Abstract
Loss of growth inhibitory responses to the cytokine transforming growth factor β (TGF-β) in cancer cells may result from mutational inactivation of TGF-β receptors or their signal transducers, the Smad transcription factors. In breast cancer, however, loss of TGF-β growth inhibition often occurs without a loss of these signaling components. A genome-wide analysis of rapid TGF-β gene responses in MCF-10A human mammary epithelial cells and MDA-MB-231 breast cancer cells shows that c-myc repression, a response that is key to the TGF-β program of cell cycle arrest, is selectively lost in the cancer cell line. Transformation of MCF-10A cells with c-Ha-ras and c-erbB2 oncogenes also led to a selective loss of c-myc repression and cell cycle arrest response. TGF-β stimulation of epithelial cells rapidly induces the formation of a Smad complex that specifically recognizes a TGF-β inhibitory element in the c-myc promoter. Formation of this complex is deficient in the oncogenically transformed breast cells. These results suggest that a Smad complex that specifically mediates c-myc repression is a target of oncogenic signals in breast cancer.
Tissue growth is kept in check by signals that limit cell division and survival. A prototypic carrier of such signals in vertebrates is transforming growth factor β (TGF-β), a ubiquitous cytokine with profound growth inhibitory effects on epithelial and other tissues, and the founding member of a large family of regulators of cell division, differentiation, adhesion, movement, and death (1–3). Alterations in TGF-β signaling can be devastating, as demonstrated by heritable disorders arising from mutations in this pathway and cancers arising partly from a loss of TGF-β signaling (4).
TGF-β family members act as ligands that assemble membrane receptor complexes that activate Smad proteins, which then assemble transcriptional complexes that control gene expression (3). The ligand brings together specific members from two families of transmembrane serine/threonine kinases known as the type I and type II receptors, respectively. In the resulting complex, the type II receptor subunits phosphorylate and activate the type I receptor subunits that, in turn, phosphorylate receptor-regulated Smads (R-Smads). This releases R-Smads from cytoplasmic anchors, allowing their accumulation in the nucleus and association with Smad4, a partner in the assembly of transcriptional complexes. Smad proteins have two conserved domains that bind DNA and transcriptional coactivators or corepressors, respectively. Smads additionally interact with diverse DNA binding cofactors that direct the resulting complex to specific target genes (5, 6). The profile of DNA binding Smad cofactors that a cell expresses as a function of its developmental state and conditions determines the response of that cell to TGF-β. This mode of signaling is highly regulated (7) and is shared by other members of the TGF-β family, including the activins, the nodals, the bone morphogenetic proteins, and the anti-Müllerian hormone in vertebrates, and homologues in Drosophila melanogaster and Caenorhabditis elegans (3).
The version of this pathway that mediates TGF-β signaling involves the type I receptor TβR-I, the type II receptor TβR-II, the R-Smads Smad2 and Smad3, and Smad4 (3). Most of these components suffer inactivating mutations in human cancer. Prominent examples are provided by TβR-II (8, 9) and SMAD4 (also known as deleted in pancreatic carcinoma locus 4, DPC4) (10–12), which are frequently mutated in gastrointestinal cancers and pancreatic and metastatic colon cancers, respectively. In contrast, breast cancer cells often lose TGF-β antimitogetic responses without inactivation of TGF-β receptors or Smad proteins (13–15). Moreover, in breast cancer cells, TGF-β stimulates invasion (16, 17) and formation of TGF-β-dependent bone metastases in model systems (18). The events that shut off the growth inhibitory response and may turn TGF-β into an oncogenic signal remain a mystery.
Progress has been made in understanding how TGF-β signals inhibit cell division (4). The cell division cycle proceeds by the action of cyclin-dependent protein kinases (cdk) (19). TGF-β action inhibits the cdks that drive the G1 phase of the cell cycle, namely, cdk2, cdk4, and cdk6 (4). This is mediated, in part, by stoichiometric inhibitory proteins. In epithelial cells from the skin, lung, and breast, TGF-β rapidly elevates expression of the cdk4/6 inhibitor p15Ink4b (20–22). p15Ink4b binding to cdk4 and cdk6 not only inhibits these kinases but also displaces from these complexes the protein p27Kip1 (21, 23). p27Kip1 is a cdk2 inhibitor that, in the proliferating cells, can stay bound to cdk4 and cdk6 complexes without causing inhibition (19, 24, 25). When mobilized by TGF-β action, p27Kip1 binds to and blocks cdk2 (26). In keratinocytes, colon and ovarian epithelial cells, TGF-β additionally elevates the expression of the p27-related inhibitor, p21Cip1 (27–29), and in mammary epithelial cells it represses the cdk-activating phosphatase Cdc25A (30). These are all rapid gene responses. Thus, in ways that may differ between different epithelial cell types, TGF-β inhibits cell division by triggering a program of cdk inhibitory gene responses.
Another important event in the TGF-β antiproliferative program is the inhibition of c-myc expression (2). A ubiquitous promoter of cell growth and proliferation, c-Myc functions as a transcriptional activator or inhibitor depending on the target gene (31, 32). c-Myc can bind to the initiator element of the p15Ink4b promoter, inhibiting p15Ink4b expression (33). Because c-Myc mRNA and protein are short-lived, their levels decrease rapidly in response to TGF-β, relieving c-Myc-mediated repression of p15Ink4b (23). In addition to removing Myc-mediated repression, TGF-β may enable activation of p15Ink4b via a specific Smad complex that recognizes this promoter. However, artificially averting c-Myc down-regulation blocks the ability of TGF-β to induce p15Ink4b and inhibit the cell cycle (23). Similar control mechanisms may operate on p21Cip1 (34, 35). Thus, c-Myc down-regulation is a key event in the TGF-β program of growth inhibition.
Comparing the genome-wide profile of TGF-β gene responses in nontumorigenic and tumor-derived human mammary cells, we noticed that the loss of TGF-β growth inhibitory effect in the latter coincides with a selective loss of the c-myc down-regulation response. Transformation of nontumorigenic mammary cells with c-Ha-ras and c-erbB2 oncogenes led to a similar phenotype. Prompted by these observations, we investigated the mechanism of c-myc down-regulation by TGF-β, and whether this process is inhibited by oncogenic alterations. We show that TGF-β stimulation of breast epithelial cells rapidly leads to the accumulation of a Smad complex that specifically recognizes a TGF-β inhibitory element in the c-myc promoter. Formation of this complex is selectively thwarted in breast cancer cells.
Materials and Methods
Cell Lines.
HaCaT, SW480.7, MDA-MB-468, and MDA-MB-231 cells were maintained in DMEM plus 10% FBS (GIBCO/BRL). MCF-10A and MCF-10A(Ras/ErbB2) cells were maintained in a 1:1 mixture of DMEM and Ham's F-12 (GIBCO/BRL) supplemented with 5% horse serum (GIBCO/BRL), 10 μg/ml insulin (Sigma), 0.5 μg/ml hydrocortisone (Sigma), and 0.02 μg/ml epidermal growth factor (Sigma). The MCF-10A(Ras/ErbB2) cell line is a derivative that was double-transfected with c-Ha-ras (G12V) oncogene and wild type c-erbB2 (36). [125I]Deoxyuridine incorporation assays were done as described (37).
Oligonucleotide Array Expression Analysis.
