Abstract
Two clinical isolates of Pseudomonas aeruginosa, TL-1 and TL-2, were isolated from a patient transferred from Bangladesh and hospitalized for osteomyelitis in Paris, France. P. aeruginosa TL-1 expressed the extended-spectrum β-lactamase VEB-1a and was susceptible only to imipenem and colistin, while P. aeruginosa TL-2 expressed only the naturally occurring blaAmpC gene at a basal level and exhibited a wild-type β-lactam resistance phenotype. In TL-1, the typical 5′-end conserved sequence (5′-CS) region of class 1 integrons usually present upstream of the blaVEB-1a gene was replaced by a truncated 3′-CS and a 135-bp repeated element (Re). Downstream of the blaVEB-1a gene, an insertion sequence, ISPa31 disrupted by ISPa30, and an orf513 sequence, belonging to a common region (conserved region 1 [CR1]) immediately upstream of the aphA-6 gene, were present. Further downstream, a second truncated 3′-CS region in direct repeat belonged to In51, an integron containing two gene cassettes (aadA6 and the OrfD cassette). Thus, the overall structure corresponded to a sul-type class 1 integron termed In121. Genetic analyses revealed that both isolates were clonally related and differed by a ca. 100-kb fragment that contained In121. Both isolates contained another integron, In122, that carried three gene cassettes: aadB, dfrA1, and the OrfX cassette. This work identifies for the first time the spread of Re-associated blaVEB genes located on a sul-type integron. It also reports for the first time a CR1 element in P. aeruginosa that is associated with an aminoglycoside resistance aphA-6 gene that is expressed from a composite promoter.
Pseudomonas aeruginosa is naturally resistant to aminopenicillins and narrow-spectrum cephalosporins due to combined mechanisms of resistance, such as AmpC cephalosporinase production (4, 45), low outer-membrane permeability, and expression of efflux systems (45). Resistance to expanded-spectrum cephalosporins results mostly from overexpression of the cephalosporinase and from acquired expanded-spectrum β-lactamases (ESBLs) (8, 11, 45). In addition to the TEM/SHV-type β-lactamases, non-TEM and non-SHV Ambler class A β-lactamases (1, 47) have been reported in P. aeruginosa, with specific geographical distributions, e.g., PER-1 widespread in Turkey and South Korea, GES-2 in South Africa, and VEB-1 in Southeast Asia (12, 13, 35, 44, 48).
The blaVEB-1 gene, initially reported from an Escherichia coli isolate from a Vietnamese patient, was plasmid and integron located (33). The gene has subsequently been detected in several gram-negative species from Thailand and Vietnam, emphasizing the spread of VEB-1 β-lactamase in East Asian countries (5, 7, 12, 13, 15, 17, 24, 25, 26, 43). In addition, VEB-1 has been detected in P. aeruginosa isolates from Kuwait (34), in Acinetobacter baumannii isolates from France (6, 32), in P. aeruginosa isolates from India (2) and, recently, in Providencia stuartii isolates from Algeria (3).
The blaVEB-1a gene has been characterized in a peculiar genetic environment in a P. aeruginosa isolate from India (2). This gene was chromosomally located, and instead of being part of a typical class 1 integron structure, it was flanked by two identical 135-bp sequences, termed repeated elements (Re), that were bracketed by two truncated 3′-end conserved sequences (3′-CS) (10) of class 1 integrons in direct repeat (DR) (2, 3). These Re carried a strong promoter that drove the expression of the downstream-located blaVEB-1a gene (2). Very recently, a similar structure was found in P. stuartii from Algeria, suggesting that these Re may be widespread.
In the present work, the genetic environment of the blaVEB-1a gene was characterized from a multidrug-resistant P. aeruginosa clinical isolate from a patient hospitalized in France who was transferred from Bangladesh. In this strain, a Re-associated blaVEB-1 gene was characterized as part of a novel sul-type integron that included an orf513 gene as part of a conserved region 1 (CR1) element next to an aminoglycoside resistance gene, aphA-6. This work further illustrates the polymorphism of the genetic background of associated blaVEB-1 genes.
MATERIALS AND METHODS
Bacterial strains, plasmids, electroporation, and culture conditions.
Clinical P. aeruginosa isolates TL-1 and TL-2 were identified by standard biochemical techniques (API-20NE; bioMérieux, Marcy-l'Etoile, France). They were recovered from a bone infection from a 23-year-old patient hospitalized at the hospital Saint Michel (Paris) for chronic osteomyelitis. Following a motor vehicle accident in October 2002, this patient was hospitalized in Dhaka (Bangladesh) for a nail osteosynthesis of a tibial fracture. In December 2002, he was rehospitalized for a clinical osteomyelitis in Madras (India), where the nail was replaced by an external fixator. Upon the patient's admission at the hospital Saint Michel in October 2003, P. aeruginosa TL-1 was isolated from pus coming out from the external fixator. Thorough cleaning of the wound and replacement of the external fixator were then performed. In February 2004, P. aeruginosa TL-1 and TL-2 were isolated from the drainage of a remaining microabscess.
The clinical strains P. aeruginosa 10.2 (2) and P. aeruginosa JES (26) were used as blaVEB-1 gene-containing control strains, and P. aeruginosa PAO1 was used as a control strain in variable-number tandem repeat analysis (28). E. coli DH10B (Invitrogen, Cergy-Pontoise, France) was used as a bacterial host in electroporation experiments. Rifampin-resistant E. coli C600 and P. aeruginosa PU21 strains were used in conjugation experiments. E. coli NCTC 50192 harboring 154-, 66-, 38-, or 7-kb plasmids was used as a plasmid-containing reference strain (16, 46). The low-copy cloning vector pBBR1MCS.3 (18) was used in cloning experiments. Electroporation into E. coli DH10B was performed as previously described (30). Bacterial cells were grown in Trypticase soy (TS) broth and onto TS agar plates (Sanofi Diagnostics Pasteur, Marnes-La-Coquette, France).
