Abstract
The T2 family of nonspecific endoribonucleases (EC 3.1.27.1) is a widespread family of RNases found in every organism examined thus far. Most T2 enzymes are secretory RNases and therefore are found extracellularly or in compartments of the endomembrane system that would minimize their contact with cellular RNA. Although the biological functions of various T2 RNases have been postulated on the basis of enzyme location or gene expression patterns, the cellular roles of these enzymes are generally unknown. In the present work, we characterized Rny1, the only T2 RNase in Saccharomyces cerevisiae. Rny1 was found to be an active, secreted RNase whose gene expression is controlled by heat shock and osmotic stress. Inactivation of RNY1 leads to unusually large cells that are temperature-sensitive for growth. These phenotypes can be complemented not only by RNY1 but also by both structurally related and unrelated secretory RNases. Additionally, the complementation depends on RNase activity. When coupled with a recent report on the effect of specific RNAs on membrane permeability [Khvorova, A., Kwak, Y-G., Tamkun, M., Majerfeld, I. & Yarus, M. (1999) Proc. Natl. Acad. Sci. USA 96, 10649–10654], our work suggests an unexpected role for Rny1 and possibly other secretory RNases. These enzymes may regulate membrane permeability or stability, a hypothesis that could present an alternative perspective for understanding their functions.
Secreted endoribonucleases are often used as model proteins because of their stability and availability in large quantities. Pancreatic RNase A, for example, was one of the first enzymes to be isolated, purified in crystalline form, and sequenced, thereby making RNase A one of the preferred enzymes for structural enzymology studies. More recently, secreted RNases from amphibians, as well as other RNases, have captured the attention of researchers because of their potential use as therapeutic agents. However, from a physiological perspective, the function of most secreted RNases remains an open question (reviewed in refs. 1–4).
The T2 RNases (EC 3.1.27.1), a family of endoribonucleases with no absolute base specificity, are found in all organisms so far examined (5). Although ubiquitous in nature, most are located where RNA is not thought to be readily available (e.g., outside the cell or in the vacuole). This idea has led researchers to propose biological functions for these enzymes other than the processing of cellular RNA. One role proposed for a number of plant T2 RNases, expressed specifically during leaf senescence or when phosphorus is limiting, is the scavenging of phosphate from ribonucleotides (reviewed in ref. 6). Other T2 RNases may have a protective function, as their expression is elevated in response to wounding or pathogen invasion (7–9). Some viral T2 RNases are thought to suppress function of the host immune system by acting as cytotoxins (10, 11). The most well-characterized members of this family of RNases, the self-incompatibility RNases (S-RNases), are associated with a genetic barrier that prevents inbreeding in three plant families (12). S-RNases are thought to prevent self-fertilization by acting as selective cytotoxins of “self” pollen.
The many functions proposed for T2 RNases contrast with the occurrence of these enzymes in a diverse range of organisms, which suggests T2 RNases could share a single, evolutionarily conserved function. As a step to elucidating this function, we characterized Rny1, the only member of the T2 RNase family present in Saccharomyces cerevisiae. We show that Rny1 is an active, secreted RNase and that RNY1 expression is rapidly regulated in response to certain environmental and stress conditions. Cells that lack Rny1 activity (rny1Δ) are larger than wild-type (WT) cells and have impaired growth at 37°C. The mutant phenotype of rny1Δ cells can be complemented by a number of secreted RNases, but not by an inactive version, indicating that a secreted RNase activity is important. We discuss these findings in the context of a hypothesis that T2 RNases may act as regulators of membrane permeability, a function that has not been considered previously for these enzymes.
Materials and Methods
Media, Growth, and Yeast Strains.
Standard growth media [rich medium, yeast extract/peptone/dextrose (YPD); minimal medium, synthetic dropout (SD)] and procedures for genetic manipulation were used (13, 14). S. cerevisiae strains used in this study are shown in Table 1. For heat shock and osmotic-stress experiments, liquid cultures were grown at 25°C to early logarithmic phase and diluted to an optical density at 600 nm of 0.1. Half the culture was incubated at 25°C, and the other half was shifted to 37°C or kept at 25°C but with the addition of NaCl to a final concentration of 0.3 M. Aliquots were harvested from each culture at the indicated time points.