RNA sample collection and generation of biotinylated cRNA probe are carried out essentially as described in the standard Affymetrix (Santa Clara, CA) genechip protocol. Briefly, total RNA was prepared from 5 × 106 cultured cells that were untreated or treated with TGF-β by using a Qiagen (Chatsworth, CA) RNeasy mini kit. Twenty-five micrograms of total RNA was used to prepare double-stranded cDNA by using a Custom Superscript Kit (GIBCO/BRL) and a T7-(dT)24 primer (Genset, San Diego). A quarter of each cDNA sample was used to prepare biotinylated cRNA probe by using the BioArray HighYield RNA Transcript Labeling Kit (Enzo, New York). Twenty micrograms of cRNA probe was fragmented and mixed with the hybridization mixture, which was described in standard Affymetrix protocol. Each sample was hybridized with an Affymetrix Human Genome U95A microarray for 16 h at 45°C. Chips are washed and stained on an Affymetrix fluid station and scanned with a Hewlett–Packard argon-ion laser confocal microscope. Fluorescence intensity was measured for each chip and normalized to the average fluorescence intensity for the entire chip. Absolute analysis of each chip and comparative analysis of TGF-β-treated samples with the untreated samples were carried out by using the Affymetrix genechip expression analysis software.
Plasmids.
A low-basal activity luciferase reporter plasmid, pBV-Luc, has been described (38). Reporter plasmids containing various restriction fragments of the human c-myc promoter, including pDel-1, pDel-2, pDel-3, pDel-4, pFrag-A, pFrag-B, pFrag-C, pFrag-D, and pFrag-E (illustrated in Fig. 1A) were generously provided by B. Vogelstein and K. Kinzler (Johns Hopkins, Baltimore, MD) (38). pDel-5 and pDel-6 were generated by excising KpnI/SmaI and KpnI/XhoI fragments, respectively, from pDel-2. DNA fragments −26/+334 and −109/−3 (relative to the P2 transcription initiation site) of the c-myc promoter were amplified by PCR and subcloned into the KpnI/EcoRI site of pBV-Luc (38) to generate pDel-7 and pFrag-F, respectively. Wild-type and mutant forms of the TGF-β inhibitory element (TIE) sequence (−92/−63 relative to the P2 transcription initiation site of c-myc) were synthesized as oligonucleotides with flanking BamHI and BglII sites, annealed, and then cloned directionally in triple copies into the BamHI site of pGL2 reporter plasmid (Promega) to generate p3xTIE-luc and p3xTIE-mut-luc. pFrag-F-TIEmut, pDel-1-TIEmut, pDel-2-TIEmut, and pDel-6-TIEmut were generated by site-directed mutagenesis with the primer set 5′GGGCTTCTCAGAGGCAATTCGGGAAAAAGAACGG3′ and 5′CCGTTCTTTTTCCCGAATTGCCTCTGAGAAGCCC3′. The mutated sequence in TIE-mut is shown in Fig. 2A. Mammalian expression vectors encoding human Smad2, Smad3, Smad4, and Smad4(1–514) (39, 40), and the reporters 4xSBE-Luc (41) and −339/+641Smad7Luc (42) have been described.
Figure 1.
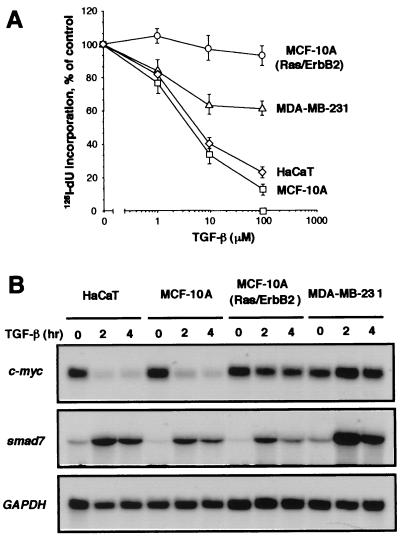
Loss of c-Myc down-regulation in breast cancer cells resistant to TGF-β growth inhibition. (A) Inhibition of [125I]deoxyuridine incorporation into DNA by TGF-β in three cell lines used in DNA microarray analysis and in HaCaT cells. Exponentially growing cultures were incubated for 20 h with the indicated concentrations of TGF-β. Data are the average of triplicate determinations ± SD. (B) Endogenous gene responses in various human cell lines. Exponentially growing cultures of HaCaT, MCF-10A, MCF-10A(Ras/ErbB2), or MDA-MB-231 cells were incubated with 100 pM TGF-β for the indicated time. Total RNA was isolated and subjected to Northern analysis. Blots were probed with human c-myc, smad7, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA probes.
Figure 2.
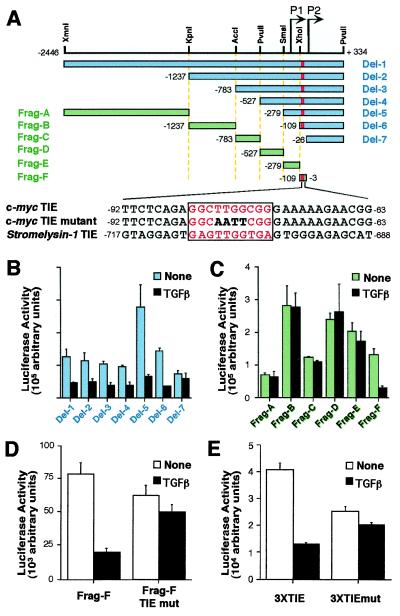
A c-myc TIE-like element mediates TGF-β repression. (A) Diagram of the human c-myc promoter/luciferase constructs and sequence of TIE in c-myc and stromelysin-1. The bars represent the inserted c-myc promoter sequence in the reporter plasmid. P1 and P2 are two transcriptional start sites at the c-myc promoter. Annotations of promoter nucleotide sequence are relative to P2 transcription starting site. TIE-like elements are highlighted by red bars. Thirty-base pair sequences encompassing the TIE element are shown, with the TIE element highlighted in red letters and boxed. The mutations introduced in the c-myc TIE are shown in bold letters. (B–E) Transient expression analysis of c-myc reporter constructs in proliferating HaCaT cells. Cells were transfected with the indicated plasmids and were left untreated or treated with TGF-β for 16 h before luciferase activities were determined. Data are mean ± SD of triplicate experiments.
Reporter Assays.
HaCaT, SW480.7, and MDA-MB-468 cells were transfected by using Lipofectamine (GIBCO/BRL), and MDA-MB-231 was transfected by using Lipofectamine 2000 (GIBCO/BRL), according to manufacturer's instructions. MCF-10A and MCF-10A(Ras/erbB2) were transfected by using DEAE-Dextran as described (30). After transfection, cells were incubated in media containing 10% serum for 16–20 h and then subjected to luciferase assays. pCMV5, an empty vector plasmid, was used as a negative control for transfection and luciferase assay. Luciferase assays were carried out by using the luciferase assay kit (Promega) and a Berthold (Nashua, NH) luminometer. A cytomegalovirus (CMV)-Rellina luciferase plasmid (Promega) was used as a control, to normalize the transfection efficiency, and was assayed as described (43).
Oligonucleotide Precipitation Assays.