Antimicrobial agents and susceptibility testing.
Routine antibiograms were determined by the disk diffusion method on Mueller-Hinton (MH) agar (Sanofi Diagnostics Pasteur). The antimicrobial agents and their sources have been described elsewhere (23). MICs of β-lactams for P. aeruginosa and E. coli DH10B (pRTL-1) were determined and interpreted as described previously (9). The double-disk synergy test was performed with cefepime and ticarcillin-clavulanic acid disks on MH agar plates. To inhibit the activity of the AmpC-type β-lactamase of P. aeruginosa, the double-disk test was also performed on cloxacillin (250 μg/ml)-containing plates (11).
Mating-out assay.
Conjugation experiments were attempted between the P. aeruginosa TL-1 isolate as the donor and rifampin-resistant E. coli C600 and P. aeruginosa PU21 as recipients in liquid and solid media at 37°C. The mating culture was plated onto TS agar plates containing ticarcillin (100 μg/ml) and rifampin (200 μg/ml).
Nucleic acid extractions.
Recombinant plasmids were extracted using QIAGEN plasmid mini-midi kits (QIAGEN, Courtaboeuf, France), whereas natural plasmids were extracted according to Kieser (16). Total DNA from P. aeruginosa isolates was extracted as described previously (30).
PCR amplification, cloning experiments, and sequencing.
Taq DNA polymerase was obtained from Roche (Roche Diagnostics, Meylan, France). Standard PCR amplification experiments (37) were attempted. Primers specific for genes coding for β-lactamases (OXA-10, TEM, SHV, PER, VEB, and GES) and the antibiotic resistance genes previously identified with the blaVEB-1 gene in class 1 integrons (aadB, arr-2, cmlA5, and aadA1) have been previously described (2, 24, 25). The orf513 primers have been described previously (31), and the Re-specific primers have been described by Aubert et al. (2, 3). The PCR products were purified using QIAquick columns (QIAGEN).
T4 DNA ligase and restriction endonucleases were used according to the manufacturer's recommendations (Amersham Biosciences, Saclay, France). The ligation products of PstI-digested total DNA of P. aeruginosa TL1 into PstI-restricted pBBR1MCS.3 were electroporated into E. coli DH10B, and selection was performed onto TS agar plates containing amoxicillin (100 μg/ml) and tetracycline (30 μg/ml).
Sequencing was performed using laboratory-designed primers on an ABI PRISM 3100 automated sequencer (Applied Biosystems, Les Ullis, France). Nucleotide and deduced protein sequence alignments were carried out on the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov).
Mapping the aphA-6 transcription start site.
Reverse transcription and rapid amplification of cDNA ends (RACE) were performed with the 5′ RACE system version 2.0 (Invitrogen). Total RNA (5 μg) extracted from P. aeruginosa TL-1 (QIAGEN RNeasy maxi kit) was used to determine the aphA-6 transcription initiation site.
After a reverse transcription step with gene-specific primer GSP1 (5′-AACTCATTCCATAGACTTAGGT-3′) and reverse transcriptase, the cDNA was tailed with cytosines by using the terminal deoxynucleotidyl transferase and was subsequently amplified with another gene-specific primer, GSP2 (5′-TAAATGGACAATCAATAATAGC-3′), combined with an oligo(dG) adapter primer provided with the kit. This PCR product was used as a template for a nested PCR assay with another adapter and GSP3 primer (5′-AACTCAACATTTTCGCTTCACG-3′). The PCR products obtained were directly sequenced. The transcription initiation site was determined as the first nucleotide following the sequence of the adapter primer.
Genotyping.
Pulsed-field gel electrophoresis (PFGE) was performed with VEB-1-producing P. aeruginosa JES, 10.2, TL-1, and TL-2 strains using XbaI and SpeI (Amersham Biosciences), as previously described (12). SpeI macrorestriction patterns were digitized and analyzed using Taxotron software (Institut Pasteur, Paris, France) and subsequently interpreted according to the recommendations of Tenover et al. (41).
Hybridization.
DNA-DNA hybridizations were performed as described by Sambrook and Russell (37), with a Southern transfer of a PFGE agarose gel that contained total DNA of P. aeruginosa isolates. The probe consisted in a 650-bp PCR-generated fragment from recombinant plasmid pRTL-1 and was internal to the blaVEB-1 gene. Labeling of the probe and signal detection were carried out by use of the ECL nonradioactive labeling and detection kit according to the manufacturer's instructions (Amersham Biosciences).
IEF analysis.
β-Lactamase extracts were prepared as described previously (12, 13) and subjected to analytical isoelectric focusing (IEF) on a pH 3.5 to 9.5 ampholine polyacrylamide gel (Amersham Biosciences), as described elsewhere (24, 30).
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this paper have been submitted to the EMBL/GenBank nucleotide sequence database under the accession numbers DQ315788 and DQ315789.
RESULTS AND DISCUSSION
Susceptibility testing of clinical isolates.
Disk diffusion susceptibility testing revealed that P. aeruginosa TL-1 was resistant to most β-lactams (except imipenem), fosfomycin, amikacin, gentamicin, kanamycin, netilmicin, tobramycin, and ciprofloxacin and was susceptible to colistin. A synergy image between cefepime- and clavulanic acid-containing disks on MH agar plates suggested the presence of an ESBL (data not shown). MIC values of β-lactams for P. aeruginosa TL-1 mirrored those obtained by disk diffusion susceptibility testing (Table 1). Susceptibility to several β-lactams was fully recovered after clavulanic acid or tazobactam addition.
TABLE 1.