Table 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| PSY316 | MATα trp1-Δ99 ade2-101 ura3-53 his3Δ200 leu2-3,-112 lys2 | 15 |
| SEY6211 | MATa ura3-52 leu2-3,-112 his3Δ200 trp1Δ901 ade2 suc2Δ9 | 16 |
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | 17 |
| YD2129 | MATa rny1Δ∷kanMX4 his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | 18 |
| PGY1 | MATα trp1-Δ99 ade2-101 ura3-53 his3Δ200 leu2-3,-112 lys2 rny1Δ∷URA3 | This study |
| PGY2 | MATa ura3-52 leu2-3,-112 his3Δ200 trp1Δ901 ade2 suc2Δ9 rny1Δ∷URA3 | This study |
| PGY3 | MATα trp1-Δ99 ade2-101 ura3-53 his3Δ200 leu2-3,-112 lys2/pG1 (TRP1) | This study |
| PGY4 | MATα trp1-Δ99 ade2-101 ura3-53 his3Δ200 leu2-3,-112 lys2/p2023 (TRP1 RNY1) | This study |
| PGY5 | MATα trp1-Δ99 ade2-101 ura3-53 his3Δ200 leu2-3,-112 lys2 rny1Δ∷URA3/pG1 (TRP1) | This study |
| PGY6 | MATα trp1-Δ99 ade2-101 ura3-53 his3Δ200 leu2-3,-112 lys2 rny1Δ∷URA3/YEpWL (TRP1 LEU2) | This study |
| PGY7 | MATα trp1-Δ99 ade2-101 ura3-53 his3Δ200 leu2-3,-112 lys2 rny1Δ∷URA3/p1270 (TRP1 LEU2 RNS1) | This study |
| PGY8 | MATα trp1-Δ99 ade2-101 ura3-53 his3Δ200 leu2-3,-112 lys2 rny1Δ∷URA3/p1130 (TRP1 LEU2 RNS2) | This study |
| PGY9 | MATα trp1-Δ99 ade2-101 ura3-53 his3Δ200 leu2-3,-112 lys2 rny1Δ∷URA3/p1258 (TRP1 LEU2 RNS3) | This study |
| PGY10 | MATα trp1-Δ99 ade2-101 ura3-53 his3Δ200 leu2-3,-112 lys2 rny1Δ∷URA3/YEpWL.RNaseA (TRP1 LEU2 RNaseA) | This study |
| PGY11 | MATα trp1-Δ99 ade2-101 ura3-53 his3Δ200 leu2-3,-112 lys2 rny1Δ∷URA3/p2023 (TRP1 RNY1) | This study |
| PGY12 | MATα trp1-Δ99 ade2-101 ura3-53 his3Δ200 leu2-3,-112 lys2 rny1Δ∷URA3/p2069 (TRP1 LEU2 H119D rnaseA) | This study |
Plasmids and Construction of rny1Δ Strains.
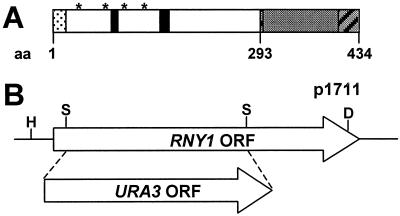
Strains lacking the Rny1 protein were produced by homologous recombination. A 1,748-bp genomic fragment corresponding to the RNY1 ORF and flanking regions was amplified by PCR. After cloning, the ORF was disrupted by replacing a 770-bp fragment with the URA3 gene from plasmid YcpLac33 (19) to make p1711. A 1,411-bp fragment of p1711 was used to transform yeast strains PSY316 and SEY6211 (Fig. 1B). Colonies were selected in SD-Ura and screened by PCR. Bona fide deletions of RNY1 (rny1Δ) were confirmed by Southern blotting. The RNY1 expression plasmid p2023 was constructed by inserting the RNY1 ORF (Research Genetics, Huntsville, AL) into the plasmid pG1 (20) between the glyceraldehyde-3-phosphate dehydrogenase promoter and the 3-phosphoglycerate kinase terminator. Plasmids p1270 (RNS1), p1130 (RNS2), and p1258 (RNS3) have been described (21, 22). Plasmid YEpWL.RNase A (23) was obtained from Ronald Raines (University of Wisconsin–Madison). Plasmid p2069 was created by introducing the mutation H119D in YEpWL.RNase A by using site-directed mutagenesis by combined chain reaction (24).
Figure 1.
RNY1 characterization. (A) Predicted Rny1 amino acid sequence reveals the presence of the two conserved sequences characteristic of the RNase T2 family of RNases (black boxes), four N-glycosylation sites (asterisk), a secretion signal (dotted line), and a C-terminal extension (gray box) containing two putative nuclear localization signals (diagonal bars). (B) RNY1 deletion strategy. Haploid yeast cells carrying a disrupted rny1 gene were produced by homologous recombination after introducing into WT cells a Hindlll–Dral fragment from plasmid p1711 that contains the selectable marker URA3 replacing part of the RNY1 ORF between the two Sful sites.
DNA, RNA, and Protein Analysis.
Southern and Northern blot analyses were performed according to standard techniques (14, 25). Extracellular proteins were obtained from the culture media of cells at late logarithmic growth phase. After pelleting the cells, the supernatants were concentrated 100-fold by using Ultrafree 15 filtration devices (Millipore). Concentrated extracts were centrifuged at 15,000 × g for 10 min, and the supernatants were analyzed for RNase activity. Intracellular extracts were obtained by using glass beads as described (26). Zymogram electrophoresis to analyze RNase activity was performed as described (27).
Deglycosylation of Extracellular Proteins.
Extracellular proteins (10 μg) were incubated at 37°C for the indicated time in 100 mM NH4HCO3 buffer (200 μl final volume) with or without the addition of 4 units of N-glycosidase F (PNGase F; Calbiochem). Samples were stored at −20°C and concentrated to 20 μl by using Microcon 10 concentrators (Millipore) before analysis of RNase activity.
Cell Microscopy.
Cells were visualized by using a differential interference contrast microscope. Staining (4′,6-diamidino-2-phenylindole) was performed as described (14).
Results
Characterization of Rny1.
