Abstract
Since the introduction of H3N2 swine influenza viruses (SIVs) into U.S. swine in 1998, H1N2 and H1N1 reassortant viruses have emerged from reassortment between classical H1N1 and H3N2 viruses. In 2004, a new reassortant H3N1 virus (A/Swine/Minnesota/00395/2004) was identified from coughing pigs. Phylogenetic analyses revealed a hemagglutinin segment similar to those of contemporary cluster III H3N2 SIVs and a neuraminidase sequence of contemporary H1N1 origin. The internal genes were of swine, human, and avian influenza virus origin, similar to those of contemporary U.S. cluster III H3N2 SIVs. The recovery of H3N1 is further evidence of reassortment among SIVs and justifies continuous surveillance.
Influenza A virus is a highly infectious respiratory pathogen in the respective natural hosts, which include birds, lower mammals, and humans (1, 13, 24). The primary clinical manifestations of viral infection in mammals are fever and acute respiratory distress (nasal discharge, coughing, and dyspnea).
Influenza A virus is an enveloped RNA virus containing eight segments of negative-sense RNA (13), which encode 10 proteins, including hemagglutinin (HA), neuraminidase (NA), matrix 1 (M1), M2, nucleoprotein (NP), nonstructural proteins (NS1 and NS2), and a polymerase complex (PA, PB1, and PB2). The virus can be further classified into 16 HA and 9 NA subtypes, all of which are maintained in aquatic birds (4, 13, 18, 23, 24).
Currently, variants of three predominant HA-NA combinations of influenza viruses (H1N1, H3N2, and H1N2) are circulating in swine populations throughout the world (2, 14). Prior to 1998, only the “classical” H1N1 virus, comprised of RNA segments that were all of swine origin, was identified in U.S. swine populations (14) following the first isolation of an influenza virus in the early 1930s (19). At present, the predominant H1N1 swine influenza viruses (SIVs) isolated in the United States are reassortant H1N1 viruses carrying RNA segments (PA and PB2) of avian origin (7). In late 1998, a novel SIV of H3N2 subtype (double-reassortant H3N2 viruses) was isolated from pigs in North Carolina demonstrating respiratory disease. The virus contained HA, NA, and PB1 segments of human influenza virus origin and other gene segments from classical H1N1 SIV (21, 28). Subsequent H3N2 isolates from other states were triple-reassortant viruses, containing HA, NA, and PB1 segments from human influenza viruses; M, NS1, and NP segments from classical H1N1 SIV; and PA and PB2 segments from avian viruses (21, 28). Additionally, reassortment between triple-reassortant and classical swine H1N1 viruses has produced H1N2 viruses (8). To date, variants of triple-reassortant H3N2 and H1N2 and reassortant H1N1 influenza viruses are circulating in U.S. swine populations (3, 8, 9, 10, 11, 15, 22). Here, we report the identification of a fourth HA-NA subtype combination—H3N1—in U.S. swine as a result of reassortment between H3N2 and H1N1 viruses.
In October 2004, a 1,000-head swine farm in Minnesota experienced a severe respiratory disease outbreak in 14-week-old pigs, which was characterized by coughing and increased mortality. The morbidity was approximately 80%, and the accumulated mortality reached 5% during the episode. Normal death loss in the index farm was less than 1% prior to the outbreak. Tissues from two dead pigs were submitted to the Veterinary Diagnostic Laboratory at the University of Minnesota for respiratory-disease diagnostic tests.
Grossly, the lungs were heavy and dark red. Microscopically, there was acute, purulent, and histiocytic bronchopneumonia with subacute pleuritis. Influenza A virus RNA, porcine reproductive and respiratory syndrome virus RNA, and type 2 porcine circovirus DNA were detected by PCR testing of the lung tissue, and Mycoplasma hyopneumoniae DNA was detected by PCR on bronchial swabs. Actinobacillus suis and Pasteurella multocida were isolated from the lungs.
