Abstract
The Saccharomyces cerevisiae genome database contains two ORFs with homology to aquaporins, AQY1 and AQY2. Aqy1p has been shown to be a functional aquaporin in some strains, such as Σ1278b. AQY2 is disrupted by a stop codon in most strains; however, Σ1278b has an intact ORF. Because Σ1278b Aqy2p has an intracellular localization in Xenopus oocytes and in yeast, other strains of yeast were examined. Aqy2p from Saccharomyces chevalieri has a single amino acid in the third transmembrane domain (Ser-141) that differs from Σ1278b Aqy2p (Pro-141). S. chevalieri Aqy2p is a functional water channel in oocytes and traffics to the plasma membrane of yeast. The Σ1278b parental strain, the aqy1-aqy2 double null yeast, and null yeast expressing S. chevalieri Aqy2p were examined under various conditions. Comparison of these strains revealed that the aquaporin null cells were more aggregated and their surface was more hydrophobic. As a result, the aquaporin null cells were more flocculent and more efficient at haploid invasive growth. Despite its primary intracellular localization, Σ1278b Aqy2p plays a role in yeast similar to Aqy1p and S. chevalieri Aqy2p. In addition, Aqy1p and Aqy2p can affect cell surface properties and may provide an advantage by dispersing the cells during starvation or during sexual reproduction.
Aquaporin water channels have been identified in nearly all life forms. Although many aquaporin homologs transport only water (orthodox aquaporins), some transport water and glycerol (aquaglyceroporins). Many microorganisms have both. For example in Escherichia coli, AqpZ functions as an aquaporin, whereas GlpF appears to function as a glycerol channel (1). In Saccharomyces, four aquaporin-related ORFs were identified in the genome database (2). Two ORFs, YFL054C and FPS1, are more closely related to the aquaglyceroporins (3). Fps1p mediates glycerol efflux, a process that is important for maintaining osmotic balance required for cell fusion during mating (4) and for tolerating hypoosmotic shock (5).
The other two ORFs, AQY1 and AQY2, are more closely related to aquaporins. Many lab strains, including the Saccharomyces database strain (S288C), have been shown to contain nonfunctional alleles of Aqy1p (3), whereas natural isolates and industrial strains often contain functional Aqy1p sequences (6, 7). Aqy1p from strain Σ1278b functions as a water channel when expressed in Xenopus oocytes (3, 7). In addition, Σ1278b aqy1 null yeast are less sensitive to repeated osmotic shocks than the parent strain (3); however, a clear positive role for Aqy1p has not yet been identified.
In the Saccharomyces genome database strain, S288C, AQY2 contains an 11-bp deletion that creates a stop codon in the middle of the AQY2 gene. When AQY2 from more than 50 strains were examined, only Σ1278b and one of its derivatives were found to contain AQY2 without the deletion (6). When Σ1278b Aqy2p was tested in oocytes, no water channel function was demonstrated because of impaired trafficking to the plasma membrane (6). Here we report an allele of Aqy2p that exhibits full water channel activity when expressed in Xenopus oocytes. Our studies also reveal that expression of Aqy1p and Aqy2p cause a reduction in surface hydrophobicity resulting in greater dispersion of yeast cells, a property that may promote a metabolic advantage during growth.
Materials and Methods
Yeast Strains and Media.
Standard yeast media and genetic manipulations were used (8). Uracil-deficient medium to select for Ura+ transformants was made by using 27 g/liter of dropout base and 0.77 g/liter of complete supplement mixture minus uracil (BIO101).
The yeast strains are described in supplemental Table 1 (which is published on the PNAS web site, www.pnas.org). Strain JC0015 (MATα aqy2Δ∷loxP-kanMX-loxP) was generated by using the one-step gene replacement (9), and the geneticin-resistance cassette (10). Primers 5′KOKanY2 and 3′KOKanY2 were used to amplify the geneticin-resistance cassette.
JC0175 (MATα aqy1Δ∷loxP) was produced by looping out the geneticin cassette through recombination at the loxP sites. JC0012 was transformed with plasmid pSH47, and expression of the Cre recombinase was induced with galactose (10). The cells were plated on YPD (yeast extract/peptone/dextrose) medium and replica plated onto YPD-geneticin plates (Life Technologies, 200 μg/ml). JC0176 (MATα aqy1Δ∷loxP aqy2Δ∷loxP-kanMX-loxP) was generated by using one-step gene replacement to replace AQY2 in JC0175.
