intracellular localization of OsRac1. (A) Increased production of momilactone A in transgenic rice expressing the constitutively active OsRac1. NT, untransformed control plants; KA1, KA4, KA5, KA7, KA8, KA10, and KA12, independent transgenic rice plants expressing the constitutively active OsRac1; KD1 and KD2, independent transgenic rice plants expressing the dominant-negative OsRac1. (B) Alterations in expression of two defense-related genes in transgenic rice plants expressing the constitutively active OsRac1. D9 is a rice gene homologous with terpenoid cyclase, and POX22.3 is a rice peroxidase induced by a bacterial pathogen (39). Total RNA was isolated from the leaves of transgenic plants and used for Northern blot analysis. Methods for RNA isolation and Northern blot analysis have been described (26). NT, untransformed control plants; and KA1, KA4, KA5, KA7, KA8, KA10, and KA12, independent transgenic rice plants expressing the constitutively active OsRac1. (C) Intracellular localization of OsRac1. (Upper) Various constructs were introduced into rice protoplasts isolated from suspension cells by electroporation. pGFP, GFP alone; pGRac1, a GFP fusion with the wild-type OsRac1; pGRac1–19V, a GFP fusion with the constitutively active OsRac1; pGRac1–212S, a GFP fusion with the constitutively active OsRac1 carrying the C212S mutation; pGRac1–1-24N, a GFP fusion with the dominant-negative OsRac1. C, cytoplasm; and V, vacuole.
We next examined the expression of two genes which are closely associated with disease resistance in rice. Expression of D9 was highly activated in all lines expressing the constitutively active OsRac1 whereas it was suppressed in controls and transgenic plants expressing the dominant-negative OsRac1 (Fig. 4B). D9 was isolated from lesion-mimic mutants of rice (26) resistant to the rice blast and found to be up-regulated in these mutants. Its deduced amino acid sequence is homologous with terpenoid synthase (T.K. and K.S., unpublished results). One of the rice peroxidase genes, POX22.3, which was originally isolated as a gene activated during the resistance reactions of rice against Xanthomonas oryzae pv. oryzae (39), was shown to be strongly down-regulated in the transgenic rice expressing the constitutively active OsRac1 that acquired resistance to the blast disease (Fig. 4B). In untransformed control plants and transgenic lines expressing dominant-negative OsRac1, it was highly expressed (Fig. 4B). These observations suggest that OsRac1 may also function to down-regulate H2O2-scavenging activities at the level of transcription and that this function may be independent of its role in the activation of NADPH oxidase.
These results indicate that constitutively active OsRac1 activates phytoalexin synthesis and alters the expression of defense-related genes to confer resistance to the blast fungus. Therefore, OsRac1 can be considered to be a molecular switch for disease resistance in rice.
OsRac1 Localizes to the Plasma Membrane of Rice Protoplasts.
H2O2 production in the leaf sheath cells of transgenic plants expressing the constitutively active OsRac1 was often localized in the periphery of the cells and the intercellular space (Fig. 1B). Moreover, membrane localization of neutrophil NADPH oxidase has been well established (11), and a plant homolog of gp91phox, the catalytic subunit of the mammalian NADPH oxidase, was shown to be localized at the plasma membrane (13). Therefore, to study the intracellular localization of the OsRac1 protein, we introduced various GFP constructs fused to the N-termini of different forms of OsRac1 (Fig. 4C) into rice protoplasts by electroporation and examined their intracellular localization by using a confocal microscope. When GFP alone was expressed in rice protoplasts, it localized to the cytoplasm (Fig. 4C, pGFP). In contrast, GFP fused to either the wild-type OsRac1 (Fig. 4C, pGRac1) or the constitutively active OsRac1 (Fig. 4C, pGRac1–19V) clearly localized to the plasma membrane in rice protoplasts. The Rho subfamily of the small GTPase contains a C-terminal motif, -CXXL, for prenylation, and the Cys residue in the motif is important for this posttranslational modification (40). Therefore, we made the mutant OsRac1, in which the C-terminal Cys was replaced by Ser and fused with GFP (Fig. 4C, pGRac1-212S). This mutation caused loss of membrane localization, indicating that the Cys residue at the C-terminal motif was essential for the membrane localization of OsRac1. This result is consistent with an observation that deletion of the C-terminal motif caused loss of membrane localization of the Arabidopsis Rac2 (41). Furthermore, we examined the intracellular localization of the dominant-negative OsRac1 and found that it localized to the plasma membrane as well (Fig. 4C, pGRac1–24N).