Virus isolation (25) was attempted on lung tissues for influenza virus to further characterize the virus, since the herd was considered to be well vaccinated for swine influenza. A cytopathic virus capable of agglutinating rooster red blood cells was isolated in MDCK cells inoculated with the sample. The culture fluid was positive for influenza A viral RNA by reverse transcription (RT)-PCR and for influenza A virus NP by antigen-capture enzyme-linked immunosorbent assay (Directgen FluA; Becton-Dickinson, Franklin Lakes, NJ). Unexpectedly, the subtype of the virus isolate, as determined by a real-time multiplex RT-PCR (16), was H3N1. The isolate was plaque cloned (27), retested, and again subtyped by RT-PCR as H3N1. The presence of H3N1 virus in the lung tissues was also confirmed by RT-PCR and was the only HA-NA combination of SIV detected in the samples. Subsequently, the virus isolate was submitted to the U.S. Department of Agriculture National Veterinary Services Laboratories (Ames, Iowa) for subtyping by hemagglutination inhibition and neuraminidase inhibition assays and confirmed as H3N1. The isolate was then designated A/Swine/Minnesota/00395/2004 (H3N1).
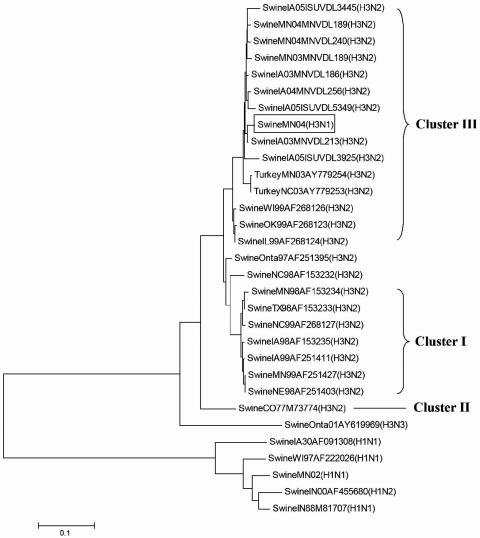
Further genotypic characterization of the virus was conducted by nucleic acid sequencing and phylogenetic analyses. Viral RNA was prepared from 200 μl of virus suspension with a QIAamp RNeasy Mini Kit (QIAGEN, Inc., Valencia, CA), using a protocol recommended by the manufacturer. PCR amplification of each gene segment was performed under standard conditions. The sequencing primers were designed based on a previous report (6) and are summarized in Table 1. PCR products were then purified using a QIAamp Gel extraction kit (QIAGEN) and sequenced using an ABI 3730 DNA Analyzer (Applied Biosystems, Inc., Foster City, CA). The gene sequences obtained were aligned and analyzed using Lasergene analysis software (DNASTAR, Madison, WI). Phylogenetic analyses of nucleotide sequences using the ClustalV method (5) and a BLAST search in the Influenza Sequence Database (12) demonstrated an HA segment with 95.9 to 99.5% nucleotide similarity to that of cluster III H3N2 SIVs (17, 21), the predominant H3 genotype circulating in U.S. swine (Fig. 1). The NA segment was close to that of classical H1N1 virus, with 92 to 93% identity among viral sequences available in GenBank (Table 2 and Fig. 2) but showed higher homology (98 to 99%) with contemporary H1N1 viruses from the Midwest. Other genes were of swine (M, NP, and NS1), avian (PA and PB1), and human (PB2) origin (Table 2), representative of the internal gene composition of contemporary triple-reassortant swine H3N2 viruses in North America (21, 28). This suggests that A/Swine/Minnesota/00395/2004 H3N1 influenza virus is a reassortant containing genes from triple-reassortant H3N2 and contemporary H1N1 SIVs. The clinical implications of this reassortant remain to be determined.
TABLE 1.