JC0009 from the Σ1278b background was mated with Σ1278b, yielding JC0022 (MATa/α his3∷hisG/HIS3 trp1Δ1/TRP1 ura3–52/URA3 leu2∷hisG/LEU2). JC0012 and JC0015 were mated with JC0009 to produce heterozygous diploids. The diploids were sporulated, tetrads were dissected, and geneticin-resistant spores were selected: JC0102 (MATα aqy2Δ∷loxP-kanMX-loxP ura3–52) and JC0145 (MATα aqy2Δ∷loxP-kanMX-loxP his3∷hisG ura3–52). Additional geneticin-resistant spores from the heterozygous diploids were selected and mated to form homozygous diploids, yielding JC0023 (MATa/α aqy1Δ∷loxP-kanMX-loxP/aqy1Δ∷loxP-kanMX-loxP HIS3/his3∷hisG TRP1/trp1Δ1 ura3–52/ura3–52). Geneticin-resistant spores from heterozygous diploids null for either AQY1 or AQY2 were mated, and aqy1-aqy2 double-null spores were identified: JC0123 (MATa aqy1Δ∷loxP-kanMX-loxP aqy2Δ∷loxP-kanMX-loxP trp1Δ1). Mating JC0123 and JC0176 yielded JC0177. Spores from dissected tetrads were screened by PCR to determine whether AQY1 or AQY2 were deleted. All strains used in the experiments were confirmed by Southern blotting (3).
Sequencing of AQY1 and AQY2.
Genomic DNA was prepared to sequence AQY1 and AQY2 from various strains (8). The ORFs were amplified by using PCR with the Expand High Fidelity System (Roche Molecular Biochemicals), and the PCR products were sequenced. Primers used to amplify AQY1, 5′AQY1 and 3′AQY1, were complementary to DNA about 50 nt upstream and 50 nt downstream. AQY2 was sequenced similarly.
Plasmid Construction.
Plasmids were constructed by using standard cloning methods (11) and are listed in supplemental Table 1. PCR amplification of AQY2 from strains GRF5, NRRL-Y-12632, FY86, and Σ1278b using primers ScAQP201 and ScAQP202, and insertion into pCR2.1 using the TA Cloning Kit (Invitrogen) resulted in plasmids p799201, pGN18201, pFY201, and p912201. Plasmid p912202 was made by amplifying AQY2 from Σ1278b by using primers ScAQP2–5′ and 2ScAQP2–3′ and by ligating the product into pCR2.1. AQY2 was excised from p912202 by using BamHI and ligated into the BglII site of the pXβG-ev1 expression vector (12) to form pX912201. Plasmid pXSchev was generated when primers ScAQP2–5′ and 2ScAQP2–3′ were used to amplify AQY2 from S. chevalieri genomic DNA. The fragment was digested with BamHI, and ligated into pXβG-ev1.
Site directed mutagenesis of pX912201 was carried out by PCR, yielding pXFPVS and pXFPAA. The same procedure was used to mutate pX912103 to pXVSFP. Sense and anti-sense PCR primers were made complementary to the target region with the desired point mutations (VSFPa and VSFPs, FPAAa and FPAAs, VSFPa and VSFPs). Each mutagenesis primer was paired with the appropriate upstream or downstream primer so that either the 5′ or 3′ half of AQY1 or AQY2 from before or after the mutation could be amplified. The two halves of the gene were combined in a PCR reaction so that they would overlap at the mutation site. After digestion with BamHI for AQY2 or BglII for AQY1, the inserts were ligated into pXβG-ev1.
For expression of Σ1278b AQY2 or S. chevalieri AQY2 in yeast, the same BamHI fragments from pXβG-ev1 were ligated into pYES2 (Invitrogen), a 2 μ plasmid with a GAL1 promoter for gene expression, yielding pGAQY2 and pYSchev.
Osmotic Water Transport Assays in Xenopus Oocytes.
Capped cRNAs were synthesized in an in vitro reaction with pXβG-ev1 plasmids linearized with XbaI (12). Oocytes from Xenopus laevis were defolliculated and injected with 25 ng of cRNA or with 50 nl of water. They were incubated for 3 days at 18°C in 200 mosM Barth's solution. Pf was determined by placing oocytes in 70 mosM Barth's solution and monitoring swelling with videomicroscopy (13).
Yeast Immunofluorescence Microscopy.