Cells were treated with TGF-β for 1 h under normal culture conditions with serum, and lysed by sonication in HKMG buffer (10 mM Hepes, pH 7.9/100 mM KCl/5 mM MgCl2/10% glycerol/1 mM DTT/0.1% Nonidet P-40) with phosphatase inhibitors and a mixture of protease inhibitors. Cell debris was removed by centrifugation twice for 5 min at 10,000 × g at 4°C. Cell extracts were incubated with 1 μg of biotinylated double-strand oligonucleotides corresponding to the wild-type or mutant c-myc TIE(−92/−63), the Smad-binding element (SBE), or the Smad-binding region of junB for 16 h. The sequences of oligonucleotides used are as follows: 5′-TGCCGTCTAGACTGCCGTCTAGACTGCCGTCTAGACTGCCGTCTAGACTGCCACGTCTAGCGAATTCGGATCC-3′ for SBE and 5′-TAATAATTACTATTTCTCAGACAGTCTGTCTGCCTGTCTTAAGTGTCTCACGTCTAGCGAATTCGGATCC-3′ for junB. The sequences of the TIE and TIE-mutant are shown in Fig. 2A. DNA–protein complexes were collected by precipitation with streptavidin-agarose beads (Pierce) for 1 h, washed three times with HKMG buffer, and subjected to Western immunoblotting analysis using previously described rabbit polyclonal antisera (44, 45).
Results
Loss of c-Myc Response in Breast Cancer Cells Resistant to TGF-β Growth Inhibition.
The relative insensitivity of breast cancer cells to the growth inhibitory effect of TGF-β while retaining other responses is represented in three cell lines that we used in the present studies. The MCF-10A cell line was derived from normal human mammary tissue (46) and exhibits a strong growth inhibitory response to TGF-β, despite having a defective Ink4a/Ink4b locus (30). Its derivative, MCF-10A(Ras/ErbB2), was generated by sequential transfection with the human oncogenes c-Ha-ras and c-ErbB2 (36). This cell line is invasive in vitro (47) and is refractory to the antiproliferative effect of TGF-β (Fig. 1A). MDA-MB-231 is a human breast cancer cell line with a hyperactive Ras pathway (48, 49). MDA-MB-231 cells are poorly growth inhibited by TGF-β (Fig. 1A) but are stimulated to form bone metastases in athymic mice (18).
Because of the central role of c-myc repression in the TGF-β growth inhibitory program, we examined this gene response in the cell lines. TGF-β addition rapidly decreased (t1/2 < 2 h) the level of c-myc mRNA in MCF-10A cells, as reported (30), and in HaCaT keratinocytes, another cell line that is profoundly growth inhibited by TGF-β (21) (Fig. 1B). However, TGF-β had little or no effect on c-myc expression in MCF-10A(Ras/ErbB2) or MDA-MB-231 cells, as determined by Northern analysis (Fig. 1B). The absence of c-myc response to TGF-β in the transformed cell lines is in contrast to their ability to respond with induction of Smad7 (Fig. 1B). Smad7 is an antagonistic Smad whose expression is rapidly induced by TGF-β in many cell types and provides feedback regulation of TGF-β signaling (50). These results showed that, in these two transformed cell lines, the TGF-β pathway remains competent to induce some gene responses but is unable to down-regulate c-myc.
The Loss of c-Myc Response Is Selective.
To determine how extensive the loss of TGF-β gene responses was in these two oncogenically transformed cell lines, we conducted a genome-wide analysis of using high density DNA oligonucleotide microarrays. The three mammary cell lines were incubated with or without TGF-β for 2 h (two experiments) or 4 h (two experiments). Complementary RNAs preparations obtained from these cultures were hybridized to oligonucleotide microarrays containing 12,000 probe sets (Affymetrix Human Genome U95-A Microarray).
Analysis of the MCF-10A data revealed 24 transcripts whose signal was increased >2.0-fold and 10 whose signal was decreased >2.0-fold in at least three of four experiments (Table 1 and Table 2, which is published as supplemental data on the PNAS web site, www.pnas.org). The average change (increase or decrease) of each of these signals was >2.5-fold (Table 1). This group includes 11 previously reported TGF-β-induced genes and only one previously reported TGF-β-repressed gene, c-myc (Table 1). All of these are rapid gene responses because they scored at least once in the samples that received TGF-β for 2 h. To identify TGF-β gene responses of slightly slower kinetics, we searched for genes whose signal did not score after a 2-h incubation with TGF-β but was consistently increased or decreased >2.5-fold after 4 h. This yielded three genes, one that was up-regulated and two that were repressed by TGF-β (Table 1). Interestingly, genes encoding transcription factors, extracellular matrix components, signal transduction components, and cytokines account for over three-quarters of all of the TGF-β-responsive genes observed in MCF-10A and the other cell lines (Table 1).
Table 1.
Summary of genes regulated by TGF-β
| GenBank accession no. | Description | Class | Cell lines
|
||
|---|---|---|---|---|---|
| MCF-10A | MCF-10A (Ras/ErbB2) | MDA-MB-231 | |||
| Up-regulated transcripts | |||||
| X58377 | Adipogenesis inhibitory factor* | CK | 3.1 | 3.1 | 16.8 |
| M14083 | β-Migrating plasminogen activator inhibitor-1* | EM | 3.5 | 3.3 | 3.7 |
| J04111 | c-Jun* | TF | 4.4 | 2.4 | 3.3 |
| AB004066 | DEC-1, helix-loop-helix transcription factor | TF | 2.6 | 2.4 | 3.8 |
| J04102 | Erythroblastosis virus oncogene homolog 2 (ets-2) | TF | 3 | 6 | 4.7 |
| X04327 | Erythrocyte 2,3-bisphosphoglycerate mutase† | 4 | 2.6 | 3.1 | |
| X04430 | IFN-β2a | CK | 6.3 | 2.8 | 2.9 |
| U77914 | Jagged | CK | 2.5 | 3.2 | 2.7 |
| M29039 | Jun-B* | TF | 2.6 | 2.7 | 10.4 |
| J03764 | Plasminogen activator inhibitor-1* | EM | 4.5 | 3.5 | 4.5 |
| AF010193 | SMAD7* | ST | 3 | 2.6 | 4.5 |
| AF024710 | Vascular endothelial growth factor (VEGF)* | CK | 2.6 | 2.5 | 2.3 |
| M26576 | Collagen IV* | EM | 2.8 | 2.5 | |
| X06374 | Platelet-derived growth factor α (PDGF-α) | CK | 3.2 | 2.4 | |
| X70683 | Sox-4 | TF | 2.8 | 2.6 | |
| M55152 | Transglutaminase (TGase) | 6.3 | 2.2 | ||
| M85169 | Cytohesin | EM | 2.5 | 2.7 | |
| L07919 | Distal-less homeobox gene (Dlx-2) | TF | 6.5 | 6.8 | |
| AF031167 | IL-15 | CK | 3.1 | 2.4 | |
| X17033 | Integrin α-2 subunit* | EM | 4.3 | 4.6 | |
| X82209 | MN1 | TF | 4.2 | 3.5 | |
| U31201 | Laminin γ2* | EM | 2.9 | 3.6 | |
| V01512 | c-Fos | TF | 2.6 | ||
| X78947 | Connective tissue growth factor* | CK | 4.6 | ||
| L20861 | Wnt-5a | CK | 3.7 | ||
| X02419 | Urokinase plasminogen activator* | EM | 2.6 | ||
| Down-regulated transcripts | |||||
| S73591 | Brain-expressed HHCPA78 homolog | −2.5 | −2.3 | −2.1 | |
| AL049471 | cDNA DKFZp586N012 | −3.4 | −3.1 | −2.9 | |
| U65093 | Msg1-related gene 1 (mrg1) | TF | −3.9 | −2.4 | −7.8 |
| Y00630 | Arg-Serpin (plasminogen activator inhibitor-2) | EM | −2.4 | −3.1 | |
| U45878 | Inhibitor of apoptosis protein-1 | ST | −2.8 | −2.6 | |
| S62539 | Insulin receptor substrate-1 | ST | −2.9 | −3.2 | |
| W29115 | Homo sapiens cDNA | −4 | −2.7 | ||
| AA522530 | Homo sapiens cDNA | −3.9 | −3.8 | ||
| L36463 | Ras interactor (RIN1) | ST | −2.9 | −2.7 | |
| M31166 | Tumor necrosis factor-inducible gene-14 (TSG-14) | CK | −4.9 | −2.5 | |
| V00568 | c-Myc* | TF | −2.8 | ||
| J05008 | Endothelin-1 | CK | −6 | ||
| AB002298 | KIAA0300† | −3 | |||
| X77956 | Id1*† | TF | −3.9 | ||
Genes whose signal was increased or decreased by TGF-β by more than 2-fold in at least three of four experiments are listed (except those labeled †). Fold changes are the average of four experiments. CK, Cytokine growth factor or receptor ligand; TF, transcription factor or modulator; ST, signal transduction molecule; EM, extracellular matrix component or modifier.