Compositions of sequencing primers
| Primer | Sequence |
|---|---|
| H3F | 5′-AGC AAA AGC AGG GGA TAA TTC T-3′ |
| H3-490F | 5′-CTG AAC GTG ACT ATG CCA-3′ |
| H3-1046F | 5′-GCG CAA TCG CAG GTT T-3′ |
| H3-1507F | 5′-GCA TAG GGT CAA TCA GAA-3′ |
| H3-362R | 5′-TAA GGG TAA CAG TTG CTG-3′ |
| H3-792R | 5′-CAG TAT GTC TCC CGG TTT-3′ |
| HR | 5′-AGT AGA AAC AAG GGT GTT TTT-3′ |
| NAF | 5′-AGC AAA AGC AGG AGT TTA AAA TG-3′ |
| N1-409F | 5′-GCA ACA ACA CTA AAC AAC AG-3′ |
| N1-1144F | 5′-ATT TGG GAT CCT AAT GGA TGG ACA G-3′ |
| N1-547R | 5′-GCA CTT GCT GAC CAA GCA ACT GAT T-3′ |
| NAR | 5′-AGT AGA AAC AAG GAG TTT TTT-3′ |
| PB1-1 | 5′-TAT TCG TCT CAG GGA GCG AAA GCA GGC A-3′ |
| PB1-F1 | 5′-GGT CGA ATG GTC TTAA CAG C-3′ |
| PB1-R1 | 5′-GTT GCA GGT CCA AGA TCA TTG-3′ |
| PB1-2341R | 5′-ATA TCG TCT CGT ATT AGT AGA AAC AAG GCA TTT-3′ |
| PB2-1 | 5′-TAT TCG TCT CAG GGA GCG AAA GCA GGT C-3′ |
| PB2-F1 | 5′CCA CTG TGG ACC ATA TGG CC-3′ |
| PB2-F2 | 5′-GAT CAA TGG TCC TGA GTC AG-3′ |
| PB2-R1 | 5′-GCC TTC ATC CGG GTC TTT AG-3′ |
| PB2-R2 | 5′-GCG ACA TCT CCG TGC TTG G-3′ |
| PB2-2233R | 5′-ATA TCG TCT CGT ATT AGT AGA AAC AAG GTA GTT-3′ |
| PA-1 | 5′-TAT TCG TCT CAG GGA GCG AAA GCA GGT AC-3′ |
| PA-F1 | 5′-GCA TTA CTG AAG CAC CGA TTT G-3′ |
| PA-F2 | 5′-CTG GAG AGG AGA TGG CTA C-3′ |
| PA-R1 | 5′-GCT CTC AAT CTG CTG AAG AG-3′ |
| PA-R2 | 5′-CAG TAG CTC TGC AAT GGG ACA C-3′ |
| PA-R3 | 5′-CTT CAT CAA GGG TGT AGT CTG C-3′ |
| NP-1 | 5′-TAT TCG TCT CAG GGA GCA AAA GCA GGG TA-3′ |
| NP-F1 | 5′-GAA GAG CAT CCC AGT GCT G-3′ |
| NP-R1 | 5′-CCC TTT CAA AGT CAT GCC C-3′ |
| NP-1565R | 5′-ATA TCG TCT CGT ATT AGT AGA AAC AAG GGT ATT TTT-3′ |
| NS-1 | 5′-TAT TCG TCT CAG GGA GCA AAA GCA GGG TG-3′ |
| NS-890R | 5′-ATA TCG TCT CGT ATT AGT AGA AAC AAG GGT GTT TT-3′ |
| M-1 | 5′-TAT TCG TCT CAG GGA GCA AAA GCA GGT AG-3′ |
| M-1027R | 5′-ATA TCG TCT CGT ATT AGT AGA AAC AAG GTA GTT TTT-3′ |
FIG. 1.