From a search of the S. cerevisiae genome database, one gene (ORF YPL123C, GenBank protein accession no. AAB68239) with a significant match to genes from the T2 RNase family of RNases was identified. The YPL123C gene is predicted to encode a 48-kDa protein with an amino acid sequence that contains the two active-site motifs that are characteristic of the T2 RNase family (Fig. 1A). The sequence of YPL123C from amino acids 1–293 can be aligned to that of T2 RNase from Aspergillus oryzae, which is the archetypal enzyme in this family (5). Overall, the two sequences were 36% identical and 66% similar at the amino acid level. T2 RNase-related enzymes are targeted to the secretory pathway, and those expressed in eukaryotic cells usually have attached carbohydrates (5). The yeast gene encodes a protein with four potential glycosylation sites and a putative secretion signal of 18 amino acids at the N terminus. In addition to the region of T2 RNase-related sequence, the last 140 amino acids () of the YPL123C protein form a C-terminal extension that is not found in other T2 RNase family members and is not related to the sequence of any other protein. Analysis of this region by using psort ii (28) indicated the presence of two putative nuclear localization signals. Based on our analysis, the putative YPL123C protein represents a new member of the T2 RNase family and the only member of this family in yeast. We renamed this RNase gene RNY1.
To analyze Rny1 function, a strain carrying an insertional knockout of the RNY1 ORF was constructed by homologous recombination by using URA3 as a selective marker (Fig. 1B). Gene replacement was confirmed by Southern blot analysis (data not shown). Initially, RNase activity in the intracellular or extracellular protein fractions of both the WT and rny1Δ strains was examined by using a gel-based assay. However, none of the secreted proteins produced by either strain exhibited sufficient RNase activity to be detected in this assay. Only one RNase, with an apparent molecular mass of about 70 kDa, was detected among the intracellular proteins (Fig. 2A). This activity is not encoded by RNY1, as it was present in intracellular extracts from rny1Δ cells.
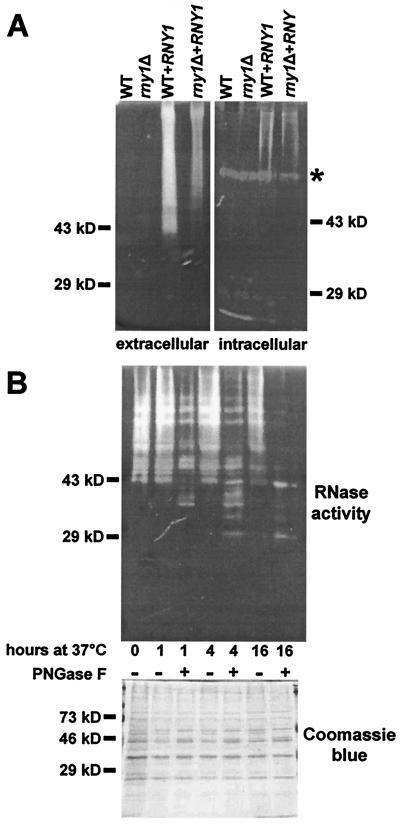
Figure 2.
Rny1 is an active RNase and is glycosylated highly. (A) Secreted proteins recovered from the growth media (12 μg per lane) or intracellular proteins (40 μg per lane) were analyzed by using a gel-based RNase assay. Proteins were prepared from WT and rny1Δ cells and from WT and rny1Δ cells transformed with the plasmid p2023 that express the RNY1 gene constitutively. The asterisk indicates the main intracellular activity (≈70 kDa). (B) Secreted proteins from an RNY1 overexpressing cell culture (10 μg) were incubated at 37°C for the indicated period of time with or without the addition of 4 units of PNGase F and then analyzed by using a gel-based RNase assay or by SDS/PAGE and Coomassie blue staining.
To show that Rny1 is an active RNase, WT and rny1Δ cells were transformed with a plasmid that contains the RNY1 gene under the control of the constitutive glyceraldehyde-3-phosphate dehydrogenase promoter. Protein extracts from cells overexpressing RNY1 had increased levels of an RNase that did not migrate as a single band on activity gels (Fig. 2A). The new RNase activity was seen as a smear in the secreted protein fraction that began at the top of the lane and ended at a point roughly level with the 43-kDa marker (Fig. 2A). A lower level of activity was also detected in the intracellular protein fraction. This activity may be a result of contamination of the intracellular fraction by secreted proteins. The RNase activity was only seen in cells transformed with the RNY1 construct and was not seen in cells transformed with either the empty vector or with other RNase genes (data not shown).
One reason for Rny1 to migrate as a heterodispersed protein in activity gels could be the presence of posttranslational modifications, such as N-glycosylation. The presence of N-glycans on Rny1 was tested by incubating the secreted proteins from WT cells overexpressing RNY1 in the presence or absence of PNGase F. After a 16-h treatment with PNGase F, the smear of RNase activity was resolved into two main bands. One band, with a mobility of ≈43 kDa, likely corresponds to the mature Rny1 polypeptide. The other band, with a mobility of ≈29 kDa, may correspond to the N-terminal “T2 RNase-related” domain of Rny1 (Fig. 2B, Upper). It is not known whether the proposed cleavage of the C-terminal extension occurred during extraction or treatment of Rny1 with PNGase F or whether it normally occurs during Rny1 secretion. A general postextraction proteolysis is unlikely, as a Coomassie blue-stained gel indicates that abundant proteins in the sample are not degraded by treatment with PNGase F (Fig. 2B, Lower). However, it is possible that deglycosylated Rny1 is more sensitive to proteolysis than other proteins in the extract. In any event, the overexpression studies show that Rny1 is an active, glycosylated RNase that is predominantly secreted into the extracellular space.