Previously described TGF-β gene response.
Gene response that occurred only after 4 h of TGF-β treatment and was more than 2.5-fold (see text for details).
Of the 37 TGF-β gene responses observed in MCF-10A cells, as many as 25 were present in at least one of the two transformed cells lines, MCF-10A(Ras/ErbB2) or MDA-MB-231 (Table 1). Furthermore, these two cell lines showed a large number of positive or negative TGF-β genes responses that were not observed in MCF-10A cells (see Table 2) Three of these were observed in both transformed cells lines (Table 1). Collectively, these data demonstrate that, despite their resistance to the growth inhibitory effect of TGF-β, these two transformed cell lines are still competent to generate many normal TGF-β gene responses.
Eight TGF-β gene responses present in MCF-10A cells were consistently absent in both transformed cell lines (Table 1 and Table 2). These eight include four TGF-β-induced genes and four TGF-β-repressed genes. c-myc down-regulation was among the latter. Three of the missing responses, namely, induction of connective tissue growth factor, and repression of Id1 and endothelin-1, could be secondary to the loss of c-Myc response, because these genes may lie downstream of c-Myc (34, 51, 52). In any event, these data demonstrate that c-myc repression is part of a small group of TGF-β gene responses that are selectively lost in MCF-10A(Ras/ErbB2) and MDA-MB-231, compared with their MCF-10A counterpart.
Mapping of the Minimal TGF-β Response Element in the c-myc Promoter.
To investigate the basis for c-myc repression by TGF-β in epithelial cells and the selective loss of this response in breast cancer cells, we searched for minimal promoter elements that may mediate this response in nontransformed epithelial cells. A series of luciferase reporter constructs containing various fragments of the human c-myc promoter (Fig. 2A) was tested in HaCaT cells in the presence or absence of TGF-β addition. A 2.8-kb region of the c-myc promoter displayed basal transcriptional activity that was repressed by TGF-β (Fig. 2B). Analysis of sequential deletion fragments of the c-myc promoter showed that sequences up to nucleotide −109 relative to the P2 transcription initiation site (53) did not significantly decrease the TGF-β responsiveness of this promoter. However, further deletion to nucleotide −26 abolished the effect of TGF-β (Fig. 2B). To confirm that a TGF-β response element is located between nucleotides −109 and −26, we performed reporter analysis on several restriction fragments spanning the c-myc promoter (Fig. 2A). Among them, only fragment F, which encompasses the −109/−26 region, was susceptible to inhibition by TGF-β (Fig. 2C). We were not able to assign TGF-β responsiveness to the region −279/−109 of c-myc (Fig. 2 B and C), which was previously reported to contain a TGF-β control element (54). Thus, a regulatory element mediating TGF-β repression may be located between nucleotides −109 and −26 of the c-mycpromoter.
A c-myc TIE-like Element Mediates TGF-β Repression.
The −109 and −26 region of c-myc contains a sequence (−84GGCTTGGCGG−75) that closely conforms to the sequence of a previously described TIE in stromelysin-1 (55) (Fig. 2A). To determine whether the TIE-like element from c-myc could play a role in the TGF-β response, the conserved core sequence TTGG in this element was mutated to AATT in the −109 to −26 (Frag-F) reporter construct (Fig. 2A). Although this mutation did little to the basal activity of the reporter, it almost completely abolished the TGF-β inhibitory response (Fig. 2D). Mutation of the TIE in the context of the −109 promoter segment (Del-6) or the 2.8-kb promoter region (Del-1) also prevented inhibition of these reporters by TGF-β (data not shown). To confirm that this TIE-like element is sufficient to mediate TGF-β repression, tandem repeats of a 30-bp sequence (nucleotides −92 to −63) encompassing the TIE element were cloned into the luciferase reporter plasmid pGL2. Indeed, the resulting reporter plasmid p3xTIE-luc displayed strong inhibitory response to TGF-β whereas a similar reporter construct, p3xTIE-mut-luc, which carries the mutant forms of the TIE element, was refractory to TGF-β inhibition (Fig. 2E). These results suggest that the TIE element located within the −92/−63 region of the c-myc promoter is necessary and sufficient for the inhibitory effect of TGF-β.
Recognition of the c-myc TIE by TGF-β-Dependent Smad Complex.
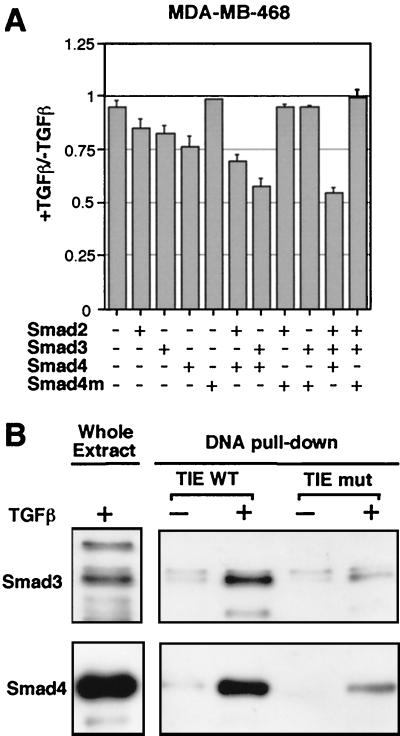
Smad proteins mediate transcriptional activation responses to TGF-β family signals (4). However, Smads are also known to associate with diverse transcriptional corepressors (45, 56, 57), implying that Smads could be involved in mediating negative transcriptional responses. To determine whether Smad proteins are required for down-regulation of c-myc, we asked whether expression of exogenous Smads would rescue this TGF-β response in human epithelial cells lacking endogenous Smad4. The human cell line MDA-MB-468 is a rare case of a breast cancer cell line lacking endogenous Smad4 (58). MDA-MB-468 cells are deficient in transcriptional responses to TGF-β (39). Although TGF-β addition to MDA-MB-468 cells failed to inhibit the transcriptional activity of the c-myc promoter, transfection of Smad vectors restored this response (Fig. 3A). Transfection of Smad4 alone or in combination with Smad2 or Smad3 restored a limited but consistent inhibitory response of the c-myc promoter in this cell line. Transfection of a tumor-derived mutant Smad4(1–514) (10, 39) failed to rescue this response (Fig. 3A). Similar results were obtained with the human colon cancer cell line SW480.7 (data not shown), which is also defective in Smad4 (43).