MICs of β-lactams for P. aeruginosa TL-1, P. aeruginosa TL-2 clinical isolate, E. coli DH10B (pRTL-1), and E. coli DH10B reference strain
| β-Lactam(s)a | MIC (μg/ml) for:
|
|||
|---|---|---|---|---|
| P. aeruginosa TL-1 | P. aeruginosa TL-2 | E. coli DH10B (pRTL-1)b | E. coli DH10B | |
| Ticarcillin | >512 | 8 | >512 | 4 |
| Ticarcillin-CLA | 8 | 8 | 32 | 4 |
| Piperacillin | 256 | 2 | >512 | 2 |
| Piperacillin-TZB | 8 | 4 | 2 | 2 |
| Cefuroxime | >512 | >512 | >512 | 4 |
| Cefoxitin | >512 | >512 | 4 | 4 |
| Cefotaxime | >512 | 32 | 512 | <0.06 |
| Cefotaxime-CLA | 64 | 32 | 0.25 | <0.06 |
| Ceftazidime | >512 | 4 | >512 | 0.5 |
| Ceftazidime-CLA | 16 | 2 | 4 | 0.25 |
| Cefepime | 256 | 2 | >512 | <0.06 |
| Aztreonam | 512 | 4 | >512 | 0.12 |
| Imipenem | 0.5 | 1 | 0.12 | 0.12 |
CLA, clavulanic acid at a fixed concentration of 2 μg/ml; TZB, tazobactam at a fixed concentration of 4 μg/ml.
The recombinant plasmid pRLT-1 contained the blaVEB-1a gene.
The P. aeruginosa TL-2 isolate had a wild-type β-lactam resistance phenotype but the same non-β-lactam resistance phenotype as P. aeruginosa TL-1, except for being intermediate to amikacin.
No plasmid DNA was detected in P. aeruginosa TL-1 and TL-2 isolates despite repeated attempts. Transfer of the ticarcillin resistance marker from P. aeruginosa TL-1 to rifampin-resistant E. coli C600 or P. aeruginosa PU21 failed, suggesting a chromosomal location of the β-lactamase genes.
Preliminary PCR screening.
PCR experiments with primers specific for blaTEM, blaSHV, blaGES, and blaPER failed. A PCR product of 650 bp was obtained using primers specific for blaVEB genes. Sequencing of this PCR product revealed 100% identity with the blaVEB-1 gene identified in E. coli MG-1 (33). The blaVEB-1 gene has been described as a gene cassette located within the variable region of class 1 integrons, often associated with the aadB, arr-2, cmlA5, blaOXA-10, and aadA1 gene cassettes (12, 24, 25). The absence of PCR products using combinations of primers specific to class 1 integron structures and to the antibiotic resistance genes suggested that the genetic environment of blaVEB-1 in P. aeruginosa TL-1 could be different from that of known blaVEB-1-containing integrons.
PCR amplification experiments using primers located within the Re located next to blaVEB-1 genes in rare VEB-expressing isolates indicated that the genetic environment of the blaVEB-1 gene was similar to that reported for P. aeruginosa 10.2 and was therefore further investigated (2).
Cloning of the β-lactamase genes.
Shotgun cloning of β-lactamase genes from TL-1 yielded several E. coli transformants. Disk diffusion antibiograms revealed two distinct β-lactam resistance profiles. The first one comprised resistance to amino-, carboxy-, and ureido-penicillins, to narrow- and expanded-spectrum cephalosporins, and to aztreonam, but not to imipenem. A synergy image between clavulanic acid and expanded-spectrum cephalosporin disks suggested the presence of an ESBL that was consistent with the expression of the clavulanate-inhibited VEB-1 β-lactamase. One of the E. coli transformants displaying an ESBL phenotype was retained for further analysis. The plasmid contained in that recombinant E. coli isolate was designated pRTL-1 and contained a 9-kb PstI insert. The plasmid pRTL-1 also conferred resistance to gentamicin, kanamycin, amikacin, and netilmicin. MICs of β-lactams for E. coli DH10B (pRTL-1) mirrored the results obtained with disk diffusion susceptibility testing (Table 1). Susceptibility to several β-lactams was recovered after clavulanic acid or tazobactam addition.
The second phenotype, which was not further studied, was consistent with an AmpC-type cephalosporinase expression (resistance to amoxicillin, amoxicillin/clavulanate, and cephalothin and reduced susceptibility to cefuroxime).
IEF analysis.
β-Lactamase extracts of cultures of P. aeruginosa TL-1 and TL-2 and E. coli DH10B (pRTL-1) were subjected to analytical IEF. P. aeruginosa TL-1 expressed two β-lactamases with pI values of 7.4 and 8.6, consistent with those of β-lactamases VEB-1 and AmpC from P. aeruginosa, respectively (2). P. aeruginosa TL-2 expressed β-lactamase with a pI value of only 8.6, consistent with that of AmpC from P. aeruginosa. The pI 7.4 enzyme was also identified in extracts of E. coli DH10B (pRTL-1).
Characterization of the blaVEB-1-like gene and of its genetic environment.
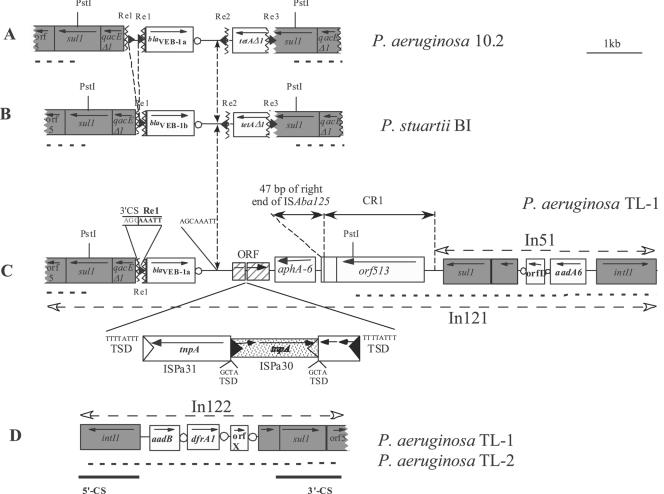
The sequence of the blaVEB-1-like gene from pRTL-1 revealed a blaVEB-1a allele identical to that initially described for a P. aeruginosa isolate from Kuwait (2, 34). Sequence analysis on both sides of the blaVEB-1a gene revealed an atypical genetic environment. Although the blaVEB-1a gene was followed by a 59-base element (59-be) sharing 100% sequence identity with that previously described for the blaVEB-1 gene cassette (25, 33), it was not preceded by a sequence for a typical recombination core site (Fig. 1C) (36). This blaVEB-1a gene cassette was truncated at its left-hand side just before the cassette-specific ribosomal binding site (RBS) that was still present. Further upstream of the blaVEB-1a gene, a qacEΔ1 gene followed by a sul1 gene, both in opposite orientation compared to blaVEB-1a, was found. The recombination core site of qacEΔ1 that is usually present at the beginning of a 3′-CS region was absent due to a truncation (Fig. 1C). Analysis of the sequence between the beginning of the truncated blaVEB-1a gene cassette and the 3′-CS region revealed a 135-bp sequence identical to that of the previously characterized Re1 (Fig. 1C) (2, 3).