These results suggest that OsRac1 primarily functions at the plasma membrane, an observation that is consistent with its function as an activator of plant NADPH oxidase, which is likely to be localized at the plasma membrane. Furthermore, recent studies on the interaction of a plant R gene product and its corresponding avirulence (avr) gene product derived from the pathogen indicate that they are both targeted to the plasma membrane and suggest that their interaction takes place at the plasma membrane (42–44). Therefore, in some plant–pathogen interactions, a complex of R and avr gene products may transduce the signal to the Rac protein at the plasma membrane and lead to the activation of NADPH oxidase.
Conclusions
The observations that the constitutively active OsRac1 conferred resistance to rice blast and bacterial blight and that OsRac1 is involved in elicitor signaling suggest that Rac GTPase plays a role in the common pathway in rice disease resistance. This conclusion was supported by increased phytoalexin synthesis and alterations of defense-related gene expression in transgenic plants expressing the constitutively active OsRac1. Combined with our previous results showing that OsRac1 functions to activate ROI production and HR cell death in rice (24), we propose a model of rice disease resistance (Fig. 5). Whether OsRac1 is required by all R genes for blast resistance remains to be tested, because more than 13 R genes corresponding to various races of the rice blast fungus are known (36). The Rac genes constitute a multigene family in plants (45), and some are involved in the tip growth of the pollen tube (41, 45). Thus, it will be important to determine which members of the gene family are actually involved in disease resistance of rice. Results of our study suggest the possibility that disease-resistant crops can be produced by introduction of a single molecular switch acting downstream of R genes in the resistance-signaling pathway.
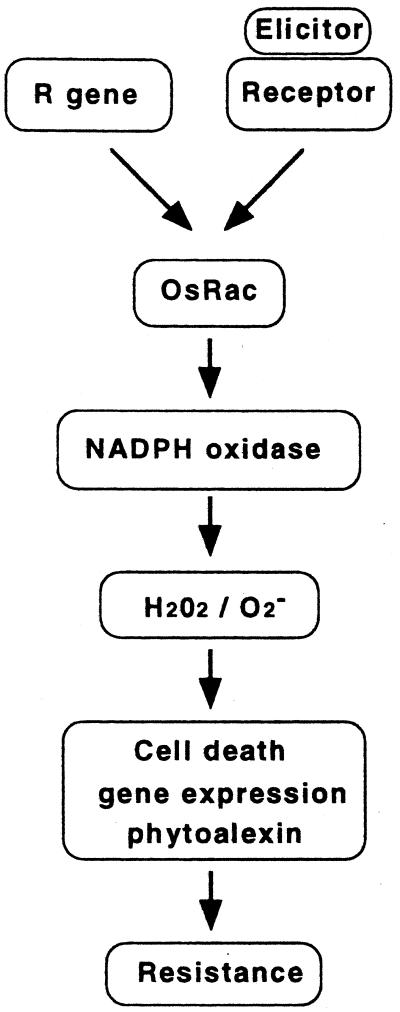
Figure 5.
Proposed model of disease resistance in rice. OsRac1 is placed downstream of R genes and functions to activate NADPH oxidase to cause an oxidative burst during resistance reactions. OsRac1 is also an intermediate in reactions N-acetylchitooligosaccharide to elicitors such as N-acetylchitooligosaccharides. ROI generated by NADPH oxidase then induce cell death, phytoalexin production, and alteration of gene expression. Production of autofluorescence is OsRac1 dependent. These coordinated changes in cellular metabolisms lead to disease resistance.
Acknowledgments
We thank Dr. Naoto Shibuya for the elicitor, Dr. Kosei Takeuchi for help in the confocal analysis of GFP expression, Ms. Mika Nobuhara for excellent technical help, and the members of our lab for suggestions and comments on the manuscript. This work was supported in part by a grant (JSPS-RFTF96R16001) from the Research for the Future program by The Japan Society for the Promotion of Science.