Indirect immunofluorescence of Aqy2p from Σ1278b and S. chevalieri was performed by using plasmids pGAQY2 and pYSM11 (8). Yeast were grown overnight in uracil-deficient medium and then diluted in YPGal (1% yeast extract/2% peptone/2% galactose) and incubated overnight to induce expression. Anti-Kar2p antibody (a gift from S. Michaelis, Johns Hopkins University School of Medicine) was used at a 1:5000 dilution. Anti-Pma1p antibodies (from J.D.B.) were used at a dilution of 1:100. Anti-Aqy2p polyclonal antiserum to a synthetic peptide from the N terminus (NH2-CSNESNDLEKNISHLDPTGVDN-COOH) was raised in rabbits (Covance Research Products, Denver, PA) and affinity purified by using a SulfoLink column (Pierce). Anti-Aqy2p antibody was used at a dilution of 1:100, and the secondary antibody Alexa 488 conjugated goat anti-rabbit (Molecular Probes) was used at 1:400. Mounting media were used to prevent fading of the specimens: 50% glycerol, 50% PBS with 10 mg/ml DABCO (1,4-diazabicyclo[2.2.2]octane) and 50 ng/ml of DAPI (4′6,-diamidino-2-phenylindole). Cells were photographed by using the ×100 objective of a Zeiss Axioplan 2 fluorescence microscope.
Osmotic Cycling.
Cultures were switched between high osmolarity growth conditions and hypoosmolar wash conditions (3). Each cycle consisted of incubating the cultures in YPD + 1.7 M sorbitol for 1 h at 30°C before pelleting the cells and washing with sterile distilled, deionized H2O. To examine phenotypes of aquaporin null cells expressing Σ1278b Aqy2p from plasmid pGAQY2, cultures were grown in galactose medium. For Σ1278b cells expressing S. chevalieri Aqy2p, cultures were grown in glucose medium. Galactose was not used because of growth inhibition of cells expressing S. chevalieri Aqy2p compared with cells containing the empty vector.
Cell Aggregation, Flocculation, and Invasive Growth.
To examine the aggregation of the cells, colonies from plates were resuspended in YPD. After vortexing of the cell suspension, the cells were visualized with a ×40 objective by using a light microscope with phase contrast. Flocculation and haploid invasive growth were tested (14). Overnight YPD cultures of equal cell density were vortexed and allowed to settle. Photographs of the cultures were taken at intervals to mark differences in flocculation. Haploid invasive growth was measured by patching fresh colonies from various strains onto a YPD plate. The plate was incubated for 3 days at 30°C. The growth of the patches was documented by scanning the plate with a flatbed scanner. The cells on the surface of the plate were washed away with H2O, and the plate was scanned again.
Polystyrene Binding Assay.
Hydrophobicity was assayed by using polystyrene beads (15). Cells from a fresh YPD plate were resuspended in 50 mM Na-phosphate buffer (pH 7.0), spun down, and resuspended with Na-phosphate buffer. An equal volume of cell suspension was mixed with 0.807-μm diameter polystyrene beads (Sigma) (3 μl of 10% bead suspension in 1 ml). The mixture was vortexed for 30 s and examined with a light microscope.
Results
Sequence and Functional Analysis of Aqy2p from Σ1278b.
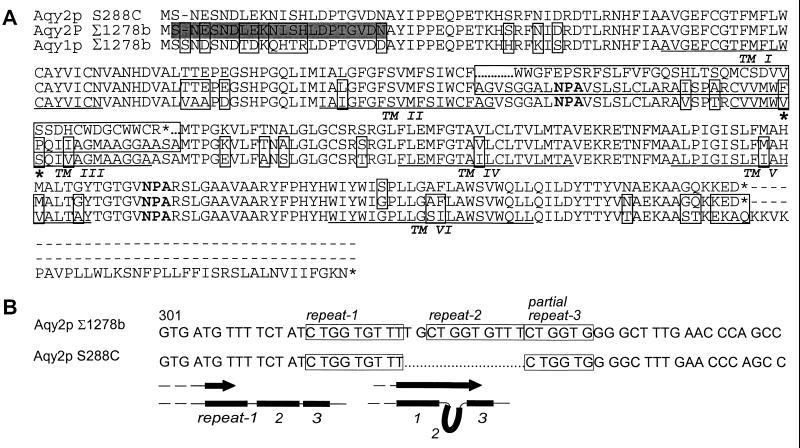
S. cerevisiae genome database strain S288C contains AQY2 fragmented into two ORFs, YLL052C and YLL053C. This pattern is also observed in strains FY86, NRRL-Y-12632, and GRF5 (data not shown). In contrast, Σ1278b AQY2 contains a single intact AQY2 ORF. The difference in the nucleotide sequence between the two AQY2 alleles corresponds to a deletion of 11 bp that creates a premature stop codon (Fig. 1A). Curiously, part of the 11-bp deletion corresponds to a sequence that is repeated three times in Σ1278b AQY2 (Fig. 1B, top). Deletion and insertion of repeats in the Saccharomyces genome have been observed (16). Possible mechanisms for the deletion may include errors during recombination or polymerase slippage during replication (17) (Fig. 1B, bottom). Σ1278b Aqy1p and Aqy2p are highly conserved with amino acid sequences which are 87% identical (Fig. 1A).