RNY1 Expression Is Regulated by Several Stress Conditions.
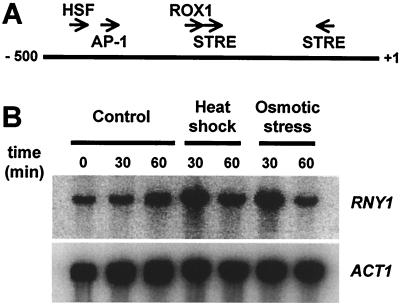
Although Rny1 is an active RNase, it cannot be detected in WT cells grown at 25°C in SD medium. This observation could indicate that the RNY1 gene is either not expressed under these conditions or is expressed at very low levels. Analysis of the RNY1 promoter showed the presence of a number of sequence elements that are recognized by stress-related transcription factors (Fig. 3A). These include elements regulated by heat shock factor (HSF; ref. 29), oxidative stress (AP-1; ref. 30), hypoxia (ROX1; ref. 31), and two stress response elements that control transcription in response to a number of stress conditions (32). These elements may indicate that RNY1 is regulated during stress conditions. This possibility was explored by Northern blot analysis of RNY1 transcript levels in WT cells after exposure to either heat shock or osmotic stress (Fig. 3B). Both treatments resulted in rapid increases in the RNY1 transcript. A 5.3-fold increase in RNY1 levels was seen 30 min after the cells were shifted from 25° to 37°C, and a 6.5-fold increase occurred after the cells were cultured in 0.3 M NaCl for 30 min. These increases compare with the 4.3-fold increase in RNY1 transcript levels observed in response to DNA damage by methyl methanesulfonate in DNA microarray analysis (33). The response to heat and osmotic shock was transient: RNY1 transcript returned to levels approaching that in untreated cells within 60 min of application of the stress conditions (Fig. 3B).
Figure 3.
(A) The RNY1 promoter contains several putative stress-regulated elements. Arrows indicate the position of the different binding elements. HSF, heat shock factor; AP-1, yeast AP-1; ROX1, Rox1-binding site; STRE, stress response element. The numbers indicate the distance from the ATG codon (+1). (B) RNY1 expression is regulated by different stress conditions. WT cells were grown to early exponential phase at 25°C and diluted to an optical density at 600 nm of 0.1. Half the culture was incubated at 25°C, and the other half was shifted to 37°C or kept at 25°C but with the addition of NaCl to a final concentration of 0.3 M. Aliquots (2 ml) were harvested at the indicated time points and total RNA was analyzed with Northern blot by using RNY1 or ACT1 probes.
Phenotype of rny1Δ Strains.
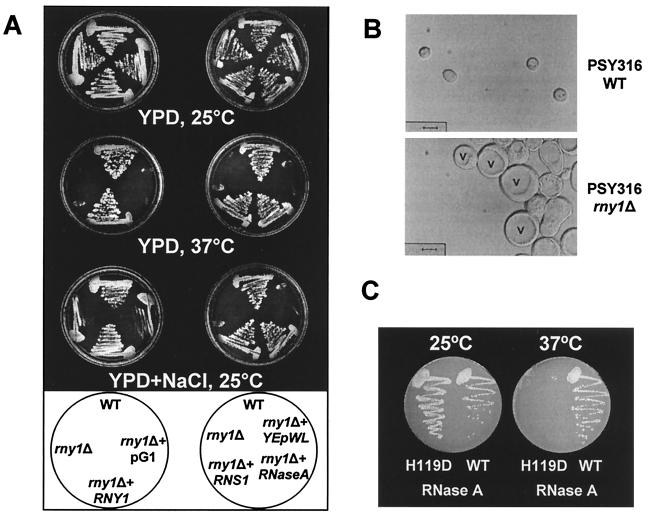
Cells of yeast strain PSY316 that had a portion of the RNY1 ORF deleted were able to grow at 25°C in YPD (Fig. 4A); therefore, Rny1 is not essential under normal growth conditions. PSY316 tends to flocculate when grown in liquid culture under suboptimal conditions, and this tendency was markedly higher in rny1Δ cells, which grew as aggregates even under optimal conditions. Differential interference contrast microscopy showed that rny1Δ cells were larger and less uniformly round than WT cells (Fig. 4B). The average diameter of WT cells was 4.2 ± 0.3 μm and that of rny1Δ cells was 11.1 ± 1.1 μm. rny1Δ cells had larger vacuoles, and 4′,6-diamidino-2-phenylindole staining showed no increase in the size or number of nuclei, which were morphologically similar to those of WT cells (data not shown). The larger cell size and bigger vacuole are features of cells that are osmotically sensitive (see Discussion).
Figure 4.