FIG. 1.
Schematic representations of the genetic environment of the blaVEB-1-like gene (A) in P. aeruginosa 10.2 (2) from India, (B) in P. stuartii BI (3) from Algeria, (C) in P. aeruginosa TL-1 from Bangladesh, and (D) of In122 found in P. aeruginosa TL-1 and TL-2. The Re are represented as black triangles, and the flanking sequence identity breakpoints in the blaVEB-1-like gene environment are designated by vertical broken lines. The coding regions are shown as boxes, with an arrow indicating the orientation of transcription and white circles indicating 59-be's. Dark-gray boxes correspond to genes or ORFs present on class 1 integron conserved sequences. Restriction sites that were used for cloning are indicated. Dashed lines in bold indicate regions that were identified using PCR (the sequences of primers are available upon request). Horizontal dashed-lined arrows indicate the homology breakpoints in In121, compared to the structures found in P. aeruginosa 10.2 and P. stuartii BI. Filled and empty triangles represent inverted repeats of insertion sequences. Target site duplications are also indicated.
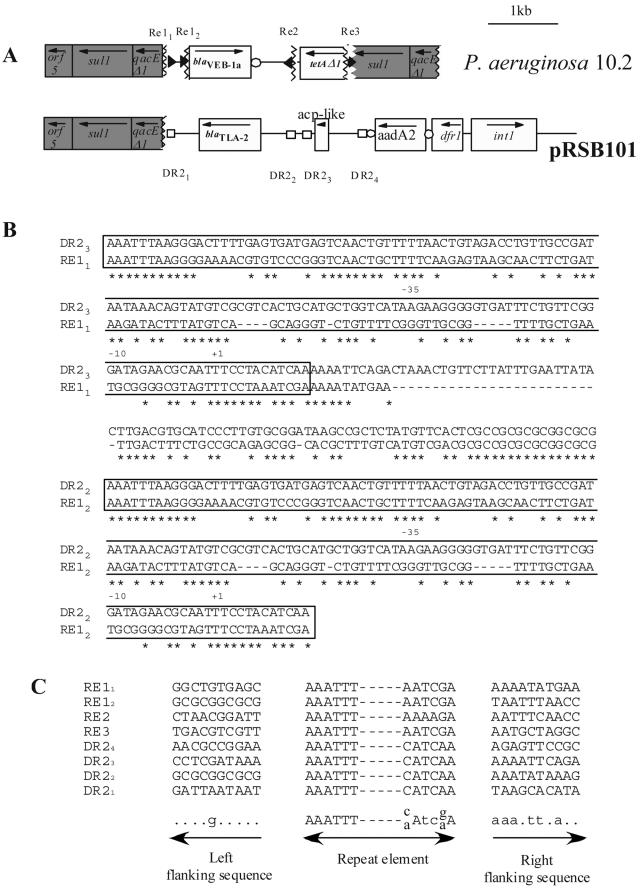
When we blasted the Re1 sequence against the GenBank database, a 37% identity hit was found with another repeat element, DR2, recently described for a plasmid isolated from an uncultivated bacterium of activated sludge (40). This DR2 sequence was found on both sides of the blaTLA-2 gene that codes for an ESBL (Fig. 2A) (14). Even though the overall identity was low, strong conservations were found at the extremities of both elements (Fig. 2B). Most interestingly, the promoter characterized in Re1 seems to be conserved in DR2 elements as well, suggesting that the DR2 elements may also serve as promoter sequences for blaTLA-2 gene expression. Moreover, the intervening sequence between the two adjacent Re1s and that between DR2.2 and DR2.3 display significant sequence identity (Fig. 2B). When we analyzed the flanking sequences of these elements, no obvious consensus sequences were obtained, with the exception of the right-hand sequence that was adenine and thymidine rich (Fig. 2C). Furthermore, as for P. aeruginosa 10.2, the qacEΔ1 gene cassette present in the 3′-CS of the class 1 integron was also truncated by the insertion of DR2 in plasmid pRSB101, suggesting that this sequence might be a hot spot for insertion of these Re or DR2 sequences.
FIG. 2.
(A) Schematic representations of the genetic environment of the blaVEB-1 gene in P. aeruginosa 10.2 and blaTLA-2 gene from plasmid pRSB101. The flanking sequence identity breakpoints in the blaVEB-1-like gene environment are designated by vertical broken lines. The homology breakpoints are designated by broken lines. The Re of the blaVEB-1 gene environment and the DR2 elements are represented by black triangles and white squares, respectively. The coding regions are shown as boxes, with an arrow indicating the orientation of transcription. Dark-gray filled boxes correspond to genes or ORFs present on class 1 integron conserved sequences. White circles indicate 59-be's. (B) Nucleotide alignment of the Re1-Re1 region of P. aeruginosa 10.2 and the DR2.3-DR2.2 region from plasmid pRSB101 (40). Re and DR2 elements are boxed. Dashes represent gaps that have been introduced into the alignment. Stars represent identical positions between the two sequences. The +1, −10, and −35 written below the sequences correspond to promoter sequences identified by Aubert et al. (2). (C) Nucleotide alignment of the flanking sequences of the different Re and DR2 elements. A consensus sequence is written below the alignment. Capital letters represent highly conserved positions (eight out of eight), and small letters represent positions with moderate conservations (five out of eight). Dots represent polymorphic positions.
