Abstract
Mammals possess multiple, closely linked β-globin genes that differ in the timing of their expression during development. These genes have been thought to be derived from a single ancestral gene, by duplication events that occurred after the separation of the mammals and birds. We report the isolation and characterization of an atypical β-like globin gene (ω-globin) in marsupials that appears to be more closely related to avian β-globin genes than to other mammalian β-globin genes, including those previously identified in marsupials. Phylogenetic analyses indicate that ω-globin evolved from an ancient gene duplication event that occurred before the divergence of mammals and birds. Furthermore, we show that ω-globin is unlinked to the previously characterized β-globin gene cluster of marsupials, making this the first report of an orphaned β-like globin gene expressed in a vertebrate.
The globin genes and their homologues have been studied in a wide range of species including bacteria, plants, and animals; as a consequence there exists a wealth of information on the structure, expression, and function of these genes (1). The availability of such an extensive data set, particularly that involving DNA sequences, has enabled the evolutionary history of the globin genes to be reconstructed to a level of detail that is probably unsurpassed by any other gene family.
As originally postulated by Ingram (2) on the basis of partial amino acid sequences from several globin chains, it is now clear that primordial globin genes underwent a series of gene duplication events that led, in vertebrates at least, to a number of coordinately expressed genes that make up the present myoglobin, α-globin, and β-globin gene families (1). These duplication events have enabled specialized forms of globin to evolve that are physiologically adapted to the oxygen transport requirements in different tissues, at different developmental stages.
We have been interested in exploiting the ancient evolutionary separation between monotremes, marsupials, and eutherian mammals (3) to determine the timing and nature of gene duplications that occurred in the β-globin gene family. It has become clear from our previous work on the dunnart (Sminthopsis crassicaudata; refs. 4–6), and from that reported by Koop and Goodman on the North American opossum (Didelphis virginiana; ref. 7), that the first duplication event that gave rise to the early- and late-expressed globin genes in mammals (as typified by the embryonically expressed ɛ-globin genes and the late-expressed β-globin genes) occurred before the separation of marsupials and eutherians, approximately 120 million years ago. In contrast to the eutherian mammals that have a complex β-globin locus consisting, in humans for example, of five expressed genes and a pseudogene, marsupials, as exemplified by the dunnart and opossum, appeared to have a relatively simple β-globin gene cluster consisting of only two genes: ɛ-globin, expressed in embryos and newborn pouch young, and β-globin, expressed soon after birth and into adult life (4, 5, 7). Both marsupials and eutherians appear to have a locus control region (LCR) upstream of the β-globin locus (1). In newborn pouch young of another marsupial species, Macropus eugenii (tammar wallaby, hereafter referred to as the tammar), ɛ-globin is expressed in three hemoglobin proteins, one with adult α-globin and two with separate ζ-like globin chains (8).
Recent studies in our laboratories of blood from neonatal tammars uncovered a form of hemoglobin containing a β-like globin chain (ω-globin) whose partial amino acid sequence more closely resembled that of avian globins than of any other mammalian globins, including those previously characterized in marsupials and monotremes (8, 9). At no stage of pre- or postnatal development did this form of hemoglobin constitute more than 25% of the total hemoglobin present. These observations were of particular interest because we found previously that the embryonic blood in marsupials has unusual respiratory properties when compared with the embryonic blood of eutherian mammals (10–12). These properties include values of nH (Hill coefficient, or index of cooperativity) greater than 4, indicating that the functional unit is larger than a tetramer, and a lower affinity for oxygen. In the tammar and another marsupial, the brushtail possum (Trichosurus vulpecula), the oxygen equilibrium curve (OEC) of the embryonic-type blood is actually to the right of the OEC for the maternal blood (11), this unusual position being unfavorable to the transfer of oxygen from the mother to the embryo. A further interesting feature of gas exchange in marsupials is provided by the recent finding that in a newborn marsupial of the dasyurid family, Sminthopsis douglasi, 90% of the neonatal gas exchange occurs through the skin rather than the lungs (13).
The aim of the research documented in this paper was to characterize the ω-globin gene of marsupials and to investigate the phylogenetic relationship of ω-globin with other members of the β-globin gene family. We report here that the ω-globin gene is widespread, if not universal, in marsupials and is transcriptionally active in marsupial neonates. Furthermore, we show that the ω-globin gene lineage evolved from a gene duplication event that probably predated the separation of birds and mammals. Of particular interest is our finding that the ω-globin gene is unlinked to the main β-globin gene cluster in at least two marsupials species.