Figure 3.
Recognition of the c-myc TIE by TGF-β-dependent Smad complex. (A) MDA-MB-468 cells were cotransfected with c-myc-luciferase reporter (Del-2, 1 μg) and the indicated combinations of constructs [200 ng of pCMV5-Smad2, 30 ng of pCMV5-Smad3, 60 ng of pCMV5-Smad4, or pCMV5 vector encoding the Smad4(1–514) mutant]. Transfected cells were left untreated (−) or treated (+) with TGF-β for 16 h. Luciferase activity was determined and is represented as the ratio relative to the values without TGF-β treatment. The amount of DNA used in the transfections was kept equal by using empty pCMV5 plasmid. (B) Binding of a TGF-β-induced endogenous Smad complex to the TIE. Cell extracts from HaCaT cells untreated or pretreated with 100 pM TGF-β for 1 h were incubated with biotinylated wild-type TIE or mutant TIE oligos and streptavidin-agarose beads. Protein-DNA complexes were subjected to Western blotting analysis and probed with anti-Smad3 and anti-Smad4 polyclonal antibodies.
To determine whether Smad proteins physically interact with the TIE element on TGF-β induction, we used biotinylated double-stranded oligonucleotides corresponding to the TIE sequence to precipitate binding proteins from cell extracts. This type of assay has been previously used to detect the interaction of specific Smad complexes with specific promoter elements (45, 59). When subjected to this assay, extracts from HaCaT cells incubated with TGF-β yielded TIE-binding complexes containing Smad3 and Smad4, as determined by Western immunoblotting using specific antibodies (Fig. 3B). Because Smad3 and Smad4 form a complex in response to TGF-β (60), it is likely that Smad3 and Smad4 are in the same TIE-binding complex. Accumulation of this TIE-interacting Smad complex required cell incubation with TGF-β, and occurred rapidly within 1 h of TGF-β addition (Fig. 3B). Mutations in the core sequence of the TIE element in the oligonucleotide (see Fig. 2A) greatly decreased the recognition of this oligonucleotide by the TGF-β-induced Smad complexes (Fig. 3B). Thus, TGF-β rapidly induces the formation of a Smad complex, in HaCaT cells, that specifically recognizes the TIE element of c-myc.
The TIE element in the c-myc promoter partly overlaps a consensus binding site for the transcription factor E2F (nucleotides −77 to −69 relative to the P2 transcription initiation site). E2F has been shown to repress gene expression by binding the hypophosphorylated form of tumor suppressor retinoblastoma (pRb). Because TGF-β can inhibit the phosphorylation of pRb, it was possible that E2F secondarily contributes to c-myc down-regulation by TGF-β. However, the wild-type and the mutant c-myc TIE oligonucleotides precipitated endogenous E2F4 from HaCaT cells equally well, as determined by anti-E2F4 Western immunoblotting (data not shown). Because all known E2F family members have equivalent DNA binding activity, this suggests that the binding of E2F to TIE sequence may not contribute significantly to transcriptional repression by TGF-β. In similar oligonucleotide precipitation experiments, we could not detect any binding of the Smad corepressors TGIF (45) or Ski (56, 61) to the TIE element, suggesting that these repressor(s) may not mediate the effect of TGF-β on the c-myc promoter.
Attenuation of Smad Binding to the c-myc TIE in Transformed Cells.
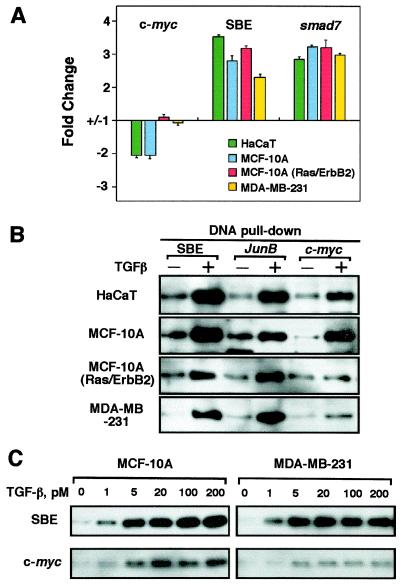
Because c-myc repression by TGF-β was present in HaCaT and MCF-10A cells but missing in MCF-10A(Ras/ErbB2) and MDA-MB-231 cells (refer to Fig. 1), we sought to determine whether this correlated with a loss in the ability of these cells to mediate repression of the c-myc promoter. We transfected HaCaT cells with reporter plasmids expressing luciferase gene under the control of c-myc 1.5-kb promoter region (Del-2), the TGF-β responsive region of the smad7 promoter (42), or a multimeric version of the Smad cognate sequence GTCT (SBE) that is recognized with high affinity by activated Smads (41). Transcription from the smad7 promoter or the SBE multimer was consistently activated by TGF-β in these four cell lines, whereas the c-myc promoter was repressed only in HaCaT and MCF-10A cells (Fig. 4A). This provides additional evidence that the c-myc reporter constructs used here recapitulate the response of the endogenous gene.
Figure 4.
Attenuation of Smad binding to the c-myc TIE in cells resistant to growth inhibition by TGF-β. (A) Reporter responses in various cell lines. HaCaT, MCF-10A, MCF-10A(Ras/ErbB2), and MDA-MB-231 cells were transfected with the indicated reporter constructs with (+) or without (−) TGF-β treatment for 16 h before luciferase activity was determined, and the change relative to the basal activity was calculated and plotted. (B and C) Binding of TGF-β-induced Smad complexes to the c-myc TIE element. Cell extracts prepared from HaCaT, MCF-10A, MCF-10A(Ras/ErbB2), and MDA-MB-231 untreated (−) or pretreated with indicated concentration of TGF-β for 1 h were incubated with biotinylated oligonucleotides corresponding to the TGF-β response elements of c-myc or junB, or a multimer of the Smad binding element GTCT (SBE). Precipitates were subjected to Smad4 Western immunoblotting analysis.
Next we investigated whether oncogenic transformation of these mammary epithelial cells specifically altered the ability of Smad proteins to bind to the c-myc TIE. Extracts from HaCaT, MCF-10A, MCF-10A(Ras/ErbB2), and MDA-MB-231 cells incubated with or without TGF-β were subjected to TIE oligonucleotide precipitation assays. Biotinylated oligonucleotides corresponding, respectively, to the TGF-β response element of the junB promoter (62), a TGF-β-activated gene (see Table 1 and refs. 63 and 64), or four copies of the SBE efficiently precipitated TGF-β-induced complexes containing Smad4 from all four cell lines (Fig. 4B). Oligonucleotides containing the TIE element of c-myc promoter were recognized by a TGF-β-induced Smad4 complex in HaCaT and MCF-10A cells. However, the level of TIE-binding Smad complex in TGF-β-treated MCF-10A(Ras/ErbB2) cells and MDA-MB-231 cells was consistently much lower, reaching no more than one quarter of the level observed in MCF-10A and HaCaT cells (Fig. 4B). This was not due to a general decrease in the TGF-β sensitivity of the transformed cells, because this profile of decreased accumulation of TIE-binding Smad complex was observed over a range of TGF-β concentrations (Fig. 4C). These results suggest that the TGF-β-dependent formation of a Smad complex that specifically recognizes the inhibitory element of c-myc is defective in breast cancer cells that have an otherwise active TGF-β/Smad pathway.