Figure 1.
Sequence alignment of Aqy1p and Aqy2p. (A) Comparison of Aqy1p from Σ1278b, Aqy2p from S288C (database strain), and Aqy2p from Σ1278b. Residues that differ between sequences are boxed. The deletion in S288C AQY2 is marked with dots. Aqy2p residues 141 and 142 are identified with asterisks. Putative transmembrane domains are identified (TM1–6). The shaded residues correspond to the sequence of the peptide used to make anti-Aqy2p antibodies. (B) The deletion in S288C and Σ1278b AQY2 with nucleotide repeats is boxed. Schematic diagram shows a possible mechanism for repeat removal that could explain the S288C deletion.
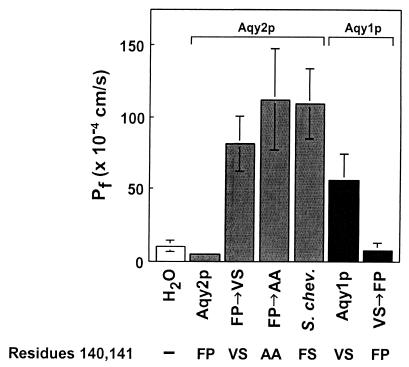
To determine the coefficient of osmotic water permeability (Pf), Xenopus oocytes were injected with cRNA corresponding to Σ1278b Aqy2p and the rate of swelling was measured in hypoosmolar buffer. Oocytes injected with Σ1278b AQY2 cRNA exhibited low water permeabilities (Fig. 2), although immunoblots indicated that the polypeptide was expressed by the oocytes (data not shown). Indirect immunofluorescence of oocytes expressing Σ1278b Aqy2p revealed that the polypeptide did not traffic to the plasma membrane (data not shown).
Figure 2.
Osmotic water permeability (Pf) measurements of Aqy2p and Aqy1p mutants. Oocytes were injected with water or with cRNA to express Σ1278b Aqy2p, Aqy2p with mutations F140V and P141S (FP→VS), Aqy2p with mutations F140A and P141A (FP→AA), S. chevalieri Aqy2p, Σ1278b Aqy1p, and Aqy1p with mutations V141F and S142P (VS→FP). After 3 days in culture, oocytes were transferred from 200 mosM Barth's solution to 70 mosM, and swelling was measured.
Sequence Analysis of AQY2 and AQY1 from Multiple Strains of Saccharomyces.
Searching for functional homologs, AQY2 was sequenced from strains with different origins, including strains that experience only limited laboratory culturing, industrial strains, and clinical isolates of S. cerevisiae (supplemental Table 2; see www.pnas.org). Most strains contain either the 11-bp deletion or have other frameshift mutations. In addition to Σ1278b, only two strains were found to contain intact AQY2 ORFs. Aqy2p from S. diastaticus is identical to Σ1278b; Aqy2p from S. chevalieri has a serine at residue 141 rather than a proline. This single amino acid substitution was also noted in Aqy2p from some strains with frameshift mutations.
AQY1 was also sequenced from the same strains. Aqy1p from Σ1278b was previously noted to require both a valine at residue 121 and proline at residue 255 to exhibit high water permeability when expressed in oocytes (3, 7). In contrast, almost half of the Aqy1p polypeptides have a methionine at position 121 and threonine at residue 255 that abolish water channel function in oocytes.
Functional Analysis of Aqy2p Polymorphisms at Residue 141.
To learn more about residue 141 of Aqy2p, site directed mutagenesis was undertaken to change residues in the third transmembrane domain. Aqy2p from Σ1278b contains Phe-140 and Pro-141, and the corresponding residues in Aqy1p are Val-141 and Ser-142. Oocytes injected with cRNA for Aqy2p double mutant Phe-140→Val, Pro-141→Ser exhibited high osmotic water permeabilities as did oocytes injected with the mutant Phe-140→Ala, Pro-141→Ala (Fig. 2). Conversely, oocytes injected with cRNA for the corresponding Aqy1p mutant Val-141→Phe, Ser-142→Pro exhibited low water permeability. Finally, when S. chevalieri Aqy2p was tested (contains Phe-140 and Ser-141), the oocytes also exhibited high water permeability (Fig. 2). Thus, only one strain of Saccharomyces was found to have Aqy2p that was functional when expressed in oocytes.
Immunofluorescence of Aqy2p.