Methods
PCR primer design was based on partial amino acid sequences and on regions of high sequence identity revealed by sequence alignments of a range of β-like globin genes. PCR reactions were carried out in a final volume of 50 μl of PCR buffer (GeneWorks, Adelaide, Australia) which contained 100 ng template DNA, 0.4 μM of each primer, 0.2 mM dNTP, and 2 mM MgCl2. PCR was used to amplify a partial ω-globin product from the tammar genomic DNA (gDNA) (adult male F87ABE) with the primers RogF and CDR3 (see later for primer sequences). The PCR product obtained was cloned into pGEM-T vector (Promega). The missing 5′ and 3′ regions of the ω-globin gene were obtained by using Inverse PCR (14), performed with self-ligated EcoRI fragments of gDNA. Automated sequencing was carried out by using a Prism Dye terminator kit (Applied Biosystems) and a 377 DNA sequencer. Primers, drsf and G290, were used to PCR-amplify and sequence partial ω-globin sequences from S. crassicaudata (dunnart) gDNA. The 3′ region of the dunnart ω-globin gene was PCR-amplified by using the primers SCUTR and G314. The 5′ region of the gene was obtained by using the following technique. A linker primer PstA was annealed to PstB and ligated to PstI digested dunnart gDNA. Fragments in the size range 3- to 5-kb were purified from an agarose gel and used to seed a PCR amplification (55°C annealing) with the primers G291 and PstA, followed by a second amplification using a 4:1 ratio of primers G290 (1.0 μM) to PstA (0.25 μM). An approximately 400-bp fragment was amplified and sequenced. PCR was used to amplify and sequence partial ω-globin sequences from gDNA of Dasyurus viverrinus (native cat), Macropus giganteus (gray kangaroo), Trichosurus vulpecula (brushtail possum), Perameles gunnii (bandicoot), and Monodelphis domestica (South American opossum). The following primer combinations were used for each species (primer identification shown in parentheses): native cat (G287/G291), gray kangaroo (drsf/IR1), brushtail possum (drsf/IR1), bandicoot (G287/IR1), and South American opossum (G287/G290). Reverse transcription (RT)–PCR was carried out by using RNA isolated from one-day-old marsupial neonates as previously described (5) and the Superscript II preamplification system (GIBCO) with the primers G287 and G291.
Phylogenetic analyses were carried out by using PAUP* Version 4b2 (15) and a heuristic search option. Analysis of the partial marsupial ω-globin and dunnart and native cat ɛ- and β-globin sequences (coding regions only) used maximum parsimony and a branch and bound search option treating all characters as equally weighted. The Xenopus laevis β-globin gene (GenBank accession no. J00978) was used as an outgroup to root the tree (6). The reliability of phylogenetic groupings was tested by using bootstrap analysis based on 1,000 replications (16). Maximum likelihood (ML) and distance approaches used the HKY-85 model (17) with an ML-estimated transition/transversion ratio of 0.784 and discrete gamma rate variation with an estimated shape parameter of 0.6423. ML trees were generated by using a heuristic search option and distance trees were generated by using the Neighbor Joining algorithm in PAUP*. Dot plot analyses were carried out by using the computer programs compare and dotplot (Genetics Computer Group, Madison, WI; window size = 21; stringency = 12) available on-line from the Australian National Genomic Information Service (http://www.angis.org.au/).
Genomic Southern analysis was performed by using standard techniques (18). R-banded chromosomes were prepared from 3-day cultures of peripheral blood lymphocytes from an adult tammar wallaby. Cultures were treated with BrdUrd (20 mg/ml) for 7.5 h and colchicine for 45 min. Radioactive in situ hybridization was carried out as previously described (19). Slides were exposed to 1:1 diluted L4 emulsion (Ilford) for 27–33 days.