Phenotype of rny1Δ strain in the PSY316 background. (A) Cells (PSY316 background) were grown in YPD plates or YPD plus 50 g/liter NaCl plates at 25° or 37°C, as indicated. For complementation experiments, rny1Δ cells were transformed with plasmid containing RNY1, the A. thaliana RNS1 gene, the mammalian gene for pancreatic RNase A, or empty vector (pG1 or YEpWL). (B) WT or rny1Δ cells (PSY316 background) were grown to exponential phase in SD medium at 25°C. Cells were viewed by using differential interference contrast microscopy. v, vacuole. (Bar = 5 μm.) (C) rny1Δ cells were transformed with plasmid containing the mammalian gene for pancreatic RNase A (WT) or the RNase A mutant H119D(H119D) and grown in SD plates at 25° or 37°C, as indicated.
As heat shock and osmotic stress regulate RNY1 expression, we tested the ability of rny1Δ cells to grow under these conditions. The mutant strain did not grow when it was incubated at 37°C and grew slower than WT when plated on YPD plus 50 g/liter NaCl (Fig. 4A). This observation indicated that rny1Δ cells were temperature sensitive and osmosensitive. Both phenotypes were complemented by transforming the rny1Δ strain with the WT RNY1 gene under the control of the constitutive promoter GDPp (Fig. 4A). No complementation was seen with the empty vector pG1. Deleting RNY1 in two other yeast strains, SEY6211 and BY4741, did not result in either a temperature-sensitive or osmosensitive phenotype (data not shown). The dependence of the rny1Δ phenotype on genetic background indicates that other factors may compensate for the lack of Rny1 function, and that at least one of these factors is missing in strain PSY316.
Several RNases Can Complement the rny1Δ Phenotype.
We tested the ability of other RNases to complement the rny1Δ phenotype to determine whether the phenotype was caused by a lack of Rny1 RNase activity or a lack of another feature of the protein, for example its C-terminal extension. Three different RNase genes from the plant Arabidopsis thaliana were used in these experiments. These genes, RNS1, RNS2, and RNS3, were chosen because they are also members of the T2 RNase family (34), and their expression is regulated in response to various developmental cues and stresses (21, 22, 35).
Because overexpressed Rny1 seemed to be secreted into the extracellular space, we targeted the other RNases to this compartment as well by fusing the DNA sequence for the α-factor signal peptide to the RNS1 and RNS3 coding regions. The RNS2 coding region was unmodified; secretion was achieved by using its own signal peptide. These constructs were transformed into rny1Δ cells. In each case, correct expression and extracellular localization was confirmed by analyzing extracellular and intracellular protein extracts with RNase activity gels (data not shown).
The ability of rny1Δ cells transformed with RNS1, RNS2, or RNS3 to grow under stress conditions was tested. Each gene was able to restore the ability of the cells to grow at 37°C and under osmotic stress (Fig. 4A and data not shown), indicating that the catalytic activity of Rny1 probably plays an important role in the response of yeast cells to these stress conditions. However, as the three plant RNases belong to the same family as Rny1, it was also possible that some structural feature shared by all four enzymes was responsible for the complementation we observed.
To test this possibility, we expressed an unrelated RNase in rny1Δ cells. The enzyme chosen was bovine pancreatic RNase A, also a secreted RNase, but unrelated in sequence to T2 RNase and belonging to another family (4). The α-factor signal peptide was used again in this experiment. Transformed rny1Δ cells expressing RNase A also were able to grow at 37°C and on high concentrations of NaCl (Fig. 4 A and C). The empty vector (YEpWL) did not complement the mutant phenotype (Fig. 4A). To demonstrate further that the activity, not a structural feature of the RNase, was necessary for complementation, a mutant version of RNase A, H119D, was also tested. It has been shown that residue H119 is critical for activity within the RNase A family (36) and that this specific mutation (H119D) preserves the conformational stability of the protein at pH 6.0 (37). As shown in Fig. 4C, the inactive RNase A mutant was unable to complement the temperature-sensitive phenotype, confirming that yeast cells require RNase activity to grow under these stress conditions. Activity (or lack thereof) for WT and mutant proteins was confirmed by zymogram and evidence of expression was obtained by Western blot analyses (data not shown).
Discussion
This report advances understanding of the biology of T2 RNases by characterizing the only member of this RNase family in S. cerevisiae, RNY1. As discussed below, some yeast cells that do not produce Rny1 have a phenotype indicative of a change of either the permeability or stability of membranes. This finding, coupled with the regulation of RNY1 by a number of environmental stresses, suggests that the yeast T2 RNase, and perhaps secreted RNases in other organisms, may be involved in maintaining cellular homeostasis in entirely unexpected ways.
RNY1 encodes an active RNase with a signal sequence that, when overexpressed, was found primarily outside the cell. The enzyme is glycosylated and may be processed proteolytically. We could not identify Rny1 activity in WT cells on our RNase activity gels, presumably because its activity was below detection. The only RNase activity evident was an intracellular protein of about 70 kDa that may correspond to an enzyme characterized previously, RNase YI* (38). This activity was not encoded by RNY1.
We found that endogenous RNY1 transcript levels were low under normal growth conditions and increased in response to multiple stresses. Interestingly, a recent genome-wide expression analysis found that yeast lacking the cyclin-dependent kinase Srb10 had 5-fold higher levels of RNY1 transcripts than WT cells (39). Srb10 is part of a complex that can repress the expression of genes involved in cell-type specificity, meiosis, sugar utilization, diauxic shift, foraging morphology, and stress responses (39, 40).