Abbreviations
- ROI
reactive oxygen intermediates
- GFP
green fluorescent protein
- HR
hypersensitive response
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.021273498.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.021273498
References
- 1.Baker B, Zambryski P, Staskawicz B, Dinesh-Kumar S P. Science. 1997;276:726–733. doi: 10.1126/science.276.5313.726. [DOI] [PubMed] [Google Scholar]
- 2.Staskawicz B J, Ausubel F M, Baker B J, Ellis J G, Jones J D G. Science. 1995;268:661–667. doi: 10.1126/science.7732374. [DOI] [PubMed] [Google Scholar]
- 3.Hammond-Kosack K E, Jones J D G. Plant Cell. 1996;8:1773–1791. doi: 10.1105/tpc.8.10.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin G B. Curr Opin Plant Biol. 1999;2:273–279. doi: 10.1016/S1369-5266(99)80049-1. [DOI] [PubMed] [Google Scholar]
- 5.Dangl J L, Dietrich R A, Richberg M H. Plant Cell. 1996;8:1793–1807. doi: 10.1105/tpc.8.10.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamb C, Dixon R A. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:251–275. doi: 10.1146/annurev.arplant.48.1.251. [DOI] [PubMed] [Google Scholar]
- 7.Jabs T. Biochem Pharmacol. 1999;57:231–245. doi: 10.1016/s0006-2952(98)00227-5. [DOI] [PubMed] [Google Scholar]
- 8.Levine A, Tenhaken R, Dixon R, Lamb C. Cell. 1994;79:583–593. doi: 10.1016/0092-8674(94)90544-4. [DOI] [PubMed] [Google Scholar]
- 9.Jabs T, Dietrich R A, Dangl J L. Science. 1996;273:1853–1856. doi: 10.1126/science.273.5283.1853. [DOI] [PubMed] [Google Scholar]
- 10.Mittler R, Herr E H, Orvar B L, van Camp W, Willekens H, Inze D, Ellis B E. Proc Natl Acad Sci USA. 1999;96:14165–14170. doi: 10.1073/pnas.96.24.14165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Segal A W, Abo A. Trends Biochem Sci. 1993;18:43–47. doi: 10.1016/0968-0004(93)90051-n. [DOI] [PubMed] [Google Scholar]
- 12.Groom Q J, Torres M A, Fordham-Skelton A P, Hammond-Kosack K E, Robinson N J, Jones J D G. Plant J. 1996;10:515–522. doi: 10.1046/j.1365-313x.1996.10030515.x. [DOI] [PubMed] [Google Scholar]
- 13.Keller T, Damude H G, Werner D, Doerner P, Dixon R A, Lamb C. Plant Cell. 1998;10:255–266. doi: 10.1105/tpc.10.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin G B, Brommonschenkel S H, Chunwongse J, Frary A, Ganal M W, Spivey R, Wu T, Earle E D, Tanksley S D. Science. 1993;262:1432–1436. doi: 10.1126/science.7902614. [DOI] [PubMed] [Google Scholar]
- 15.Song W-Y, Wang G-L, Chen L-L, Kim H-S, Pi L-Y, Holsten T, Garder J, Wang B, Zhai W-X, Zhu L-H, et al. Science. 1995;270:1804–1806. doi: 10.1126/science.270.5243.1804. [DOI] [PubMed] [Google Scholar]
- 16.Ligterink W, Kroj T, zur Nieden U, Hirt H, Sheel D. Science. 1997;276:2054–2057. doi: 10.1126/science.276.5321.2054. [DOI] [PubMed] [Google Scholar]
- 17.Zhang S, Klessig D F. Proc Natl Acad Sci USA. 1998;95:7433–7438. doi: 10.1073/pnas.95.13.7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Romeis T, Piedras P, Zhang S, Klessig D F, Hirt H, Jones J D G. Plant Cell. 1999;11:273–287. doi: 10.1105/tpc.11.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romeis T, Piedras P, Jones J D G. Plant Cell. 2000;12:803–815. doi: 10.1105/tpc.12.5.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glazebrook J. Curr Opin Plant Biol. 1999;2:280–286. doi: 10.1016/S1369-5266(99)80050-8. [DOI] [PubMed] [Google Scholar]
- 21.Century K S, Shapiro A D, Repetti P P, Dahlbeck D, Holub E, Staskawicz B J. Science. 1997;278:1963–1965. doi: 10.1126/science.278.5345.1963. [DOI] [PubMed] [Google Scholar]