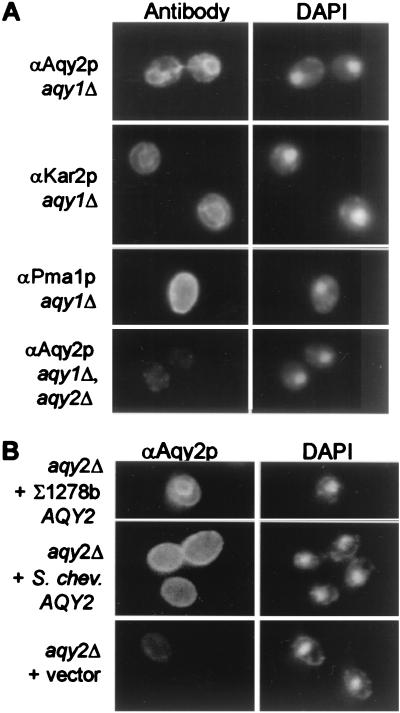
Although Aqy1p and Aqy2p differ at 8 of 21 N-terminal residues (Fig. 1A), antibodies raised to the N-terminal peptides yield significant cross reactivity (data not shown). Thus, immunofluorescence was studied in cells expressing single aquaporin genes. When aqy1 null Σ1278b cells were probed with anti-Aqy2p antibody, perinuclear staining appeared similar to staining with anti-Kar2p antibody, a marker for endoplasmic reticulum (Fig. 3A). The pattern of staining was very different from the pattern seen when anti-Pma1p was used to decorate the plasma membrane. When Σ1278b cells lacking both Aqy1p and Aqy2p were stained with anti-Aqy2p, minimal background staining appeared (Fig. 3A).
Figure 3.
Indirect immunofluorescence localization of Aqy2p from Σ1278b and S. chevalieri. (A) aqy1 null cells (JC0023) were fixed, probed with antibodies to Aqy2p, Kar2p (ER resident protein), or Pma1p (plasma membrane protein), and then incubated with fluorescence-conjugated goat anti-rabbit IgG. aqy1-aqy2 double-null cells (JC0177) were labeled with anti-Aqy2p antibodies as a negative control. DAPI staining of the same cells is shown on the right. (B) aqy2 null cells (JC0145) expressing Σ1278b Aqy2p (plasmid pGAQY2), S. chevalieri Aqy2p (pYSchev), or vector without an insert (pYES2) as a negative control. DAPI staining of the cells is shown on the right.
To compare the localization of Aqy2p from Σ1278b and S. chevalieri, aqy2 null cells were transformed with a plasmid with S. chevalieri AQY2. As a positive control, aqy2 null cells overexpressing Σ1278b Aqy2p showed the perinuclear staining pattern with the anti-Aqy2p antibody, similar to cells endogenously expressing Σ1278b Aqy2p (Fig. 3B). When the same aqy2 null cells expressed S. chevalieri Aqy2p, a distinctive plasma membrane staining pattern was observed that appeared identical to the pattern that was seen when Pma1p was labeled (Fig. 3B). Thus, Aqy2p from S. chevalieri is functional in oocytes and traffics to the plasma membrane of yeast, whereas Σ1278b Aqy2p does not.
Effect of Aqy2p on Survival During Osmotic Stress.
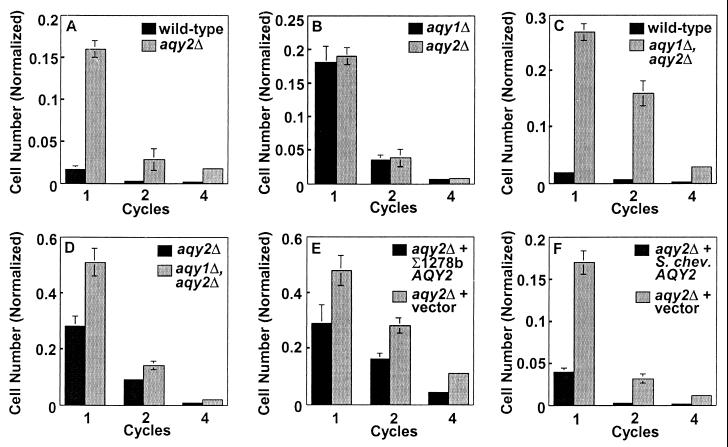
When parent strain Σ1278b and aqy2 null cells were grown in YPD + 1.7 M sorbitol and washed in H2O, the null cells were less sensitive to the osmotic stress (Fig. 4A). A similar effect was previously noted for aqy1 null cells (3). When aqy1 null cells were compared side-by-side with aqy2 null cells, no difference in survival was observed during the osmotic cycles (Fig. 4B). The aqy1-aqy2 double-null cells were much less sensitive to osmotic cycles than the parent Σ1278b strain and somewhat less sensitive than aqy2 null cells (Fig. 4 C and D). To determine the effect of S. chevalieri Aqy2p on the sensitivity to osmotic cycles, Σ1278b aqy2 null cells were transformed with S. chevalieri AQY2. As a positive control, the aqy2 null cells transformed with Σ1278b AQY2 were more sensitive to osmotic cycling than the null cells transformed with vector alone (Fig. 4E). Similarly, aqy2 null cells that expressed S. chevalieri Aqy2p were more sensitive to osmotic stress than aqy2 null cells containing vector alone (Fig. 4F).