The lack of Rny1 in the PSY316 rny1Δ strain causes changes to cell size and shape and to vacuolar morphology. Rny1 is also necessary for cell growth at high salt concentrations or at high temperatures. Similar changes in vacuolar morphology also are found in cells with a deletion of FAB1. In these cells, however, sensitivity to elevated temperatures can be suppressed partially by including high amounts of KCl or sorbitol in the medium (41). Fab1 is a kinase that converts phosphatidylinositol 3-phosphate to phosphatidylinositol 3,5-bisphosphate (42) and is likely to be activated by hyperosmotic stress (43). This enzyme has been implicated in the regulation of vacuolar homeostasis, perhaps by controlling the efflux or turnover of vacuolar membranes (41, 42). A similar role for Rny1 is unlikely because this enzyme is primarily secreted outside the cell (see below). This suggests that any changes in vacuolar morphology that were seen in rny1Δ cells were an indirect consequence of this mutation.
That other RNases can complement the growth deficiencies of rny1Δ cells suggests it is the lack of RNase activity, rather than structural properties of Rny1, that are responsible for the observed phenotype. This idea is supported by the ability of WT RNase A, but not the H119D mutant, to complement the rny1Δ phenotype, even though this enzyme belongs to a family of endoribonucleases that are unrelated in sequence to the T2 RNases.
Our observations raise the question how the mutant phenotype is connected to the absence of Rny1 from the cell. A possible role for Rny1 in Pi or nucleotide recycling is unlikely because RNY1 expression increases in response to stress conditions that do not involve nutrient limitation. Furthermore, our preliminary data show that RNY1 expression is not higher under Pi-limiting conditions (G.C.M. and P.J.G., unpublished results).
Although some Rny1 was present in cytoplasmic extracts of yeast that overexpress RNY1, this activity, like the extracellular activity, appeared on RNase gels as a smear. Treatment with PNGase F resolved the extracellular smear into two bands, indicating the presence on Rny1 of endoplasmic reticulum (ER)-attached glycans. Thus, the smear seen in the cytoplasmic extracts was probably the result of contamination by Rny1 that had passed through the ER and had been targeted either outside the cell or to the vacuole. Therefore, we can also rule out a role for Rny1 in degradation of cytoplasmic RNA during stress responses.
Consequently, we hypothesize that Rny1 has an extracellular role. The cell wall is an important osmotic barrier and also plays a key role in morphogenesis and cell–cell recognition (44). Possibly an effect of the RNase, such as modifying or loosening the cell wall through an as-yet-unknown mechanism, could explain some of the observed phenotypes. However, a more appealing connection between RNY1 and its mutant phenotype is suggested by the recent finding that specific RNAs can bind to biological membranes and affect their permeability to ions (45). This effect is postulated to be one of the ancient functions of RNA in the hypothetical prebiotic “RNA world.” If some RNAs still carry out this ancient function, then secreted RNases could have an important role in controlling membrane permeability or stability. Accordingly, RNY1 expression would be expected to increase in response to changing environmental conditions, such as osmotic stress and heat shock, that have important membrane-related implications. In this scenario, specific RNAs would maintain a given degree of membrane permeability under normal growth conditions. In the presence of heat shock or osmotic stress, membrane permeability or stability must change to maintain cell homeostasis. Yeast may regulate membrane permeability or stability during stress conditions by secreting an RNase to degrade membrane-bound RNAs. As a consequence, yeast cells lacking RNY1 would be osmosensitive, because they cannot secrete such RNases. Because other systems (such as ion channels and transporters) also contribute to maintaining cell homeostasis at the level of membrane permeability, one explanation for the alternative effects of genetic background on the rny1Δ phenotype is that many strains have systems that compensate for the loss of Rny1 function. In strain PSY316, for some reason, these systems are unable to compensate for the loss of Rny1. Other observations also point to the presence of mutations in strain PSY316 that increase its sensitivity to a lack of Rny1. For example, PSY316 cells have a tendency to aggregate when grown under suboptimal conditions. In rny1Δ cells, this flocculation phenotype is seen even under normal growth conditions. Flocculation and cell aggregation have been correlated with mutations in cell-wall synthesis genes and in regulatory genes that affect cell-wall integrity directly or indirectly (46) and are also associated with specific defects in osmotic stability (47, 48).
Although extracellular RNases are not usually thought to have a role in modifying membrane permeability, this suggestion has been made previously. Addition of RNase Bi, a secreted RNase from Bacillus intermedius and a member of the T1 RNase family, to a culture of Candida utilis affected the length of the cell cycle by shortening G1 (49). These RNase-induced changes in DNA replication and cell division are thought to be secondary effects resulting from modifications in the permeability of cell membranes (49). RNase Bi induces cell differentiation when added to human blood lymphocytes (50) and the SOS response when added to cultures of Escherichia coli (51). In each case, only catalytically active RNase Bi was effective, and membrane-bound RNAs have been proposed to be one potential substrate for this enzyme (49–51). Although there is currently no direct proof that this type of RNA exists in vivo, the results of our study provide molecular genetic evidence for a role of RNases as regulators of membrane permeability in vivo.