Phylogenetic relationship of the HA gene from newly identified H3N1 swine influenza virus [SwineMN04(H3N1)] to HA genes of other well-characterized H3N2 viruses of swine origin as illustrated by the unrooted neighbor-joining method (MEGA software 3.1).
TABLE 2.
Influenza A viruses with highest nucleotide sequence identity to H3N1 swine influenza virus (A/Swine/Minnesota/00395/2004) as determined by BLAST search in the Influenza Sequence Databasea
| Gene | Identity (%) | Virus designation | Subtype | GenBank accession no. |
|---|---|---|---|---|
| HA | 99.4 | A/Swine/Iowa/00213/2003 | H3N2 | ISDN49163 |
| 98.4 | A/Swine/Iowa/00186/2003 | H3N2 | ISDN49100 | |
| 98.0 | A/Turkey/North Carolina/12344/03 | H3N2 | AY779253 | |
| NA | 93.2 | A/Swine/Quebec/5393/91 | H1N1 | U86145 |
| 92.4 | A/Wisconsin/4754/94 | H1N1 | U53166 | |
| PB1 | 98.7 | A/Wisconsin/10/98 | H1N1 | AF342823 |
| 98.2 | A/Swine/Minnesota/593/99 | H3N2 | AF251429 | |
| 98.2 | A/Duck/North Carolina/91347/01 | H1N2 | AY233388 | |
| PB2 | 97.5 | A/Swine/Korea/CY02/02 | H1N2 | AY129163 |
| 97.4 | A/Swine/Illinois/100084/01 | H1N2 | AF455738 | |
| PA | 97.0 | A/Swine/Minnesota/593/99 | H3N2 | AF251433 |
| 96.7 | A/Duck/North Carolina/91347/01 | H1N2 | AY233389 | |
| NP | 98.3 | A/Swine/Ohio/891/01 | H1N2 | AY129159 |
| 98.3 | A/Swine/Korea/CY02/02 | H1N2 | AF455699 | |
| M | 98.8 | A/Swine/Indiana/14810-S/01 | H1N2 | AY060071 |
| 98.6 | A/Turkey/Minneasota/764-2/03 | H3N2 | AY779258 | |
| 98.4 | A/Wisconsin/10/98 | H1N1 | AF342818 | |
| NS | 98.4 | A/Swine/Indiana/14810-S/01 | H1N2 | AY060136 |
| 98.2 | A/Swine/Korea/CY02/01 | H1N2 | AY129160 |
http://www.flu.lanl.gov/.
FIG. 2.
Phylogenetic relationship of the NA gene from newly identified H3N1 swine influenza virus [SwineMN04(H3N1)] to NA genes of other well-characterized N1 viruses of human, swine, or avian origin as illustrated by the unrooted neighbor-joining method (MEGA software 3.1).
At both Iowa State University and Minnesota Veterinary Diagnostic Laboratories, subtypes of SIVs implicated in clinical cases have been closely monitored using molecular diagnostics, as well as classical virologic testing, since 1998. To our knowledge, this is the first report of H3N1 virus in U.S. swine populations. Outside the United States, the emergence of H3N1 SIV from reassortment between classical H1N1 and human-type H3N2 SIVs was reported from Taiwan in 2003 (20). Taiwanese H3N1 viruses share approximately 83% and 90% identity with HA and NA segments, respectively, of the U.S. H3N1 virus. The recovery of an H3N1 subtype is further evidence of reassortment and antigenic shift in SIVs, justifying ongoing surveillance of animal populations for influenza A viruses that could represent a direct health threat, not only to swine populations, but also to humans. This study also demonstrated the usefulness of molecular technologies for such monitoring.