Discussion
Cancer cells often lack the ability to be growth inhibited by TGF-β (4, 65), supporting the idea that loss of growth constraints is a common and necessary step in the malignant progression of cancer cells (66). In the breast, loss of TGF-β antiproliferative and apoptotic responses may compromise the turnover of the mammary epithelium, thus favoring tumor formation (67, 68). In principle, this loss could result from inactivating mutations in TGF-β receptor or Smad genes, as it happens in cancers of the gastrointestinal track (8, 9, 12), pancreas (10, 11), or ovary (69). However, mutations in TGF-β receptors or Smads are rare in breast cancer (14, 15). The lost capacity of breast cancer cells to be growth inhibited by TGF-β coincides with the presence of a hyperactive Ras pathway in these cells. Ras signaling and Smad signaling are linked at several levels, and these links allow both synergies and antagonisms between these two pathways (3). Depending on the cell type and conditions, Ras signaling has been reported to decrease TGF-β receptor expression (70), attenuate the accumulation of Smad proteins in the nucleus (44), or increase the level of the Smad transcriptional corepressor T cell growth inhibitory factor (TGIF; ref. 71). It is clear, however, that these effects do not cause a blanket inhibition of TGF-β signaling. As shown by our transcript profiling results, breast cancer cells that are no longer growth inhibited by TGF-β still display many TGF-β gene responses typical of nontumorigenic mammary epithelial cells. In fact, breast cancer cells appear to derive an advantage from keeping the TGF-β signaling engine while losing antiproliferative responses, as shown by the ability of MDA-MB-231 cells to form TGF-β-dependent bone metastases in athymic mice (18).
How is the TGF-β antiproliferative response lost in breast cancer cells in the first place? And, how selective is this loss? To begin to address these questions, here we have conducted a comparative profiling of TGF-β gene responses in nontumorigenic versus oncogenically transformed or tumor-derived human mammary epithelial cell lines. Our data indicate that a 2-h treatment of mammary epithelial cells consistently alters the expression of approximately 0.3% of the genes represented in the microarrays that we used. Despite a paucity of previously described TGF-β-repressed genes, as many as one-third of the short-term TGF-β gene responses that we observed in the mammary epithelial cell line MCF-10A are inhibitory responses, and a similar proportion is observed in the oncogenically transformed MCF-10A(Ras/ErbB2) and MDA-MB-231 mammary cell lines. Most of the TGF-β-responsive genes identified here encode transcription factors, extracellular matrix components, signal transduction components, and cytokines.
MCF-10A(Ras/ErbB2) and MDA-MB-231 cells show a typical resistance to the growth inhibitory effect of TGF-β. However, this resistance is not because of a general failure of the TGF-β pathway. These two oncogenically transformed cell lines display a majority of the TGF-β gene responses present in the growth-inhibited MCF-10A cell line. However, c-myc is part of a small group of TGF-β gene responses present in MCF-10A cells that are selectively lost in both MCF-10A(Ras/ErbB2) and MDA-MB-231 cells. Down-regulation of c-myc in MCF-10A cells, HaCaT keratinocytes, and other epithelial cell types in response to TGF-β is rapid and, because the protein is short-lived, it results in a rapid depletion of c-Myc from the cell (30). Other TGF-β gene responses missing in the two transformed cell lines, including connective tissue growth factor induction and repression of Id1 and endothelin-1, might be secondary to the loss of the c-Myc response (34, 51, 52), further narrowing the range of primary TGF-β gene responses whose loss is linked to the loss of growth inhibition. In any case, down-regulation of c-myc lies at the core of the TGF-β cdk inhibitory program (23), and its selective loss in MCF-10A(Ras/ErbB2) and MDA-MB-231 cells may be central to their resistance to growth inhibition.
The loss of c-myc down-regulation in MCF-10A(Ras/ErbB2) and MDA-MB-231 cells observed by Northern analysis and microarray analysis coincides with a loss of c-myc promoter responsiveness to TGF-β in these cells. The TGF-β-responsive region of c-myc maps within the −92 to −63 segment of the promoter. This segment contains a sequence resembling that of a TIE previously identified in the extracellular matrix protease gene stromelysin-1 (matrix metalloproteinase 3; ref. 55).We show that the c-myc TIE recapitulates the regulatory properties of the endogenous gene. The TIE is necessary and sufficient to mediate TGF-β repression of the c-myc promoter in transcriptional reporter assays in MCF-10A and HaCaT cells. TGF-β repression of the c-myc promoter is mediated by the Smad pathway. The TIE is specifically recognized by an endogenous Smad complex that is rapidly formed in response to TGF-β. The TIE identified here lies −92 to −63 relative to the P2 transcription initiation site of c-myc. This region does not contain the Smad cognate sequence CAGAC. However, several examples of Smad-responsive genes are know that lack a CAGAC sequence (72, 73). Under our conditions, little or no transcriptional repression or Smad binding activity was detected in a previously described TGF-β-responsive region upstream of the P1 transcription initiation site (54).
The TIE-binding Smad complex that is induced by TGF-β not only has properties of a relevant mediator of c-myc regulation but also appears to be a target of oncogenic action. In MCF-10A(Ras/ErbB2) and MDA-MB-231 cells, TGF-β is able to induce the formation of activated Smad complexes, as determined by the binding of endogenous Smad4 to the cognate SBE element. When the activity of a specific Smad complex was examined, namely, the Smad complex that recognizes the TGF-β response element in the junB promoter, we found that MCF-10A(Ras/ErbB2) and MDA-MB-231 cells retain an unaltered ability to form this complex in response to TGF-β. However, these cells have a markedly decreased ability to generate the TIE-binding Smad complex in response to TGF-β. This defect could lie in as yet unidentified component(s) that collaborate with Smads in the recognition of the TIE. Importantly, even though the genetic background of MCF-10A cells may be drastically different from that of MDA-MB-231, the only major difference between MCF-10A and MCF-10A(Ras/ErbB2) is that the latter has a hyperactive Ras-signaling pathway. Therefore, we hypothesize that oncogenic signals from ErbB2 and Ras could inhibit the expression or activity of component(s) that enable recognition of the c-myc TIE by an activated Smad complex. Identification of components that enable Smad binding to, and repression of, the c-myc TIE will be of interest, because it may uncover a mechanism for the selective disruption of the TGF-β growth inhibition program in breast cancer and other disorders.
Supplementary Material
Acknowledgments
We thank N. Rosen and D. Salomon for kindly providing the MCF10A(Ras/ErbB2) cell line; B. Vogelstein and K. Kinzler for c-myc promoter constructs; J. Nevins for helpful discussions; J. Calonge, C. Pouponnot, J. Seoane, and P. Siegel for valuable advice; and M. Lindor for technical assistance. We also acknowledge the use of the Howard Hughes Medical Institute Genomic Facility at The Rockefeller University. This work was supported by National Institutes of Health grants to J.M. and to Memorial Sloan–Kettering Cancer Center. C.-R.C. and Y.K. are Research Associates and J.M. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations
- TGF-β
transforming growth factor β
- R-Smad
receptor-regulated Smad
- TIE
TGF-β inhibitory element
- SBE
Smad-binding element
Footnotes
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on May 2, 2000.