RNases are essential to a variety of biological processes for reasons that are not yet understood fully. If other members of the T2 RNase family, and possibly members of other families of secreted RNases as well, can affect membrane permeability and/or stability, then a number of RNase-requiring phenomena, such as cytotoxicity, can be considered from a new perspective. The hypothesis put forward here that RNases can act as modifiers of biological membranes could therefore have implications for understanding the mechanism through which other extracellular RNases act.
Acknowledgments
We thank J. Golz (formerly of the University of Melbourne) for help with constructing the deletion strains, T. Lithgow (University of Melbourne) for comments on the manuscript, P. Liu and D. Thielle (University of Michigan) for helpful discussions and the gifts of strain SEY6211 and the ACT1 probe, A. Sanderfoot (Michigan State University) for help with microscopy, S. Triezenberg (Michigan State University) for strain PSY316, and R. Raines (University of Wisconsin–Madison) for plasmid YEpWL.RNaseA and antibodies. We also thank L. Danhof for technical assistance and N. LeBrasseur for editorial assistance. This work was funded by National Science Foundation Grant IBN9408052; Department of Energy Grant FG0291-ER200210 (to P.J.G.); and a Department of Industry, Science, and Resources bilateral grant from the Australian government (to E.N.).
Abbreviation
- PNGase F
N-glycosidase F
- WT
wild type
- YPD
yeast extract/peptone/dextrose
- SD
synthetic dropout
References
- 1.Irie M, Nitta K, Nonaka T. Cell Mol Life Sci. 1998;54:775–784. doi: 10.1007/s000180050206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benner S A, Ciglic M I, Haugg M, Jermann T M, Opitz J G, Raillard-Yoon S-A, Soucek J, Stackhouse J, Trabesinger-Rüf N, Trautwein K, et al. In: Ribonucleases: Structures and Functions. D'Alessio G, Riordan J S, editors. New York: Academic; 1997. pp. 213–243. [Google Scholar]
- 3.Cuchillo C M, Vilanova M, Nogués V. In: Ribonucleases: Structures and Functions. D'Alessio G, Riordan J S, editors. New York: Academic; 1997. pp. 271–304. [Google Scholar]
- 4.Beintema J J. Cell Mol Life Sci. 1998;54:763–765. doi: 10.1007/s000180050204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Irie M. In: Ribonucleases: Structures and Functions. D'Alessio G, Riordan J S, editors. New York: Academic; 1997. pp. 101–130. [Google Scholar]
- 6.Bariola P A, Green P J. In: Ribonucleases: Structures and Functions. D'Alessio G, Riordan J S, editors. New York: Academic; 1997. pp. 163–190. [Google Scholar]
- 7.Ye Z-H, Droste D L. Plant Mol Biol. 1996;30:697–709. doi: 10.1007/BF00019005. [DOI] [PubMed] [Google Scholar]
- 8.Galiana E, Bonnet P, Conrod S, Keller H, Panabières F, Ponchet M, Poupet A, Ricci P. Plant Physiol. 1997;115:1557–1567. doi: 10.1104/pp.115.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lers A, Khalchitski A, Lomaniec E, Burd S, Green P J. Plant Mol Biol. 1998;36:439–449. doi: 10.1023/a:1005993024161. [DOI] [PubMed] [Google Scholar]
- 10.Schneider R, Unger G, Stark R, Schneider-Scherzer E, Thiel H-J. Science. 1993;261:1169–1171. doi: 10.1126/science.8356450. [DOI] [PubMed] [Google Scholar]
- 11.Hulst M M, Himes G, Newbigin E, Moormann R J. Virology. 1994;200:558–565. doi: 10.1006/viro.1994.1218. [DOI] [PubMed] [Google Scholar]
- 12.Golz J F, Clarke A E, Newbigin E. Curr Opin Genet Dev. 1995;5:640–645. doi: 10.1016/0959-437x(95)80033-6. [DOI] [PubMed] [Google Scholar]
- 13.Sherman F. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 14.Rose M D, Winston F, Hieter P. Methods in Yeast Genetics. A Laboratory Course Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1990. [Google Scholar]
- 15.Berger S L, Piña B, Silverman N, Marcus G A, Agapite J, Regier J F, Triezenberg S J, Guarente L. Cell. 1992;70:251–265. doi: 10.1016/0092-8674(92)90100-q. [DOI] [PubMed] [Google Scholar]
- 16.Robinson J S, Klionsky D J, Banta L M, Emr S D. Mol Cell Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brachmann C B, Davies A, Cost G J, Caputo E, Li J, Hieter P, Boeke J D. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 18.Winzeler E A, Shoemaker D D, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke J D, Bussey H, et al. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 19.Gietz R D, Sugino A. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 20.Schena M, Picard D, Yamamoto K R. Methods Enzymol. 1991;194:389–398. doi: 10.1016/0076-6879(91)94029-c. [DOI] [PubMed] [Google Scholar]