Primer sequences were as follows (reading from 5′ to 3′):
| RofF: | ATGGTNCATTGGCANGAGGARAA |
| CDR3: | ATGGTNCATTGGCANGAGGARAA |
| drsf: | GGAGAAACAGATCATTTTAGC |
| G287: | GCACTGGACAGCAGAAGACA |
| G290: | CCCAAAGTTGCTGAAGTACC |
| IR1: | AGGTTCTTCACTGCCTCACC |
| 291: | CACCAAATGAGGTCAGCAC |
| SCUTR: | ATTGTATTAACCATAATCACTATG |
| G314: | GGAATCATGGCAAGAAGGTG |
| PstA: | GGCCAGAGACCCCAAGCTTCGTGCA |
| PstB: | CGAAGCTTGGGGTCTCTGGCC |
Results
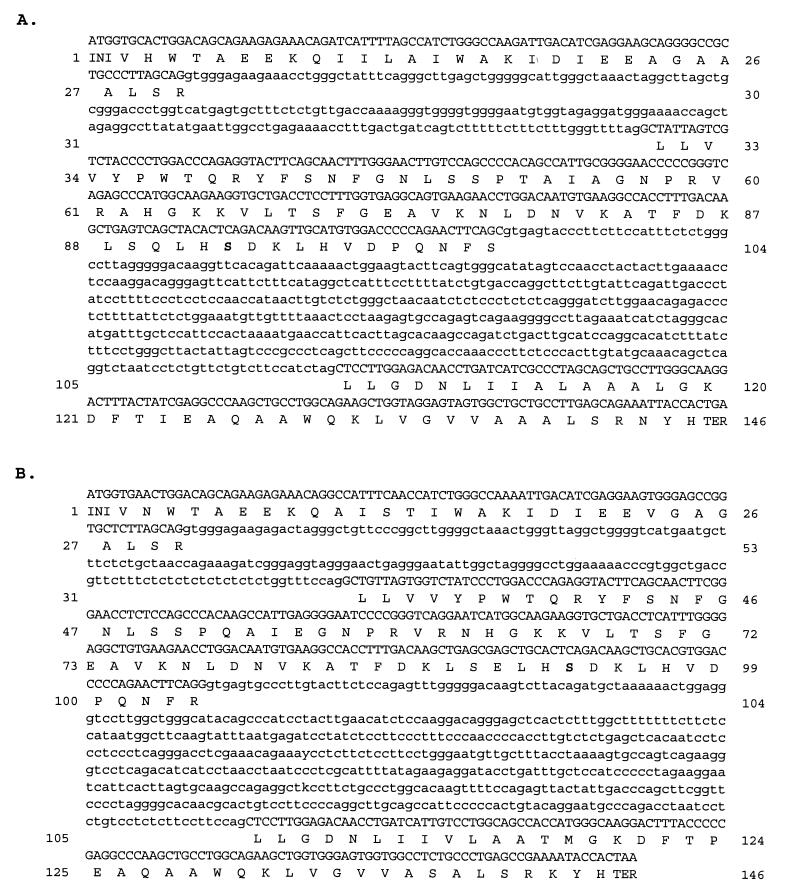
By using standard PCR and cloning techniques, the complete DNA sequences of the ω-globin gene were determined from two species of marsupial, the tammar and dunnart (Fig. 1). These genes were found to contain three exons and two introns located at the positions that are typical of all vertebrate β-globin genes. The exon sequence of each gene contains an ORF that encodes a protein of 146 amino acids (Fig. 1). The N-terminal region of tammar ω-globin is identical to the partial (54 residues) ω-globin protein sequence obtained from tammar embryonic-type blood (8). The lengths of the first intron of the tammar and dunnart ω-globin genes are 219 bp and 203 bp, respectively, and the lengths of the second intron are 548 bp and 565 bp, respectively. These introns are significantly different in length from all previously studied marsupial β-globin genes, which range in size from 109 bp to 120 bp for the first intron and 1,438 bp to 1,857 bp for the second intron (4, 5, 7).
Figure 1.
DNA sequences for tammar (A) and dunnart (B) ω-globin genes, and corresponding predicted amino acid sequences in single letter code. Noncoding DNA is shown in lowercase. The sequences have been deposited in GenBank (accession nos. AY014769 and AY014770). Amino acid residues specifically referred to in the text are shown in bold.
By using RT-PCR amplification of mRNA isolated from one-day-old neonates, we confirmed that the tammar and dunnart ω-globin genes are transcriptionally active (data not shown). In each case, the RT-PCR product had an identical sequence to the putative coding region of the ω-globin gene.