References
- 1.Roberts A B, Sporn M B. In: Peptide Growth Factors and Their Receptors. Sporn M B, Roberts A B, editors. Heidelberg: Springer; 1990. pp. 419–472. [Google Scholar]
- 2.Alexandrow M G, Moses H L. Cancer Res. 1995;55:1452–1457. [PubMed] [Google Scholar]
- 3.Massagué J. Nat Rev Mol Cell Biol. 2000;1:169–181. doi: 10.1038/35043051. [DOI] [PubMed] [Google Scholar]
- 4.Massagué J, Blain S W, Lo R S. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 5.Derynck R, Zhang Y, Feng X H. Cell. 1998;95:737–740. doi: 10.1016/s0092-8674(00)81696-7. [DOI] [PubMed] [Google Scholar]
- 6.Massagué J, Wotton D. EMBO J. 2000;19:1745–1759. doi: 10.1093/emboj/19.8.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Massagué J, Chen Y G. Genes Dev. 2000;14:627–644. [PubMed] [Google Scholar]
- 8.Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan R S, Zborowska E, Kinzler K W, Vogelstein B, et al. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 9.Grady W M, Rajput A, Myeroff L, Liu D F, Kwon K, Willis J, Markowitz S. Cancer Res. 1998;58:3101–3104. [PubMed] [Google Scholar]
- 10.Hahn S A, Schutte M, Hoque A T M S, Moskaluk C A, da Costa L T, Rozenblum E, Weinstein C L, Fischer A, Yeo C J, Hruban R H, et al. Science. 1996;271:350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 11.Goggins M, Shekher M, Turnacioglu K, Yeo C J, Hruban R H, Kern S E. Cancer Res. 1998;58:5329–5332. [PubMed] [Google Scholar]
- 12.Miyaki M, Iijima T, Konishi M, Sakai K, Ishii A, Yasuno M, Hishima T, Koike M, Shitara N, Iwama T, et al. Oncogene. 1999;18:3098–3103. doi: 10.1038/sj.onc.1202642. [DOI] [PubMed] [Google Scholar]
- 13.Reiss M, Barcellos-Hoff M H. Breast Cancer Res Treat. 1997;45:81–95. doi: 10.1023/a:1005865812918. [DOI] [PubMed] [Google Scholar]
- 14.Tomita S, Deguchi S, Miyaguni T, Muto Y, Tamamoto T, Toda T. Breast Cancer Res Treat. 1999;53:33–39. doi: 10.1023/a:1006167210269. [DOI] [PubMed] [Google Scholar]
- 15.Anbazhagan R, Bornman D M, Johnston J C, Westra W H, Gabrielson E. Cancer Res. 1999;59:3363–3364. [PubMed] [Google Scholar]
- 16.Welch D R, Fabra A, Nakajima M. Proc Natl Acad Sci USA. 1990;87:7678–7682. doi: 10.1073/pnas.87.19.7678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oft M, Peli J, Rudaz C, Schwarz H, Beug H, Reichmann E. Genes Dev. 1996;10:2462–2477. doi: 10.1101/gad.10.19.2462. [DOI] [PubMed] [Google Scholar]
- 18.Yin J J, Selander K, Chirgwin J M, Dallas M, Grubbs B G, Wieser R, Massague J, Mundy G R, Guise T A. J Clin Invest. 1999;103:197–206. doi: 10.1172/JCI3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sherr C J, Roberts J M. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 20.Hannon G J, Beach D. Nature (London) 1994;371:257–261. doi: 10.1038/371257a0. [DOI] [PubMed] [Google Scholar]
- 21.Reynisdóttir I, Polyak K, Iavarone A, Massagué J. Genes Dev. 1995;9:1831–1845. doi: 10.1101/gad.9.15.1831. [DOI] [PubMed] [Google Scholar]
- 22.Sandhu C, Garbe J, Bhattacharya N, Daksis J, Pan C H, Yaswen P, Koh J, Slingerland J M, Stampfer M R. Mol Cell Biol. 1997;17:2458–2467. doi: 10.1128/mcb.17.5.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Warner B J, Blain S W, Seoane J, Massagué J. Mol Cell Biol. 1999;19:5913–5922. doi: 10.1128/mcb.19.9.5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soos T J, Kiyokawa H, Yan J S, Rubin M S, Giordano A, DeBlasio A, Bottega S, Wong B, Mendelsohn J, Koff A. Cell Growth Differ. 1996;7:135–146. [PubMed] [Google Scholar]
- 25.Blain S W, Montalvo E, Massagué J. J Biol Chem. 1997;272:25863–25872. doi: 10.1074/jbc.272.41.25863. [DOI] [PubMed] [Google Scholar]
- 26.Reynisdóttir I, Massagué J. Genes Dev. 1997;11:492–503. doi: 10.1101/gad.11.4.492. [DOI] [PubMed] [Google Scholar]
- 27.Li C Y, Suardet L, Little J B. J Biol Chem. 1995;270:4971–4974. doi: 10.1074/jbc.270.10.4971. [DOI] [PubMed] [Google Scholar]
- 28.Datto M B, Yu Y, Wang X-F. J Biol Chem. 1995;270:28623–28628. doi: 10.1074/jbc.270.48.28623. [DOI] [PubMed] [Google Scholar]
- 29.Elbendary A, Berchuck A, Davis P, Havrilesky L, Bast J R C, Iglehart J D, Marks J R. Cell Growth Differ. 1994;5:1301–1307. [PubMed] [Google Scholar]
- 30.Iavarone A, Massagué J. Nature (London) 1997;387:417–422. doi: 10.1038/387417a0. [DOI] [PubMed] [Google Scholar]
- 31.Facchini L M, Penn L Z. FASEB J. 1998;12:633–651. [PubMed] [Google Scholar]
- 32.Dang C V. Mol Cell Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Staller, P., Peukert, K., Kiermaier, A., Seoane, J., Lukas, J., Karsunky, H., Möröy, T., Bartek, J., Massagué, J., Hänel, F. & Eilers, M. (2001) Nat. Cell Biol., in press. [DOI] [PubMed]
- 34.Coller H A, Grandori C, Tamayo P, Colbert T, Lander E S, Eisenman R N, Golub T R. Proc Natl Acad Sci USA. 2000;97:3260–3265. doi: 10.1073/pnas.97.7.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Claassen G F, Hann S R. Proc Natl Acad Sci USA. 2000;97:9498–9503. doi: 10.1073/pnas.150006697. . (First Published August 1, 2000; 10.1073/pnas.150006697) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ciardiello F, Gottardis M, Basolo F, Pepe S, Normanno N, Dickson R B, Bianco A R, Salomon D S. Mol Carcinog. 1992;6:43–52. doi: 10.1002/mc.2940060108. [DOI] [PubMed] [Google Scholar]
- 37.Laiho M, DeCaprio J A, Ludlow J W, Livingston D M, Massagué J. Cell. 1990;62:175–185. doi: 10.1016/0092-8674(90)90251-9. [DOI] [PubMed] [Google Scholar]