Figure 4.
Phenotypic analysis of Aqy2p from Σ1278b and S. chevalieri. Cells derived from Σ1278b were grown in YPD + 1.7 M sorbitol for 1 h at 30°C, spun down and washed with water, and resuspended in YPD + 1.7 M sorbitol. Cycles were repeated four times. Plating the cultures and normalizing to the starting number determined survival of the strains. Wild-type (Σ1278b), aqy2 null cells (JC0015), aqy1 null cells (JC0012), and aqy1-aqy2 double-null cells (JC0176) were compared in pairs. aqy2 null cells (JC0145) were transformed with plasmid without an insert (pYES2), plasmid with Σ1278b AQY2 (pGAQY2), or plasmid with S. chevalieri AQY2 (pYSchev), and their survival was compared as described above.
Effect of Aqy1p and Aqy2p on Surface Properties.
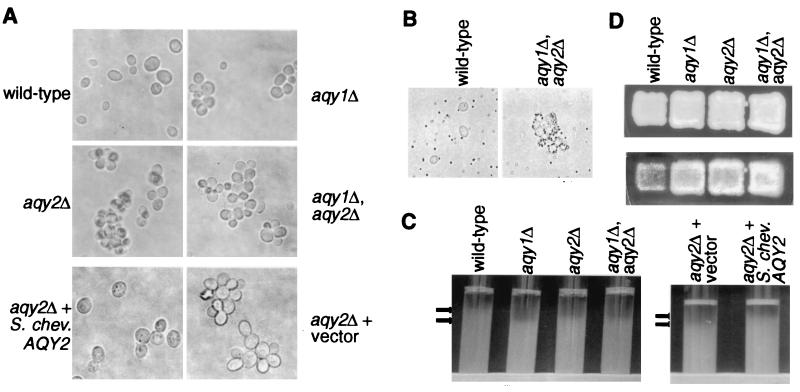
When yeast were taken from a fresh plate and were resuspended in YPD, the wild-type Σ1278b cells were more dispersed than the aqy1 null, aqy2 null, or aqy1-aqy2 double-null cells. In addition, Σ1278b aqy2 null cells expressing S. chevalieri Aqy2p were more dispersed than null cells containing vector alone (Fig. 5A). To eliminate Ca2+-mediated carbohydrate interactions, the cells were resuspended with 10 mM EDTA before vortexing, but this had no effect on aggregation (data not shown).
Figure 5.
Effect of Aqy1p and Aqy2p on cell surface properties of cells. Wild-type (Σ1278b), aqy1 null (JC0012), aqy2 null (JC0015), aqy1-aqy2 double-null cells (JC0176) were compared. (A) Effect of Aqy1p and Aqy2p on aggregation of cells after growth on YPD plate. Cells were grown on a YPD plate and resuspended in YPD media. After vortexing, cells were examined with a light microscope. In addition, aqy2 null cells (JC0145) were transformed with plasmid without an insert (pYES2) or plasmid with S. chevalieri AQY2 (pYSchev) and examined by using the same procedure. (B) Effect of Aqy1p and Aqy2p on ability to adhere to polystyrene. Wild-type (Σ1278b) and aqy1-aqy2 double null cells (JC0176) were mixed with polystyrene beads and examined with a light microscope. (C) Effect of Aqy1p and Aqy2p on flocculation of yeast cultures. Cultures were vortexed and allowed to settle. Arrows mark the top of the sinking cultures. The same procedure was followed with aqy2 null cells (JC0145) transformed with vector (pYES2) or S. chevalieri AQY2 (pYSchev). (D) Effect of Aqy1p and Aqy2p on haploid invasive growth. Cells were patched on a YPD plate and incubated at 30°C for 3 days. The cells were removed from the surface of the plate by washing the plate with water. The top panel shows the plate before washing, and the bottom panel shows the cells that remain and have invaded the agar.
To assess hydrophobicity of the strains, polystyrene beads were vortexed with the cells, and the mixture was examined under the microscope. A much larger proportion of the aqy1-aqy2 double-null strain cells were decorated with many more beads than the wild-type cells, indicating that they are more hydrophobic than wild-type cells (Fig. 5B).