Sequencing of the region located downstream of the blaVEB-1a gene cassette revealed sequence identity with those found in P. aeruginosa 10.2 and P. stuartii BI over 500 bp, up to an 8-bp sequence (AGCAAATT) (Fig. 1A, B, and C). Interestingly, these base pairs are also present in front of Re1 and thus could be reminiscent of blaVEB-1a insertion. Unlike what was found in P. aeruginosa 10.2 and P. stuartii BI (Fig. 1A and B), no other Re sequence was found, but instead, several open reading frames (ORFs) were found, some of which shared significant sequence identity with transposase genes. The first ORF of 528 bp that coded for a putative 176-amino-acid polypeptide of unknown function was interrupted by a 2,436-bp-long insertion sequence, ISPa31, which belongs to the IS66 family of insertion elements (Fig. 1C). ISPa31 was itself disrupted by another 1,671-bp-long insertion element, ISPa30, which is an IS3 family member (Fig. 1C). Just downstream, the aph-A6 gene was present in opposite orientation, immediately followed by an orf513 recombinase gene that is part of the common region CR1 (Fig. 1) (38). The aminoglycoside phosphotransferase Aph-A6 mediates resistance to kanamycin and structurally related aminoglycosides, including amikacin (21). The expression of AphA-6 in P. aeruginosa TL-1 could explain the high level of resistance to amikacin in that isolate. A 47-bp stretch corresponding to the right end of ISAba125 was detected between the aphA-6 gene and the orf513 gene (22).
In order to investigate the sequences located on either side of the cloned PstI insert, PCR primers located within the PstI insert and in presumed flanking sequences were used. At the left-hand side, the end of the sul1 gene, followed by the orf5 sequence, was found as expected. At the right-hand side of the cloned PstI insert, the end of orf513, followed by a sul1 and qacEΔ1 gene, was also present (Fig. 1C). In order to investigate whether these two latter genes belong to a class 1 integron, orf513-, sul1-, and 3′-CS-specific primers were used together with 5′-CS primer. The 5′-CS primer in combination with any of the other primers allowed for partial amplification of a class 1 integron that contained two gene cassettes. The sequence of the variable region of that integron was identical to that of In51, an integron previously identified in P. aeruginosa JES that also harbored In50, a blaVEB-1-containing integron (26, 27). The first gene cassette corresponded to aadA6, an aminoglycoside adenylyltransferase gene, and the second gene cassette, the OrfD cassette, codes for a polypeptide of unknown function (Fig. 1C). Thus, this novel structure was termed In121, encompassing the In51 structure up to the second 3′-CS.
The presence of two truncated 3′-CS has already been described in several works, especially for In34 by Partridge and Hall (29). The central region of In34 contained an ORF (orf513) designated CR1 and a trimethoprim resistance gene (dfrA10) that may have been acquired by homologous recombination, generating two truncated 3′-CS, called 3′-CS1 and 3′-CS2, on each side (29). However, in In34, no Re-like sequences have been characterized between the 3′-CS1 and 3′-CS2. In P. aeruginosa 10.2 and P. stuartii BI, two 3′-CS have been found in direct repeat, but no orf513 or CR-like sequences have been found. In In121, both orf513 with CR1 sequences and two CR1 sequences in direct-repeat 3′-CS are present (2, 3). Thus, In121 fulfilled the requirements of a sul-type integron.
In121 is the first CR1-borne sul-type integron characterized in P. aeruginosa. Another CR element, CR4, which has ca. 50% identity with CR1, has been described once for a P. aeruginosa strain from Brazil (31). Since CR1 has been characterized essentially in enterobacterial species (29), the finding in P. aeruginosa suggested gene transfer between these bacterial species.
Characterization of other integron-borne resistance genes.
Using 5′-CS and 3′-CS primers, we obtained two amplicons from P. aeruginosa TL-1 of 1.3 kb and 1.8 kb, respectively, while from P. aeruginosa TL-2, we obtained only the 1.8-kb fragment (Fig. 1C and D). The smaller product corresponded to the variable region of In51, while the 1.8-kb fragment contained three ORFs that have never been characterized together on the same integron: (i) aadB, coding for a 2′′-O-aminoglycoside nucleotidyltransferase (25, 42), (ii) dfrA1, a dihydrofolate reductase gene (40), and (iii) OrfX, an ORF of unknown function (39). This novel integron was termed In122.
Mapping of aphA-6 transcription start site.
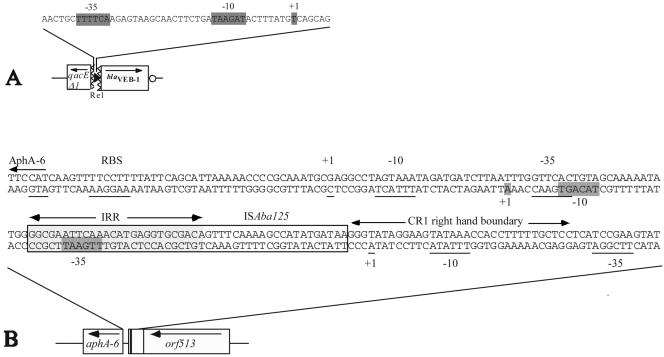
In the 5′ RACE PCR experiments, the site of initiation of transcription of the aphA-6 gene in P. aeruginosa TL-1 was mapped to be 67 bp upstream of the translational start codon (Fig. 3B). Upstream of this transcriptional start site, a −35 promoter sequence (TTGAAT) was found, and this was separated by 17 bp from a −10 promoter sequence (TACAGT) (Fig. 3B). This promoter was a composite promoter made up of a −35 sequence located in the right inverted repeat of ISAba125 (22) and of a −10 sequence located in the flanking sequence of the aphA-6 gene (21). Unlike what was found for expression of the plasmid-mediated quinolone resistance qnr gene in E. coli (20), no PCR product specific for the CR1-located promoter and no amplification product specific for the presumed aphA-6 promoter were found.