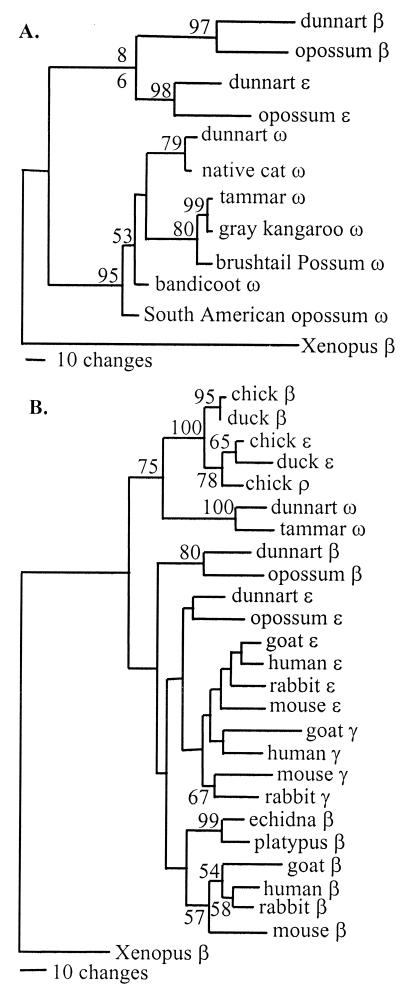
We used PCR and sequencing to determine whether ω-globin orthologues (genes that trace their most recent common ancestry to a speciation event) exist in other marsupial species. For these analyses we chose species that represented five marsupial families, including the Australian Dasyuridae (native cat), Macropodidae (gray kangaroo), Phalangeridae (brushtail possum), and Peramelidae (bandicoot), and the American Didelphidae (South American opossum). Primers for PCR were designed from conserved regions of the first and second exon sequences of tammar and dunnart ω-globin (see Methods) to amplify partial coding sequences, and the first intron, of ω-globin. Maximum parsimony phylogenetic analysis shows that these partial sequences from representative marsupial species form a monophyletic group with the tammar and dunnart ω-globin genes, supported by a 95% bootstrap value, to the exclusion of a second group containing dunnart and Didelphis virginiana (opossum) ɛ- and β-globin genes (Fig. 2A). This analysis confirms that the sequences we have amplified are orthologous to tammar and dunnart ω-globin, and indicates that ω-globin sequences are likely to be present in all marsupial species. Pairwise nonsynonymous divergence levels between taxa ranged from 0% (between tammar and gray kangaroo) and 19% (between the tammar and the South American opossum). In contrast, pairwise synonymous (silent) levels were considerably higher, ranging from 4.9% (between the tammar and gray kangaroo) to 127.6% (between South American opossum and the bandicoot). These data suggest that amino acid changing sites of marsupial ω-globin genes are under strong selective constraints that have slowed their rate of evolution relative to synonymous sites. Such a rate difference would be unlikely if these ω-globin genes were pseudogenes.
Figure 2.
(A) Most parsimonious tree of length 532 steps from an analysis of marsupial ɛ, β, and ω-globin sequences. Bootstrap values (>50%) are shown above branches. GenBank locus codes or accession nos. for sequences are: dunnart β (SCHBBHEMO), ɛ (SCHBBEGN), ω (AY014770); opossum β (OPOHBBB), ɛ (OPOHBBE); native cat ω (AY014773); gray kangaroo ω (AY014772); brushtail possum ω (AY014775); bandicoot ω (AY014774); South American opossum ω (AY014771); Xenopus (Xenopus laevis) adult β-globin (XELHBBC). (B) One of two most parsimonious trees of length 1,025 steps resulting from an analysis of aligned globin coding sequences (second tree not shown). This tree differed from the second tree only by the branching order of goat and mouse β-globin. Bootstrap values (>50%) are shown above branches. GenBank locus codes for sequences not given in A are: chick (Gallus gallus) β (bH), ɛ, ρ (GGHBBRE); duck (Cairina moschata) β (CMBGA2B2), ɛ (CMEGA2E2); human β, γ, ɛ (HSHBB); mouse (Mus musculus) β, γ (βh0), ɛ (ɛy), (MMBGCXD); goat (Capra hircus) βA (CHHBBAA), ɛI (CHEBGLI); rabbit (Oryctolagus cuniculus) β, γ, ɛ, (OCBGLO01); echidna (Tachyglossus aculeatus) β (TGLHBB) and platypus (Ornithorhynchus anatinus) β (TGLHBB).