Cultures of wild-type Σ1278b, aqy1 null, aqy2 null, and aqy1-aqy2 double-null strains were vortexed and allowed to settle. The wild-type culture consistently sank toward the bottom of the tube at a slower pace than the aqy1, aqy2, and the aqy1-aqy2 double-null strains (Fig. 5C). To examine the effect of S. chevalieri Aqy2p on culture flocculence, Σ1278b aqy2 null cells expressing S. chevalieri Aqy2p were compared with the same cells expressing vector alone. The cells expressing S. chevalieri Aqy2p were less flocculent than the cells without Aqy2p (Fig. 5C).
The aqy1 null, aqy2 null, and aqy1-aqy2 double-null strains were plated as a patch on a YPD plate. After 3 days of growth, the cells were washed from the surface of the plate with water. Although the patches looked comparable before washing (Fig. 5D, top), after washing the patch of wild-type cells that had invaded the agar was smaller and less dense than the patches of the aqy1 null, aqy2 null, and the aqy1-aqy2 double-null strains (Fig. 5D, bottom).
Discussion
Aqy1p and Aqy2p are highly homologous polypeptides. Both are selected against in laboratory strains. While the functional allele of AQY1 is more prevalent in strains that have not been cultivated in the laboratory, only one allele of AQY2 was found to be functional in oocytes.
Strain Σ1278b is unusual, having a functional Aqy1p and an intact AQY2 ORF. The two polypeptides are extremely similar yet oocytes expressing them behave very differently; Aqy2p does not function in oocytes because of intracellular retention. Surprisingly, the sequence that prevents Σ1278b Aqy2p from trafficking to the plasma membrane of oocytes is not located in the either of the termini but in one of the transmembrane domains. This location presumably results from slight misfolding of the protein attributable to a bend of the membrane-spanning α-helix and retention in the endoplasmic reticulum. However, it remains possible that this protein actually traffics and functions properly under some as yet unidentified physiologic state.
The difference in localization of Σ1278b Aqy2p and S. chevalieri Aqy2p in oocytes correlates with membrane trafficking in yeast. Σ1278b Aqy2p is localized primarily in the endoplasmic reticulum, whereas S. chevalieri Aqy2p is primarily in the plasma membrane of yeast. However, the results from the osmotic cycling experiments for Σ1278b Aqy1p and Aqy2p, and S. chevalieri Aqy2p are very similar. The similarity of the phenotypes suggests that some Σ1278b Aqy2p still traffics to the plasma membrane of yeast and functions despite its primary location being intracellular.
Yeast aquaporins may play a positive role for some strains by enhancing their dispersion. The Σ1278b wild-type strain was much more frequently found as single cells than the aquaporin null yeast. This may possibly confer an advantage when energy sources are scarce and during sexual reproduction, when separation from a group may promote survival. The well controlled environment of the laboratory and the osmotic shock of laboratory washing may explain why so many laboratory strains lack a functional Aqy1p and Aqy2p.
Our cell aggregation data also suggest that the absence or presence of aquaporins in yeast can change surface properties of the cell. Interestingly, even after addition of EDTA, no change in cell aggregation was observed, suggesting that Ca2+ dependent lectin-carbohydrate interactions are not involved. Cell surface hydrophobicity is important in cell aggregation (18). The polystyrene binding assays suggest that the aqy1-aqy2 double-null yeast can bind polystyrene more tightly because they may be more hydrophobic than the parent strains.
The structural basis for increased hydrophobicity of the aquaporin null strains is not clear. Evidence suggests that decreased mannosylation of a protein on the cell surface may increase hydrophobicity or exposure of more hydrophobic proteins on the cell surface (15). Possible glycosylation of Aqy1p or Aqy2p has not yet been explored. For unknown reasons, strains of Saccharomyces cerevisiae vary widely in flocculence and in hydrophobicity (18, 19). Because cell aggregation is related to culture flocculence and because hydrophobicity increases flocculence of yeast cultures (18), we examined the effect of aquaporins on this property. Our studies demonstrate that aqy1 null, aqy2 null, and aqy1-aqy2 double-null cultures flocculate more quickly than wild-type cells do, which is probably because the aquaporin null yeast has an increased cell surface hydrophobicity.
Other processes have been linked to flocculation such as haploid invasive growth and pseudohyphal growth. For instance, a mutation in the FLO8 gene reduces flocculence but also abolishes haploid invasive growth and pseudohyphal growth (14). In addition, strain Σ1278b is unique among strains often studied in the laboratory because of its ability to form pseudohyphae and to undergo haploid invasive growth. We have found that haploid aquaporin null cells invade agar much more efficiently than the wild-type Σ1278b; however, diploid aqy1-aqy2 double-null yeast and wild-type yeast form pseudohyphae equivalently (data not shown). This result suggests that flocculence and haploid invasive growth may be influenced by cell surface hydrophobicity, although hydrophobicity may not play a primary role in pseudohyphal growth.