- 38.He T C, Sparks A B, Rago C, Hermeking H, Zawel L, da Costa L T, Morin P J, Vogelstein B, Kinzler K W. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 39.Lagna G, Hata A, Hemmati-Brivanlou A, Massagué J. Nature (London) 1996;383:832–836. doi: 10.1038/383832a0. [DOI] [PubMed] [Google Scholar]
- 40.Ulloa L, Doody J, Massagué J. Nature (London) 1999;397:710–713. doi: 10.1038/17826. [DOI] [PubMed] [Google Scholar]
- 41.Zawel L, Dai J L, Buckhaults P, Zhou S, Kinzler K W, Vogelstein B, Kern S E. Mol Cell. 1998;1:611–617. doi: 10.1016/s1097-2765(00)80061-1. [DOI] [PubMed] [Google Scholar]
- 42.Denissova N G, Pouponnot C, Long J, He D, Liu F. Proc Natl Acad Sci USA. 2000;97:6397–6402. doi: 10.1073/pnas.090099297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Calonge M J, Massagué J. J Biol Chem. 1999;274:33637–33643. doi: 10.1074/jbc.274.47.33637. [DOI] [PubMed] [Google Scholar]
- 44.Kretzschmar M, Doody J, Timokhina I, Massagué J. Genes Dev. 1999;13:804–816. doi: 10.1101/gad.13.7.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wotton D, Lo R S, Lee S, Massagué J. Cell. 1999;97:29–39. doi: 10.1016/s0092-8674(00)80712-6. [DOI] [PubMed] [Google Scholar]
- 46.Soule H D, Maloney T M, Wolman S R, Peterson W D, Jr, Brenz R, McGrath C M, Russo J, Pauley R J, Jones R F, Brooks S C. Cancer Res. 1990;50:6075–6086. [PubMed] [Google Scholar]
- 47.Giunciuglio D, Culty M, Fassina G, Masiello L, Melchiori A, Paglialunga G, Arand G, Ciardiello F, Basolo F, Thompson E W, et al. Int J Cancer. 1995;63:815–822. doi: 10.1002/ijc.2910630612. [DOI] [PubMed] [Google Scholar]
- 48.Davidson N E, Gelmann E P, Lippman M E, Dickson R B. Mol Endocrinol. 1987;1:216–223. doi: 10.1210/mend-1-3-216. [DOI] [PubMed] [Google Scholar]
- 49.Sepp-Lorenzino L, Ma Z, Rands E, Kohl N E, Gibbs J B, Oliff A, Rosen N. Cancer Res. 1995;55:5302–5309. [PubMed] [Google Scholar]
- 50.Nakao A, Afrakhte M, Morén A, Nakayama T, Christian J L, Heuchel R, Itoh S, Kawabata M, Heldin N E, Heldin C H, ten Dijke P. Nature (London) 1997;389:631–635. doi: 10.1038/39369. [DOI] [PubMed] [Google Scholar]
- 51.Shichiri M, Adachi S, Sedivy J M, Marumo F, Hirata Y. Endocrinology. 1997;138:4584–4590. doi: 10.1210/endo.138.11.5538. [DOI] [PubMed] [Google Scholar]
- 52.Lasorella A, Noseda M, Beyna M, Iavarone A. Nature (London) 2000;407:592–598. doi: 10.1038/35036504. [DOI] [PubMed] [Google Scholar]
- 53.Marcu K B, Bossone S A, Patel A J. Annu Rev Biochem. 1992;61:809–860. doi: 10.1146/annurev.bi.61.070192.004113. [DOI] [PubMed] [Google Scholar]
- 54.Pietenpol J A, Munger K, Howley P M, Stein R W, Moses H L. Proc Natl Acad Sci USA. 1991;88:10227–10231. doi: 10.1073/pnas.88.22.10227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kerr L D, Miller D B, Matrisian L M. Cell. 1990;61:267–278. doi: 10.1016/0092-8674(90)90807-q. [DOI] [PubMed] [Google Scholar]
- 56.Luo K, Stroschein S L, Wang W, Chen D, Martens E, Zhou S, Zhou Q. Genes Dev. 1999;13:2196–2206. doi: 10.1101/gad.13.17.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun Y, Liu X, Eaton E N, Lane W S, Lodish H F, Weinberg R A. Mol Cell. 1999;4:499–509. doi: 10.1016/s1097-2765(00)80201-4. [DOI] [PubMed] [Google Scholar]
- 58.Schutte M, Hruban R H, Hedrik L, Cho K R, Nadasdy G M, Weinstein C L, Bova G S, Isaacs W B, Cairns P, Nawroz H, et al. Cancer Res. 1996;56:2527–2530. [PubMed] [Google Scholar]
- 59.Hata A, Seoane J, Lagna G, Montalvo E, Hemmati-Brivanlou A, Massagué J. Cell. 2000;100:229–240. doi: 10.1016/s0092-8674(00)81561-5. [DOI] [PubMed] [Google Scholar]
- 60.Wong C, Rougier-Chapman E M, Frederick J P, Datto M B, Liberati N T, Li J M, Wang X F. Mol Cell Biol. 1999;19:1821–1830. doi: 10.1128/mcb.19.3.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun Y, Liu X, Ng-Eaton E, Lodish H F, Weinberg R A. Proc Natl Acad Sci USA. 1999;96:12442–12447. doi: 10.1073/pnas.96.22.12442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jonk L J, Itoh S, Heldin C H, ten Dijke P, Kruijer W. J Biol Chem. 1998;273:21145–21152. doi: 10.1074/jbc.273.33.21145. [DOI] [PubMed] [Google Scholar]
- 63.Pertovaara L, Sistonen L, Bos T J, Vogt P K, Keski-Oja J, Alitalo K. Mol Cell Biol. 1989;9:1255–1262. doi: 10.1128/mcb.9.3.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Laiho M, Rönnstrand L, Heino J, DeCaprio J A, Ludlow J W, Livingston D M, Massagué J. Mol Cell Biol. 1991;11:972–978. doi: 10.1128/mcb.11.2.972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fynan T M, Reiss M. Crit Rev Oncog. 1993;4:493–540. [PubMed] [Google Scholar]
- 66.Hanahan D, Weinberg R A. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 67.Barcellos-Hoff M, Ewan K. Breast Cancer Res. 2000;2:92–99. doi: 10.1186/bcr40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nguyen A V, Pollard J W. Development (Cambridge, UK) 2000;127:3107–3118. doi: 10.1242/dev.127.14.3107. [DOI] [PubMed] [Google Scholar]
- 69.Wang D, Kanuma T, Mizunuma H, Takama F, Ibuki Y, Wake N, Mogi A, Shitara Y, Takenoshita S. Cancer Res. 2000;60:4507–4512. [PubMed] [Google Scholar]
- 70.Zhao J, Buick R N. Cancer Res. 1995;55:6181–6188. [PubMed] [Google Scholar]
- 71.Lo, R. S., Wotton, D. & Massagué, J. (2001) EMBO J., in press. [DOI] [PMC free article] [PubMed]
- 72.Kim J, Johnson K, Chen H J, Carroll S, Laughon A. Nature (London) 1997;388:304–308. doi: 10.1038/40906. [DOI] [PubMed] [Google Scholar]
- 73.Labbé E, Silvestri C, Hoodless P A, Wrana J L, Attisano L. Mol Cell. 1998;2:109–120. doi: 10.1016/s1097-2765(00)80119-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.