FIG. 3.
Characterization of transcriptional start sites. The −10 and −35 promoter sequences as well as the +1 transcriptional initiation site determined by 5′ RACE are indicated by shaded boxes. Panel A represents the promoter sequence upstream of blaVEB-1a and located in Re1 as determined by Aubert et al. (2). Panel B represents the promoter region of the aphA-6 gene. The +1, −10, and −35 written above the sequence correspond to promoter sequences suggested by Martin et al. (21), whereas those written below the sequence and located in the CR1 right-hand boundary are as described by Mammeri et al. (20).
Strain typing.
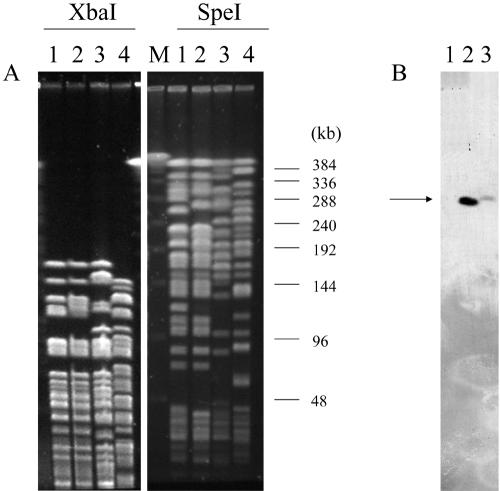
PFGE analysis using XbaI revealed only slight differences between the two P. aeruginosa TL-1 and TL-2 isolates, while they differed significantly from the two other VEB-producing P. aeruginosa isolates (Fig. 4A). However, using SpeI-restricted genomic DNA, we found that P. aeruginosa TL-1 differed by five bands from P. aeruginosa TL-2, while the two other P. aeruginosa isolates were unrelated (Fig. 4A). Similar results were obtained using two other typing techniques: multiple-locus variable number of tandem repeats (28) and random amplified polymorphic DNA (19; data not shown).
FIG. 4.
Molecular comparison of P. aeruginosa isolates. PFGE with XbaI-restricted (left panel) and SpeI-restricted (right panel) DNA (A) and blaVEB hybridization of the SpeI PFGE gel (B). Lane 1, P. aeruginosa TL-2; lane 2, P. aeruginosa TL-1; lane 3, P. aeruginosa 10.2 (2); and lane 4, P. aeruginosa JES (26). Molecular weight markers correspond to the lambda ladder (Bio-Rad).
Comparative analysis of the SpeI-restricted chromosomal DNA separated by PFGE revealed that the size of the P. aeruginosa TL-1 genome was ca. 100 kb larger than that of P. aeruginosa TL-2. These results indicated that both isolates were structurally related and that P. aeruginosa TL-2 lacked a ca. 100-kb fragment containing several antibiotic resistance genes.
The SpeI-restricted DNAs of P. aeruginosa TL1, TL2, and 10.2 were transferred onto a nylon membrane and hybridized with an internal blaVEB-1-specific probe. A hybridization signal of high molecular weight (ca. 290 kb) (Fig. 4B) was detected with P. aeruginosa TL-1 and 10.2. This result is in agreement with a likely chromosomal location of the blaVEB-1 gene.
Conclusions.
This is the first report describing a blaVEB-1a gene located on a sul-type class 1 integron in a P. aeruginosa clinical isolate. This work further underlines the global spread of blaVEB-1-like genes on different genetic structures (integrons, Re, and sul-type integrons). The presence of single 135-bp Re sequences upstream of the blaVEB-1a gene was responsible for blaVEB-1 expression (Fig. 3A) but might also be implicated in mobilization, since an 8-bp target duplication was found on both sides of the inserted fragment. This hypothesis is further supported by the structural similarity between Re and DR elements.
Finally, this work identified a novel combination of genetic elements associated with antibiotic resistance genes: sul-type integrons, CR1 elements, and Re. Most interestingly, this combination of genetic elements resulted in resistance to β-lactams and to aminoglycosides, the main classes of antibiotics used for treating clinically significant gram-negative infections.
Acknowledgments
We thank L. Poirel for helpful discussion.
T.N. and D.A. contributed equally to this work.
This work was funded by a grant from the Ministère de la Recherche (grant UPRES-EA 3539), Université Paris XI, Paris, France, and mostly by the European Community (6th PCRD, LSHM-CT-2003-503-335).