Phylogenetic analyses were also carried out to investigate the evolutionary relationships of the marsupial ω-globin lineage with the avian and other mammalian β-globin genes. For these analyses partial coding sequences and third codon positions were excluded. The latter are likely to be saturated at the level of interclass divergences, as previous studies have shown divergences of over 100% at synonymous sites for pairs of β-like globin genes from mammals and avians (6). In the maximum parsimony tree (Fig. 2B) it can be seen that the marsupial ω-globin lineage forms a sister group with the β-like globin genes of avians, supported by a 75% bootstrap value, to the exclusion of a second group containing all other mammalian β-like globin genes. A similar sister group relationship was found in trees derived by using both maximum likelihood and distance approaches. All of the analyses support the conclusion that the ω-globin lineage is more closely related to the avian β-like globin genes than mammalian β-like globin genes.
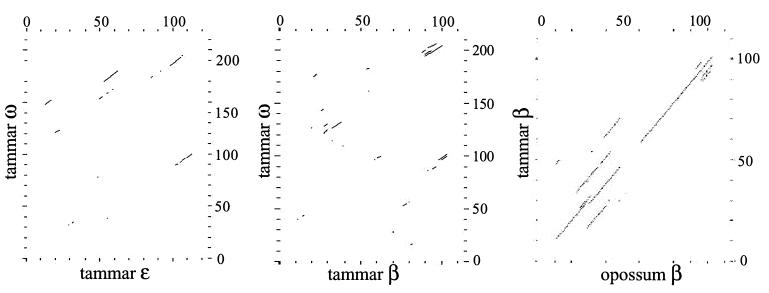
The results of dot plot analysis (Fig. 3) indicate that orthologous β-globin intron 1 sequences from two distantly related marsupial species share observable levels of sequence identity. In contrast, comparisons between intronic sequences of the tammar ω-globin and of the tammar β- and ɛ-globins show no such identity. These results indicate that it is unlikely ω-globin has evolved from a recent β-like globin gene duplication within the marsupial lineage.
Figure 3.
dotplot comparison of intron 1 sequence from marsupial β-like globin genes. Source of intronic sequence is indicated on the axes. For details on dotplot parameters see Methods.
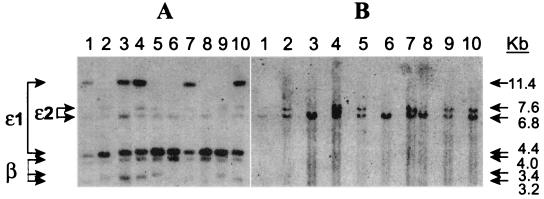
A previous study (5) of the pattern of inheritance of restriction fragment length variants in captive bred dunnart families showed the adult and embryonic β-globin genes to be closely linked to each other, but unlinked to a “third locus” provisionally named ɛ2 that hybridized on Southern blots to a dunnart ɛ-globin probe. By using a portion of dunnart ω-globin gene as a probe in Southern hybridizations, we detected an identical set of polymorphic restriction fragments to those representing the ɛ2 locus (Fig. 4). These results indicate that the locus previously named ɛ2 is, in fact, ω-globin, and that this gene is unlinked to the ɛ-globin/β-globin gene cluster in this species.
Figure 4.
Southern blot analysis of genomic DNA of individuals from a family of dunnarts raised in captivity (family M16285) by using in A an ɛ-globin probe (pSG-2H) (4) and in B a ω-globin specific probe (see Methods). Note hybridization of the ω-globin probe to an equivalent set of polymorphic fragments (6.8-kb and 7.6-kb) as locus e2. The lod scores for this family (ω-globin vs. ɛ-β globin) are Z(5:3) = −1.91 at θ = 0.05 and Z = 0 at θ = 0.45, indicating that the loci are unlikely to be closely linked. Data from additional families (5) shows ɛ2 (and hence ω-globin) is unlinked to ɛ-globin–β-globin.
To test whether ω-globin genes are unlinked to ɛ-globin and β-globin in another marsupial species we determined the chromosomal location of tammar ω-globin by in situ hybridization. Results indicated that the tammar ω-globin gene mapped to the short arm of chromosome 1 (Fig. 5). The adult β-globin gene has been previously mapped to chromosome 3 (20). Therefore, the ω-globin and β-globin genes are also unlinked in the tammar.