It is likely that aquaporins have additional roles in yeast. Increased cell surface hydrophobicity has been shown to increase virulence of yeast (15). Therefore, it is possible that aquaporin null yeast may be more virulent than wild-type yeast with functional aquaporins. Interestingly, four of five clinical S. cerevisiae isolates that we examined contained disabling mutations in AQY1 and AQY2. Aqy1p has been shown to be up-regulated almost four-fold during early sporulation (20). Although attempts to find defects in sporulation and spore germination of aqy1 null cells have not led to phenotypic differences (unpublished data), aquaporins may have subtle roles during sporulation or in other life processes of yeast that are not reproduced during laboratory culture conditions.
Supplementary Material
Acknowledgments
We thank Stefan Hohmann for critical suggestions and collegial discussions and Bettina Brown for help with sequencing. This work was supported by the National Institutes of Health.
Note Added in Proof.
Functional studies of Aqy2p in yeast have been reported (21).
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database [accession no. AF321111 (S. chevalieri AQY2)].
References
- 1.Borgnia M, Nielsen S, Engel A, Agre P. Annu Rev Biochem. 1999;68:425–458. doi: 10.1146/annurev.biochem.68.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Goffeau A, Barrell B G, Bussey H, Davis R W, Dujon B, Feldmann H, Galibert F, Hoheisel J D, Jacq C, Johnston M, et al. Science. 1996;274:546–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 3.Bonhivers M, Carbrey J M, Gould S J, Agre P. J Biol Chem. 1998;273:27565–27572. doi: 10.1074/jbc.273.42.27565. [DOI] [PubMed] [Google Scholar]
- 4.Philips J, Herskowitz I. J Cell Biol. 1997;138:961–974. doi: 10.1083/jcb.138.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tamas M J, Luyten K, Sutherland F C, Hernandez A, Albertyn J, Valadi H, Li H, Prior B A, Kilian S G, Ramos J, et al. Mol Microbiol. 1999;31:1087–1104. doi: 10.1046/j.1365-2958.1999.01248.x. [DOI] [PubMed] [Google Scholar]
- 6.Laize V, Tacnet F, Ripoche P, Hohmann S. Yeast. 2000;16:897–903. doi: 10.1002/1097-0061(200007)16:10<897::AID-YEA583>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 7.Laize V, Gobin R, Rousselet G, Badier C, Hohmann S, Ripoche P, Tacnet F. Biochem Biophys Res Commun. 1999;257:139–144. doi: 10.1006/bbrc.1999.0425. [DOI] [PubMed] [Google Scholar]
- 8.Adams A, Gottschling D E, Kaiser C A, Stearns T. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. [Google Scholar]
- 9.Orr-Weaver T L, Szostak J W, Rothstein R J. Proc Natl Acad Sci USA. 1981;78:6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guldener U, Heck S, Fielder T, Beinhauer J, Hegemann J H. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 12.Preston G M, Carroll T P, Guggino W B, Agre P. Science. 1992;256:385–387. doi: 10.1126/science.256.5055.385. [DOI] [PubMed] [Google Scholar]
- 13.Zhang R B, Logee K A, Verkman A S. J Biol Chem. 1990;265:15375–15378. [PubMed] [Google Scholar]
- 14.Liu H, Styles C A, Fink G R. Genetics. 1996;144:967–978. doi: 10.1093/genetics/144.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hazen K C, Lay J G, Hazen B W, Fu R C, Murthy S. Infect Immun. 1990;58:3469–3476. doi: 10.1128/iai.58.11.3469-3476.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gozalbo D, Hohmann S. Mutat Res. 1989;215:89–94. doi: 10.1016/0027-5107(89)90221-2. [DOI] [PubMed] [Google Scholar]
- 17.Henderson S T, Petes T D. Mol Cell Biol. 1992;12:2749–2757. doi: 10.1128/mcb.12.6.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smit G, Straver M H, Lugtenberg B J, Kijne J W. Appl Environ Microbiol. 1992;58:3709–3714. doi: 10.1128/aem.58.11.3709-3714.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masy C L, Henquinet A, Mestdagh M M. Can J Microbiol. 1992;38:1298–1306. doi: 10.1139/m92-214. [DOI] [PubMed] [Google Scholar]
- 20.Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown P O, Herskowitz I. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- 21.Meyrial, V., Laizé, V., Gobin, R., Ripoche, P., Hohmann, S. & Tacnet, F. Eur. J. Biochem., in press. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.