REFERENCES
- 1.Ambler, R. P., A. F. Coulson, J.-M. Frere, J. M. Ghuysen, B. Joris, M. Forsman, R. C. Levesque, G. Tiraby, and S. G. Waley. 1991. A standard numbering scheme for the class A beta-lactamases. Biochem. J. 276:269-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aubert, D., D. Girlich, T. Naas, S. Nagarajan, and P. Nordmann. 2004. Functional and structural characterization of the genetic environment of an extended spectrum β-lactamase blaVEB gene from a Pseudomonas aeruginosa isolate obtained in India. Antimicrob. Agents Chemother. 48:3284-3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aubert, D., T. Naas, M.-F. Lartigue, and P. Nordmann. 2005. Novel genetic structure associated with an extended-spectrum β-lactamase blaVEB gene in a Providencia stuartii clinical isolate from Algeria. Antimicrob. Agents Chemother. 49:3590-3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao, V., T. Lambert, D. Q. Nhu, H. K. Loan, N. K. Hoang, G. Arlet, and P. Courvalin. 2002. Distribution of extended-spectrum β-lactamases in clinical isolates of Enterobacteriaceae in Vietnam. Antimicrob. Agents Chemother. 46:3739-3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carbonne, A., T. Naas, K. Blanckaert, C. Couzigou, C. Cattoen, J. L. Chagnon, P. Nordmann, and P. Astagneau. 2005. Investigation of a nosocomial outbreak of extended-spectrum beta-lactamase VEB-1-producing isolates of Acinetobacter baumannii in a hospital setting. J. Hosp. Infect. 60:14-18. [DOI] [PubMed] [Google Scholar]
- 7.Chanawong, A., F. H. M'Zali, J. Heritage, A. Lulitanond, and P. M. Hawkey. 2001. SHV-12, SHV-5, SHV-2a and VEB-1 extended-spectrum beta-lactamases in Gram-negative bacteria isolated in a university hospital in Thailand. J. Antimicrob. Chemother. 48:839-852. [DOI] [PubMed] [Google Scholar]
- 8.Chen, H. Y., M. Yuan, and D. M. Livermore. 1995. Mechanisms of resistance to β-lactam antibiotics amongst Pseudomonas aeruginosa isolates collected in the UK in 1993. J. Med. Microbiol. 43:300-309. [DOI] [PubMed] [Google Scholar]
- 9.Clinical and Laboratory Standards Institute. 2005. Performance standards for antimicrobial susceptibility testing; 15th informational supplement. CLSI/NCCLS M100-S15. Clinical and Laboratory Standards Institute, Wayne, Pa.
- 10.Collis, C. M., and R. M. Hall. 1995. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob. Agents Chemother. 39:155-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Champs, C., L. Poirel, R. Bonnet, D. Sirot, C. Chanal, J. Sirot, and P. Nordmann. 2002. Prospective survey of β-lactamases produced by ceftazidime-resistant Pseudomonas aeruginosa isolated in a French hospital in 2000. Antimicrob. Agents Chemother. 46:3031-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Girlich, D., T. Naas, A. Leelaporn, L. Poirel, M. Fennewald, and P. Nordmann. 2002. Nosocomial spread of the integron-located veb-1-like cassette encoding an extended-spectrum β-lactamase in Pseudomonas aeruginosa in Thailand. Clin. Infect. Dis. 34:603-611. [DOI] [PubMed] [Google Scholar]
- 13.Girlich, D., L. Poirel, A. Leelaporn, A. Karim, C. Tribuddharat, M. Fennewald, and P. Nordmann. 2001. Molecular epidemiology of the integron-located VEB-1 extended-spectrum β-lactamase in nosocomial enterobacterial isolates in Bangkok, Thailand. J. Clin. Microbiol. 39:175-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Girlich, D., L. Poirel, A. Schlüter, and P. Nordmann. 2005. TLA-2, a novel Ambler class A expanded-spectrum β-lactamase. Antimicrob. Agents Chemother. 49:4767-4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang, X., Y. Ni, Y. Jiang, F. Yuan, L. Han, M. Li, H. Liu, L. Yang, and Y. Lu. 2005. Outbreak of infection caused by Enterobacter cloacae producing the novel VEB-3 beta-lactamase in China. J. Clin. Microbiol. 43:826-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kieser, T. 1984. Factors affecting the isolation of cccDNA from Streptomyces lividans and Escherichia coli. Plasmid 12:19-36. [DOI] [PubMed] [Google Scholar]
- 17.Kim, J. Y., Y. J. Park, S. I. Kim, M. W. Kang, S. O. Lee, and K. Y. Lee. 2004. Nosocomial outbreak by Proteus mirabilis producing extended-spectrum beta-lactamase VEB-1 in a Korean university hospital. J. Antimicrob. Chemother. 54:1144-1147. [DOI] [PubMed] [Google Scholar]
- 18.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop II, and K. M. Peterson. 1994. PBBR1MCS: a broad-host-range cloning vector. BioTechniques 16:800-802. [PubMed] [Google Scholar]
- 19.Mahenthiralingam, E., M. E. Campbell, J. Foster, J. S. Lam, and D. P. Speert. 1996. Random amplified polymorphic DNA typing of Pseudomonas aeruginosa isolates recovered from patients with cystic fibrosis. J. Clin. Microbiol. 34:1129-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mammeri, H., M. Van De Loo, L. Poirel, L. Martinez-Martinez, and P. Nordmann. 2005. Emergence of plasmid-mediated quinolone resistance in Escherichia coli in Europe. Antimicrob. Agents Chemother. 49:71-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin, P., E. Jullien, and P. Courvalin. 1988. Nucleotide sequence of Acinetobacter baumannii aphA-6 gene: evolutionary and functional implications of sequence homologies with nucleotide-binding proteins, kinases and other aminoglycoside-modifying enzymes. Mol. Microbiol. 2:615-625. [DOI] [PubMed] [Google Scholar]
- 22.Mussi, M. A., A. S. Limansky, and A. M. Viale. 2005. Acquisition of resistance to carbapenems in multidrug-resistant clinical strains of Acinetobacter baumannii: natural insertional inactivation of a gene encoding a member of a novel family of β-barrel outer membrane proteins. Antimicrob. Agents Chemother. 49:1432-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naas, T., D. Aubert, N. Fortineau, and P. Nordmann. 2002. Cloning and sequencing of the beta-lactamase gene and surrounding DNA sequences of Citrobacter braakii, Citrobacter murliniae, Citrobacter werkmanii, Escherichia fergusonii and Enterobacter cancerogenus. FEMS Microbiol. Lett. 215:81-87. [DOI] [PubMed] [Google Scholar]