Figure 5.
Scores of silver grains generated by in situ hybridization of a tritiated 530-bp genomic clone of the tammar ω-globin gene (containing the first and second exons and first intron) to R-banded chromosome 1 of the tammar. The tallest column is over the dark band arbitrarily numbered 3. The central region of arm 1p is regarded as the probable location of the ω-globin gene.
Discussion
Our results confirm the existence of a new β-like globin gene in marsupials that we have called ω-globin. Unlike all previously studied β-like globin genes in marsupials, which were shown to be orthologous to eutherian ɛ- and adult β-globins (4, 5, 7), our analyses suggest (see 75% boot strap value, Fig. 2B) that ω-globin is orthologous to avian β-like globin genes and is clearly of ancient evolutionary origin. Remarkably, we have also found that ω-globin is unlinked or “orphaned” from the main β-globin cluster in two marsupials, the tammar and dunnart. In the dunnart this cluster consists of two genes, ɛ-globin and β-globin, linked in the order 5′-ɛ-β-3′ (5). To our knowledge, this is the first report of an expressed β-globin gene that is unlinked to the main gene cluster in any vertebrate species, and raises interesting questions about both the evolutionary origin of ω-globin and the developmental regulation of ω-, ɛ-, and β-globin genes in marsupials.
Phylogenetic Analyses and a New Model of β-Globin Gene Evolution.
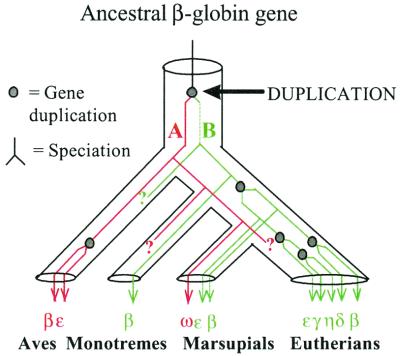
In setting the root of the tree depicted in Fig. 2B, we have assumed Xenopus β-globin forms an appropriate outgroup. A tree of very similar topology was derived if human α-globin is used as an outgroup sequence (S. Spargo, unpublished data). Therefore, based on the available sequence data, we are confident that the tree is an accurate depiction of evolutionary events. To account for our phylogenetic results we propose a new model for the evolution of β-like globin genes in mammals (Fig. 6). A feature of this model is the occurrence of an ancient gene duplication event that produces two lineages, one leading to the β-like globin genes of avians and marsupial ω-globin and a second leading to all other mammalian β-like globin genes. Although we cannot be certain of the timing of this gene duplication, our analysis indicates that it occurred before the avian/mammalian split. This model has two main implications.
Figure 6.
A model for the evolution of β-globin genes in mammals based on the phylogeny in Fig. 2B. The gene tree is drawn within the constraints of a species tree (21). The ancient gene duplication event (indicated by an arrow) gave rise to two ancestral genes, A and B. A was the progenitor of marsupial ω-globin and the β-like globin genes of birds. B gave rise to the β-like globin genes of mammals. Genes or pseudogenes that may be expected to occur are indicated by ?. To simplify the diagram, not all of the avian β-like globin genes (as exemplified by the chicken) are shown.
(i) Genes or pseudogenes that are orthologous to marsupial ω-globin may be present in monotremes and eutherian mammals. Although the presence of such genes in these species would lend support to the model, their absence would not negate the model. This is because progenitor ω-like globin genes that our model predicts to have been present in the common ancestor of monotremes and eutherians may have evolved rapidly in sequence to the point where their presence has become undetectable by using sequence homology searches. Such an outcome would be expected if the ω-globin chain was not under positive selection in these lineages. To date, we have been unable to detect ω-globin orthologues in monotremes and eutherians by using both extensive PCR analyses and genomic database searches, and currently are exploring alternative approaches to search for these genes/pseudogenes.
(ii) The bird and mammalian β-like globin clusters are not orthologous. This latter point may explain the observation that surprisingly little sequence similarity exists in comparisons of bird and mammalian β-like globin noncoding regions (1).