- 22.Falk A, Feys B J, Frost L N, Jones J D G, Daniels M J, Parker J E. Proc Natl Acad Sci USA. 1999;96:3292–3297. doi: 10.1073/pnas.96.6.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shirasu K, Lahaye T, Tan M W, Zhou F, Azevedo C, Schulze-Lefert P. Cell. 1999;99:355–366. doi: 10.1016/s0092-8674(00)81522-6. [DOI] [PubMed] [Google Scholar]
- 24.Kawasaki T, Henmi K, Ono E, Hatakeyama S, Iwano M, Satoh H, Shimamoto K. Proc Natl Acad Sci USA. 1999;96:10922–10926. doi: 10.1073/pnas.96.19.10922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hiei Y, Ohta S, Komari T, Kumashiro T. Plant J. 1994;6:271–282. doi: 10.1046/j.1365-313x.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- 26.Takahashi A, Kawasaki T, Henmi K, Shii K, Kodama O, Satoh H, Shimamoto K. Plant J. 1999;17:535–545. doi: 10.1046/j.1365-313x.1999.00405.x. [DOI] [PubMed] [Google Scholar]
- 27.Koga H. Can J Bot. 1994;72:1463–1477. [Google Scholar]
- 28.Kauffman H E, Reddy A P K, Hsieh S P H, Merca S D. Plant Dis Rep. 1973;57:537–541. [Google Scholar]
- 29.Davis J S, Vierstra D R. Weeds World. 1996;3:43–48. [Google Scholar]
- 30.Kyozuka J, Shimamoto K. In: Plant Tissue Culture Manual B2. Lindsey K, editor. Dordrecht, The Netherlands: Kluwer; 1991. pp. 1–17. [Google Scholar]
- 31.Sekizawa Y, Haga M, Hirabayashi E, Takeuti N, Takino Y. Agric Biol Chem. 1987;51:763–770. [Google Scholar]
- 32.Yamada A, Shibuya N, Kodama O, Akatsuka T. Biosci Biotechnol Biochem. 1993;57:405–409. [Google Scholar]
- 33.Kuchitsu K, Kosaka H, Shiga T, Shibuya N. Protoplasma. 1995;188:138–142. [Google Scholar]
- 34.Minami E, Kuchitsu K, He D Y, Kouchi H, Midoh N, Ohtsuki Y, Shibuya N. Plant Cell Physiol. 1996;37:563–567. doi: 10.1093/oxfordjournals.pcp.a028981. [DOI] [PubMed] [Google Scholar]
- 35.He Z, Wang Z-Y, Li J, Zhu Q, Lamb C, Ronald P, Chory J. Science. 2000;288:2360–2363. doi: 10.1126/science.288.5475.2360. [DOI] [PubMed] [Google Scholar]
- 36.Notteghem J L, Tharreau D, Silue D, Roumen E. In: Rice Blast Disease. Ziegler R S, Leong S A, Teng P S, editors. Wallingford, U.K.: CAB International; 1994. pp. 155–165. [Google Scholar]
- 37.Jia Y, McAdams S A, Bryan G T, Hershey H P, Valent B. EMBO J. 2000;19:4004–4014. doi: 10.1093/emboj/19.15.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Z-X, Yano M, Yamonouchi U, Iwamoto M, Monna L, Hayasaka H, Katayose Y, Sasaki T. Plant J. 1999;19:55–64. doi: 10.1046/j.1365-313x.1999.00498.x. [DOI] [PubMed] [Google Scholar]
- 39.Chittoor J M, Leach J E, White F F. Mol Plant–Microbe Interact. 1997;10:861–871. doi: 10.1094/MPMI.1997.10.7.861. [DOI] [PubMed] [Google Scholar]
- 40.Glomset J A, Farnsworth C C. Annu Rev Cell Biol. 1994;10:181–205. doi: 10.1146/annurev.cb.10.110194.001145. [DOI] [PubMed] [Google Scholar]
- 41.Kost B, Lemichez E, Spielhofer P, Hong Y, Tolias K, Carpenter C, Chua N-H. J Cell Biol. 1999;145:317–330. doi: 10.1083/jcb.145.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boyes D C, Nam J, Dangl J L. Proc Natl Acad Sci USA. 1998;95:15849–15854. doi: 10.1073/pnas.95.26.15849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nimchuk Z, Marois E, Kjemtrup S, Leister R T, Katagiri F, Dangl J L. Cell. 2000;101:353–363. doi: 10.1016/s0092-8674(00)80846-6. [DOI] [PubMed] [Google Scholar]
- 44.Leister R T, Katagiri F. Plant J. 2000;22:345–354. doi: 10.1046/j.1365-313x.2000.00744.x. [DOI] [PubMed] [Google Scholar]
- 45.Valster A H, Hepler P K, Chernoff J. Trends Cell Biol. 2000;10:141–146. doi: 10.1016/s0962-8924(00)01728-1. [DOI] [PubMed] [Google Scholar]