- 24.Naas, T., F. Benaoudia, S. Massuard, and P. Nordmann. 2000. Integron-located VEB-1 extended-spectrum β-lactamase gene in a Proteus mirabilis clinical isolate from Vietnam. J. Antimicrob. Chemother. 46:703-711. [DOI] [PubMed] [Google Scholar]
- 25.Naas, T., Y. Mikami, T. Imai, L. Poirel, and P. Nordmann. 2001. Characterization of In53, a class 1 plasmid- and composite transposon-located integron of Escherichia coli which carries an unusual array of gene cassettes. J. Bacteriol. 183:235-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naas, T., L. Poirel, A. Karim, and P. Nordmann. 1999. Molecular characterization of In50, a class 1 integron encoding the gene for the extended-spectrum β-lactamase VEB-1 in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 176:411-419. [DOI] [PubMed] [Google Scholar]
- 27.Naas, T., L. Poirel, and P. Nordmann. 1999. Molecular characterisation of In51, a class 1 integron containing a novel aminoglycoside adenylyltransferase gene cassette, aadA6, in Pseudomonas aeruginosa. Biochim. Biophys. Acta 1489:445-451. [DOI] [PubMed] [Google Scholar]
- 28.Onteniente, L., S. Brisse, P. T. Tassios, and G. Vergnaud. 2003. Evaluation of the polymorphisms associated with tandem repeats for Pseudomonas aeruginosa strain typing. J. Clin. Microbiol. 41:4991-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Partridge, S. R., and R. M. Hall. 2003. In34, a complex In5 family class 1 integron containing orf513 and dfrA10. Antimicrob. Agents Chemother. 47:342-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Philippon, L. N., T. Naas, A.-T. Bouthors, V. Barakett, and P. Nordmann. 1997. OXA-18, a class D clavulanic acid-inhibited extended-spectrum β-lactamase from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 41:2188-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poirel, L., M. Magalhaes, M. Lopes, and P. Nordmann. 2004. Molecular analysis of metallo-β-lactamase gene blaSPM-1-surrounding sequences from disseminated Pseudomonas aeruginosa isolates in Recife, Brazil. Antimicrob. Agents Chemother. 48:1406-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poirel, L., O. Menuteau, N. Agoli, C. Cattoen, and P. Nordmann. 2003. Outbreak of extended-spectrum β-lactamase VEB-1-producing isolates of Acinetobacter baumannii in a French hospital. J. Clin. Microbiol. 41:3542-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poirel, L., T. Naas, M. Guibert, E. B. Chaibi, R. Labia, and P. Nordmann. 1999. Molecular and biochemical characterization of VEB-1, a novel class A extended-spectrum β-lactamase encoded by an Escherichia coli integron gene. Antimicrob. Agents Chemother. 43:573-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poirel, L., V. O. Rotimi, E. M. Mokaddas, A. Karim, and P. Nordmann. 2001. VEB-1-like extended-spectrum beta-lactamases in Pseudomonas aeruginosa, Kuwait. Emerg. Infect. Dis. 7:468-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poirel, L., G. F. Weldhagen, C. De Champs, and P. Nordmann. 2002. A nosocomial outbreak of Pseudomonas aeruginosa isolates expressing the extended-spectrum beta-lactamase GES-2 in South Africa. J. Antimicrob. Chemother. 49:561-565. [DOI] [PubMed] [Google Scholar]
- 36.Recchia, G. D., and R. M. Hall. 1995. Gene cassettes: a new class of mobile elements. Microbiology 141:3015-3027. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Stokes, H. W., C. Tomaras, Y. Parsons, and R. M. Hall. 1993. The partial 3′-conserved segment duplications in the integrons In6 from pSa and In7 from pDGO100 have a common origin. Plasmid 30:39-50. [DOI] [PubMed] [Google Scholar]
- 39.Sundstroem, L., and O. Skoeld. 1990. The dhfrI trimethoprim resistance gene of Tn7 can be found at specific sites in other genetic surroundings. Antimicrob. Agents Chemother. 34:642-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szczepanowski, R., I. Krahn, B. Linke, A. Goesmann, A. Puhler, and A. Schlüter. 2004. Antibiotic multiresistance plasmid pRSB101 isolated from a wastewater treatment plant is related to plasmids residing in phytopathogenic bacteria and carries eight different resistance determinants including a multidrug transport system. Microbiology 150:3613-3630. [DOI] [PubMed] [Google Scholar]
- 41.Tenover, F. C., R. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tenover, F. C., D. Filpula, K. L. Phillips, and J. J. Plorde. 1988. Cloning and sequencing of a gene encoding an aminoglycoside 6′-N-acetyltransferase from an R factor of Citrobacter diversus. J. Bacteriol. 170:471-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tribuddharat, C., and M. Fennewald. 1999. Integron-mediated rifampin resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:960-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vahaboglu, H., R. Ozturk, G. Aygun, F. Coskunkan, A. Yaman, A. Kaygusuz, H. Leblebicioglu, I. Balik, K. Aydin, and M. Otkun. 1997. Widespread detection of PER-1-type extended-spectrum β-lactamases among nosocomial Acinetobacter and Pseudomonas aeruginosa isolates in Turkey: a nationwide multicenter study. Antimicrob. Agents Chemother. 41:2265-2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vedel, G. 2005. Simple method to determine beta-lactam resistance phenotypes in Pseudomonas aeruginosa using the disc agar diffusion test. J. Antimicrob. Chemother. 56:657-664. [DOI] [PubMed] [Google Scholar]
- 46.Vivian, A. 1994. Plasmid expansion? Microbiology 140:2188-2195. [DOI] [PubMed] [Google Scholar]
- 47.Weldhagen, G. F., L. Poirel, and P. Nordmann. 2003. Ambler class A extended-spectrum β-lactamases in Pseudomonas aeruginosa: novel developments and clinical impact. Antimicrob. Agents Chemother. 47:2385-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yong, D., J. H. Shin, S. Kim, Y. Lim, J. H. Yum, K. Lee, Y. Chong, and A. Bauernfeind. 2003. High prevalence of PER-1 extended-spectrum β-lactamase-producing Acinetobacter spp. in Korea. Antimicrob. Agents Chemother. 47:1749-1751. [DOI] [PMC free article] [PubMed] [Google Scholar]