The emergence of H3N1 viruses in swine was expected from reassortment between H1N1 and H3N2 viruses, since more than 60% of swine farms (sow and finisher units) in the Midwest had serological evidence of infection by both H1 and H3 viruses within 6 months after H3N2 SIV was first isolated in late 1998 (26). Interestingly, the emergence of H3N1 reassortant virus was not seen until 2004, whereas isolation of H1N2 reassortant viruses from U.S. swine was reported in 2000 (8). Although the exact reason for the delayed emergence of H3N1 virus cannot be determined, it is suspected that acquisition of PA and PB2 gene segments of avian influenza virus origin by swine H1N1 viruses (8) facilitated the production of the H3N1 reassortant. However, the precise role of avian polymerase genes in the host adaptation and replication efficiencies of influenza viruses is not well known and remains an interesting area of study regarding the ecology and evolution of influenza virus in swine.
Nucleotide sequence accession numbers.
The GenBank accession numbers of the HA segment, the NA segment, and other genes are DQ145537, DQ145538, and DQ145539 to DQ145544, respectively.
REFERENCES
- 1.Alexander, D. J. 2000. A review of avian influenza in different bird species. Vet. Microbiol. 74:3-13. [DOI] [PubMed] [Google Scholar]
- 2.Brown, I. H. 2000. The epidemiology and evolution of influenza viruses in pigs. Vet. Microbiol. 74:29-46. [DOI] [PubMed] [Google Scholar]
- 3.Choi, Y. K., S. M. Goyal, M. W. Farnham, and H. S. Joo. 2002. Phylogenetic analysis of H1N2 isolates of influenza A virus from pigs in the Unites States. Virus Res. 87:173-179. [DOI] [PubMed] [Google Scholar]
- 4.Fouchier, R. A., V. Munster, A. Wallensten, T. M. Bestebroer, S. Herfst, D. Smith, G. F. Rimmelzwaan, B. Olsen, and A. D. Osterhaus. 2005. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J. Virol. 79:2814-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Higgins, D. G., and P. M. Sharp. 1989. Fast and sensitive multiple sequence alignments on a microcomputer. Comput. Appl. Biol. Sci. 5:151-153. [DOI] [PubMed] [Google Scholar]
- 6.Hoffmann, E., J. Stech, Y. Guan, R. G. Webster, and D. R. Perez. 2001. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 146:2275-2289. [DOI] [PubMed] [Google Scholar]
- 7.Janke, B. H. 2004. Swine influenza: relative prevalence of reassortants and subtypes, p. 35-39. In Proceedings of 12th Annual Swine Disease Conference for Swine Practitioners. Iowa State University, Ames.
- 8.Karasin, A. I., C. W. Olsen, and G. A. Anderson. 2000. Genetic characterization and implications of an H1N2 influenza virus isolated from a pig in Indiana. J. Clin. Microbiol. 38:2453-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karasin, A. I., J. Landgraf, S. Swenson, G. Erickson, S. Goyal, M. Woodruff, G. Scherba, G. Anderson, and C. W. Olsen. 2002. Genetic characterization of H1N2 influenza A viruses isolated from pigs throughout the United States. J. Clin. Microbiol. 40:1073-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karasin, A. I., M. M. Schutten, L. A. Cooper, C. B. Smith, K. Subbarao, G. A. Anderson, S. Carman, and C. W. Olsen. 2000. Genetic characterization of H3N2 influenza viruses isolated from pigs in North America, 1977-1999: evidence for wholly human and reassortant virus genotypes. Virus Res. 68:71-85. [DOI] [PubMed] [Google Scholar]
- 11.Landolt, G. A., A. I. Karasin, L. Phillips, and C. W. Olsen. 2003. Comparison of the pathogenesis of two genetically different H3N2 influenza A viruses in pigs. J. Clin. Microbiol. 41:1936-1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Macken, C., H. Lu, J. Goodman, and L. Boykin. 2001. The value of a database in surveillance and vaccine selection, p. 103-106. In A. D. M. E. Osterhaus, N. Cox, and A. W. Hampson (ed.), Options for the control of influenza, vol. IV. Elsevier Science, Amsterdam, The Netherlands. [Google Scholar]