The absence of linkage between ω-globin and the ɛ-globin–β-globin cluster, at least in the tammar and dunnart, might be accounted for, in an evolutionary context, by several possible explanations. The duplication event which separated the ω-globin gene progenitor from the progenitor of the other mammalian β-like globin genes involved an intrachromosomal tandem gene duplication and subsequently the two gene lineages were separated by translocation. A precedent for the separation of coregulated globin genes is provided by the observation that the α- and β-globin genes are closely linked in Xenopus, but on separate chromosomes in birds and eutherian mammals. An alternative explanation is that the duplication involved a whole chromosome or whole genome. We plan to test these possible scenarios by finding genes that are syntenic to ω-globin and the ɛ-globin–β-globin cluster in marsupials, and examine the syntenic relationships of these genes in birds and eutherian mammals.
ω-Globin Expression and Function in Marsupial Neonates.
The detection of partial DNA sequences containing ORFs that are orthologous to tammar and dunnart ω-globin, in a number of phylogenetically distinct marsupial families, including American and Australian representatives, strongly suggests that this gene is present in all marsupial species. Further, the strong selective constraints on the rate of evolution of amino acid sites suggest that ω-globin genes are also likely to be expressed in all marsupials. We confirmed the expression of ω-globin in both the dunnart and tammar by RT-PCR analysis of one-day-old neonates. These results are consistent with previous studies of hemoglobin in tammar neonates (8, 10) and together show that tammar ω-globin is expressed at least three days before birth and one day after birth, with the protein remaining in circulation for up to 8 days post partum (K. H. Gill, A.A.G., L. A. Hinds, and R.A.B.H., unpublished observation). It is likely that ω-globin plays an important role in the oxygen transport requirements of neonates just before birth and in early pouch young development, but further studies are required to determine precisely what this role is.
Some clues to the possible function of ω-globin can be obtained by amino acid sequence comparisons with other vertebrate β-like globins, and a number of features of these comparisons are worth noting. Whereas the residues Trp-3 and Gln-9 occur in both tammar and dunnart ω-globin chains, no such residues have been recorded as occurring in any other mammalian β-like globins. However, Trp-3 is found in all bird β-like globins and in all reptilian β-globins except the crocodilians. Gln-9 is common, although not universal, in birds. A surprising finding was the presence of Ser-93 in ω-globin chains from both marsupial species. In the 500 chains most similar to tammar ω-globin, the residue at position 93 is Cys in all mammals, birds, and reptiles. However, Ser-93 is common in fish and, together with Trp-3, Glu-94, Arg-144, and Gln-145, has been implicated as an important residue in the Root effect in teleost fishes (this is the property in some fish hemoglobins of unloading all their oxygen as the blood is acidified; ref. 22). Allowing for the presence of an extra amino acid at position 120 in the fish β-globin chain, identical or very similar residues are present at corresponding sites in the marsupial ω-globin chains. Because of the extremely small quantities of blood available, it has not yet been possible to carry out oxygen-binding studies on the individual hemoglobin components of marsupial neonatal blood, but the presence of key Root effect residues in marsupial ω-globin is striking.
Developmental Regulation of the ω-, ɛ-, and β-Globin Genes in Marsupials.
Current models of β-like globin gene regulation in mammals suppose that all of the genes are tightly linked and under the control of a cis-acting LCR (23, 24). In the dunnart, regions of DNA upstream of the ɛ-globin–β-globin cluster show high sequence identity to regions of the human and mouse LCR (8), so it is likely that these marsupial genes also come under the influence of an LCR. However, this LCR cannot be cis-acting on the expression of the unlinked ω-globin, and the factors that regulate the expression of this gene remain to be determined. As the ω-globin and the ɛ-globin–β-globin gene cluster trace their origin to an ancient gene duplication event, it is likely that the former has evolved a novel regulatory mechanism. Studies on the regulation of ω-globin are, therefore, likely to give insights into the evolution and function of globin LCRs.
Acknowledgments
We dedicate this paper to the late Emeritus Professor Peter Martin, Botany Department, Adelaide University. Peter had a great interest in the evolution of marsupial globins. His presence at the University is greatly missed. We thank Cindy Bottema for making facilities available to G.W. We also thank Mi-Hye Lee, Robert Shroff, Scott Spargo, Velta Vingelis, and Tim Cox for helpful discussions. Tissue from tammar wallabies was very generously provided by Russell Baudinette and Jayne Skinner (Adelaide University). This work was supported by a grant from the Australian Research Council. Funding was also provided by the South Australian Museum and the Department of Genetics, Adelaide University. D.W. was in receipt of an Australian Postgraduate Scholarship. G.C.W. and C.D.K.B. were supported by a grant from the J. S. Davies Bequest to Adelaide University.