- 21.Taylor C B, Bariola P A, DelCardayré S B, Raines R T, Green P J. Proc Natl Acad Sci USA. 1993;90:5118–5122. doi: 10.1073/pnas.90.11.5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bariola P A, Howard C J, Taylor C B, Verburg M T, Jaglan V D, Green P J. Plant J. 1994;6:673–685. doi: 10.1046/j.1365-313x.1994.6050673.x. [DOI] [PubMed] [Google Scholar]
- 23.delCardayré S B, Ribó M, Yokel E M, Quirk D J, Rutter W J, Raines R T. Protein Eng. 1995;8:261–273. doi: 10.1093/protein/8.3.261. [DOI] [PubMed] [Google Scholar]
- 24.Bi W, Stambrook P J. Anal Biochem. 1998;256:137–140. doi: 10.1006/abio.1997.2516. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 26.Jazwinski S M. Methods Enzymol. 1990;182:154–174. doi: 10.1016/0076-6879(90)82015-t. [DOI] [PubMed] [Google Scholar]
- 27.Yen Y, Green P J. Plant Physiol. 1991;97:1487–1493. doi: 10.1104/pp.97.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakai K, Horton P. Trends Biochem Sci. 1999;24:34–35. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- 29.Mager W H, De Kruijff A J J. Microbiol Rev. 1995;59:506–531. doi: 10.1128/mr.59.3.506-531.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toone W M, Jones N. Curr Opin Genet Dev. 1999;9:55–61. doi: 10.1016/s0959-437x(99)80008-2. [DOI] [PubMed] [Google Scholar]
- 31.Deckert J, Rodriguez Torres A M, Hwang S M, Kastaniotis A J, Zitomer R S. Genetics. 1998;150:1429–1441. doi: 10.1093/genetics/150.4.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morano K A, Liu P C C, Thiele D J. Curr Opin Microbiol. 1998;1:197–203. doi: 10.1016/s1369-5274(98)80011-8. [DOI] [PubMed] [Google Scholar]
- 33.Jelinsky S A, Samson L D. Proc Natl Acad Sci USA. 1999;96:1486–1491. doi: 10.1073/pnas.96.4.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor C B, Green P J. Plant Physiol. 1991;96:980–984. doi: 10.1104/pp.96.3.980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bariola P A, MacIntosh G C, Green P J. Plant Physiol. 1999;119:331–342. doi: 10.1104/pp.119.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim J-S, Souèek J, Matoušek J, Raines R T. Biochem J. 1995;308:547–550. doi: 10.1042/bj3080547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quirk D J, Park C, Thompson J E, Raines R T. Biochemistry. 1998;37:17958–17964. doi: 10.1021/bi981688j. [DOI] [PubMed] [Google Scholar]
- 38.Cannistraro V J, Kennell D. Nucleic Acids Res. 1997;25:1405–1411. doi: 10.1093/nar/25.7.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holstege F C P, Jennings E G, Wyrick J J, Lee T I, Hengartner C J, Green M R, Golub T R, Lander E S, Young R A. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 40.Hengartner C J, Myer V E, Liao S M, Wilson C J, Koh S S, Young R A. Mol Cell. 1998;2:43–53. doi: 10.1016/s1097-2765(00)80112-4. [DOI] [PubMed] [Google Scholar]
- 41.Yamamoto A, DeWald D B, Boronenkov I V, Anderson R A, Emr S D, Koshland D. Mol Biol Cell. 1995;6:525–539. doi: 10.1091/mbc.6.5.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gary J D, Wurmser A E, Bonangelino C J, Weisman L S, Emr S D. J Cell Biol. 1998;143:65–79. doi: 10.1083/jcb.143.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cooke F T, Dove S K, McEwen R K, Painter G, Holmes A W, Hall M N, Michell R H, Parker P J. Curr Biol. 1998;8:1219–1222. doi: 10.1016/s0960-9822(07)00513-1. [DOI] [PubMed] [Google Scholar]
- 44.Smits G J, Kapteyn J C, van den Ende H, Klis F M. Curr Opin Microbiol. 1999;2:348–352. doi: 10.1016/s1369-5274(99)80061-7. [DOI] [PubMed] [Google Scholar]
- 45.Khvorova A, Kwak Y-G, Tamkun M, Majerfeld I, Yarus M. Proc Natl Acad Sci USA. 1999;96:10649–10654. doi: 10.1073/pnas.96.19.10649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teunissen A W R H, Steensma H Y. Yeast. 1995;11:1001–1013. doi: 10.1002/yea.320111102. [DOI] [PubMed] [Google Scholar]
- 47.Levin D E, Bartlett-Heubusch E. J Cell Biol. 1992;116:1221–1229. doi: 10.1083/jcb.116.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang N, Gardner D C J, Oliver S G, Stateva L I. Microbiology. 1999;145:309–316. doi: 10.1099/13500872-145-2-309. [DOI] [PubMed] [Google Scholar]
- 49.Kupriyanova-Ashina F G, Kolpakov A I. Mikrobiologiya. 1999;68:155–159. [PubMed] [Google Scholar]
- 50.Kurinenko B M, Bulgakova R Sh, Davydov R E. FEMS Immunol Med Microbiol. 1998;21:117–122. doi: 10.1111/j.1574-695X.1998.tb01156.x. [DOI] [PubMed] [Google Scholar]
- 51.Ivanchenko O B, Il'inskaya O N, Karamova N S, Kipenskaya L V, Leshchinskaya I B. Mikrobiologiya. 1997;66:444–448. [PubMed] [Google Scholar]