- 13.Murphy, B. R., and R. G. Webster. 1996. Orthomyxoviruses, p. 1397-1445. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 14.Olsen, C. W. 2002. The emergence of novel swine influenza viruses in North America. Virus Res. 85:199-210. [DOI] [PubMed] [Google Scholar]
- 15.Olsen, C. W., S. Carey, L. Hinshaw, and A. I. Karasin. 2000. Virologic and serologic surveillance for human, swine and avian influenza virus infections among pigs in the north-central United States. Arch. Virol. 145:1399-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richt, J. A., K. M. Lager, D. Clouser, E. Spackman, D. Suarez, and K.-J. Yoon. 2004. Real-time RT-PCR assays for the detection and differentiation of North American swine influenza viruses. J. Vet. Diagn. Investig. 16:367-373. [DOI] [PubMed] [Google Scholar]
- 17.Richt, J. A., K. M. Lager, B. H. Janke, R. D. Woods, R. G. Webster, and R. J. Webby. 2003. Pathogenic and antigenic properties of phylogenetically distinct reassortant H3N2 swine influenza viruses cocirculating in the United States. J. Clin. Microbiol. 41:3198-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Röhm, C., N. A. Zhou, J. C. Süss, J. Mackenzie, and R. G. Webster. 1996. Characterization of a novel influenza hemagglutinin, H15: criteria for determination of influenza A subtypes. Virology 217:508-516. [DOI] [PubMed] [Google Scholar]
- 19.Shope, R. E. 1931. Swine influenza III. Filtration experiments and etiology. J. Exp. Med. 54:373-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsai, C.-P., and M.-J. Pan. 2003. New H1N2 and H3N1 influenza viruses in Taiwanese pig herds. Vet. Rec. 153:408. [PubMed] [Google Scholar]
- 21.Webby, R. J., S. L. Swenson, S. Krauss, P. J. Gerrish, S. M. Goyal, and R. G. Webster. 2000. Evolution of swine H3N2 influenza viruses in the Unites States. J. Virol. 74:8243-8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Webby, R. J., K. Rossow, G. Erickson, Y. Sims, and R. G. Webster. 2004. Multiple lineages of antigenically and genetically diverse influenza A virus co-circulating in the Unites States swine population. Virus Res. 103:67-73. [DOI] [PubMed] [Google Scholar]
- 23.Webster, R. G., K. F. Shortride, and Y. Kawaoka. 1997. Influenza: interspecies transmission and emergence of new pandemics. FEMS Immunol. Med. Microbiol. 18:275-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Webster, R. G., W. J. Bean, O. T. Gorman, T. M. Chambers, and Y. Kawaoka. 1992. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56:152-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.World Health Organization. 2002. WHO manual on animal influenza diagnosis and surveillance, p. 19-23. WHO/CDS/CSR/NCS/2002.5. WHO Global Influenza Programme, Geneva, Switzerland.
- 26.Yoon, K.-J., and B. Janke. 1999. Swine influenza viruses—Emergence of new subtype in Midwest swine and current research, p. 20-25. In Proceedings of the 7th Swine Disease Conference for Swine Practitioners. Iowa State University, Ames.
- 27.Youil, R., Q. Su, T. J. Toner, C. Szymkowiak, W.-S. Kwan, B. Rubin, L. Petrukhin, I. Kiseleva, A. R. Shaw, and D. DiStefano. 2004. Comparative study of influenza virus replication in Vero and MDCK cell lines. J. Virol. Methods 120:23-31. [DOI] [PubMed] [Google Scholar]
- 28.Zhou, N. N., D. A. Senne, J. S. Landgraf, S. L. Swenson, G. Erickson, K. Rossow, L. Liu, K.-J. Yoon, S. Krauss, and R. G. Webster. 1999. Genetic reassortant of avian, swine, and human influenza A viruses in American pigs. J. Virol. 73:8851-8856. [DOI] [PMC free article] [PubMed] [Google Scholar]