Abbreviations
- gDNA
genomic DNA
- LCR
locus control region
- RT-PCR
reverse transcription–PCR
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY014769–AY01475).
References
- 1.Hardison R. J Exp Biol. 1998;201:1099–1117. doi: 10.1242/jeb.201.8.1099. [DOI] [PubMed] [Google Scholar]
- 2.Ingram V M. Nature (London) 1961;189:704–708. doi: 10.1038/189704a0. [DOI] [PubMed] [Google Scholar]
- 3.Bromham L, Phillips M, Penny D. Tree. 1999;14:113–118. doi: 10.1016/s0169-5347(98)01507-9. [DOI] [PubMed] [Google Scholar]
- 4.Cooper S J B, Hope R M. Proc Natl Acad Sci USA. 1993;90:11777–11781. doi: 10.1073/pnas.90.24.11777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper S J, Murphy R, Dolman G, Hussey D, Hope R M. Mol Biol Evol. 1996;13:1012–1022. doi: 10.1093/oxfordjournals.molbev.a025651. [DOI] [PubMed] [Google Scholar]
- 6.Lee M H, Shroff R, Cooper S J, Hope R. Mol Phylogenet Evol. 1999;12:205–214. doi: 10.1006/mpev.1999.0610. [DOI] [PubMed] [Google Scholar]
- 7.Koop B F, Goodman M. Proc Natl Acad Sci USA. 1988;85:3893–3897. doi: 10.1073/pnas.85.11.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holland R A B, Gooley A A. Eur J Biochem. 1997;248:864–871. doi: 10.1111/j.1432-1033.1997.00864.x. [DOI] [PubMed] [Google Scholar]
- 9.Holland R A, Gooley A A, Hope R M. Clin Exp Pharmacol Physiol. 1998;25:740–744. doi: 10.1111/j.1440-1681.1998.tb02288.x. [DOI] [PubMed] [Google Scholar]
- 10.Calvert S J, Holland R A B, Hinds L A. Respir Physiol. 1993;91:99–109. doi: 10.1016/0034-5687(93)90092-o. [DOI] [PubMed] [Google Scholar]
- 11.Calvert S J, Holland R A B, Gemmell R T. Physiol Zool. 1994;67:407–417. [Google Scholar]
- 12.Holland R A B. Isr J Zool. 1994;40:401–416. [Google Scholar]
- 13.Mortola J P, Frappell P B, Woolley P A. Nature (London) 1999;397:660. doi: 10.1038/17713. [DOI] [PubMed] [Google Scholar]
- 14.Ochman H, Gerber A S, Hartl D L. Genetics. 1988;120:621–623. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swofford D L. PAUP* Sunderland, Massachusetts: Sinauer; 2000. , Version 4b2. [Google Scholar]
- 16.Felsenstein J. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 17.Hasegawa M, Kishino H, Yano T. J Mol Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning, A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 19.Webb G C, Lee J S, Campbell H D, Young I G. Cytogenet Cell Genet. 1989;50:107–110. doi: 10.1159/000132734. [DOI] [PubMed] [Google Scholar]
- 20.Sinclair A H, Graves J A. Genomics. 1991;9:581–586. doi: 10.1016/0888-7543(91)90350-n. [DOI] [PubMed] [Google Scholar]
- 21.Goodman M, Czelusniak J, Moore G W, Romero-Herrera A E, Matsuda G. System Zool. 1979;28:132–168. [Google Scholar]
- 22.Mylvaganam S E, Bonaventura C, Bonaventura J, Getzoff E D. Nat Struct Biol. 1996;3:275–283. doi: 10.1038/nsb0396-275. [DOI] [PubMed] [Google Scholar]
- 23.Tuan D, Kong S, Hu K. Proc Natl Acad Sci USA. 1992;89:11219–11223. doi: 10.1073/pnas.89.23.11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stamatoyannopoulos G. Science. 1991;252:383. doi: 10.1126/science.2017679. [DOI] [PubMed] [Google Scholar]