Abstract
Filamentous fungi are widely used for the production of homologous and heterologous proteins. Recently, there has been increasing interest in Aspergillus oryzae because of its ability to produce heterologous proteins in solid-state culture. To provide an overview of protein secretion by A. oryzae in solid-state culture, we carried out a comparative proteome analysis of extracellular proteins in solid-state and submerged (liquid) cultures. Extracellular proteins prepared from both cultures sequentially from 0 to 40 h were subjected to two-dimensional electrophoresis, and protein spots at 40 h were identified by peptide mass fingerprinting using matrix-assisted laser desorption ionization-time-of-flight mass spectrometry. We also attempted to identify cell wall-bound proteins of the submerged culture. We analyzed 85 spots from the solid-state culture and 110 spots from the submerged culture. We identified a total of 29 proteins, which were classified into 4 groups. Group 1 consisted of extracellular proteins specifically produced in the solid-state growth condition, such as glucoamylase B and alanyl dipeptidyl peptidase. Group 2 consisted of extracellular proteins specifically produced in the submerged condition, such as glucoamylase A (GlaA) and xylanase G2 (XynG2). Group 3 consisted of proteins produced in both conditions, such as xylanase G1. Group 4 consisted of proteins that were secreted to the medium in the solid-state growth condition but trapped in the cell wall in the submerged condition, such as α-amylase (TAA) and β-glucosidase (Bgl). A Northern analysis of seven genes from the four groups suggested that the secretion of TAA and Bgl was regulated by trapping these proteins in the cell wall in submerged culture and that secretion of GlaA and XynG2 was regulated at the posttranscriptional level in the solid-state culture.
The filamentous fungus Aspergillus oryzae has been used in the production of traditional fermented foods such as sake (rice wine), miso (soybean paste), and shoyu (soy sauce) for more than 1,000 years (4). Because of its long use in food production, A. oryzae is listed as GRAS (i.e., generally regarded as safe) status by the U.S. Food and Drug Administration. The safety of A. oryzae is also supported by the World Health Organization (22). A. oryzae has the ability to secrete large amounts of a wide range of different enzymes into its environment, and this characteristic has been used for commercial production of both recombinant enzyme and its own enzyme. Because of its high-level production of enzymes and high degree of safety, A. oryzae, as well as Aspergillus niger and Aspergillus sojae, has received increasing attention as a host for homologous (5, 11) and heterologous (15) protein production. Many recombinant proteins have been produced in submerged culture for easy cultivation. However, there has been growing interest in solid-state culture because the amounts of enzymes secreted by filamentous fungi in solid-state culture are large and frequently exceed those secreted in submerged culture (1, 8, 9, 25). In Japan, several commercial enzymes are produced in solid-state culture in addition to by traditional fermentation methods. In solid-state culture on wheat bran, A. oryzae produces a 500-fold-higher yield of heterologous protein (chymosin) than it does in submerged culture (34). The secretion behavior of A. oryzae in the solid state can also be used as a model for many filamentous fungi. Growth of the fungus in solid-state culture is closer to natural conditions than growth in a submerged culture. Because of this, enzyme secretion in solid-state culture more closely resembles enzyme secretion under natural conditions. Because of the importance of solid-state culture for fermentation and production of commercial enzymes, the solid-state culture has been the subject of many molecular biological studies. The solid-state-specific glucoamylase gene (glaB) has a functional cis element of the glaB promoter, which responds to solid-specific induction (13, 16). The promoter has two heat shock element motifs (f-HSE and b-HSE) and a GC-rich motif (GC box). The GC box has been suggested to have a role in regulating the solid-state-specific expression of the glaB gene.
The pepA gene, which encodes an acid protease, is another gene that is expressed specifically in solid-state culture (21). Unlike glaB gene expression, pepA expression is temperature dependent. The promoter region of pepA was not found to contain a GC box but instead contains a heat shock element. In solid-state culture, the molecular mechanism for the regulation of gene expression might be very complicated.
The expression of many genes has been shown to be affected by culture conditions. Subtractive cloning of cDNA from A. oryzae was carried out in solid-state and submerged cultures, and many genes specifically expressed in the solid state were identified (2). The expression levels of catabolic genes of the glycolytic pathway and the tricarboxylic acid (TCA) cycle in solid-state culture were found to be lower than those in submerged culture, causing a release from catabolite repression and consequently leading to high-level expression of hydrolytic enzymes (24).
Although the secretion of individual proteins in submerged culture has been well analyzed, the secretion of extracellular proteins specifically produced in solid-state culture has not been examined. Therefore, little is known about extracellular proteins in solid-state culture.
To date, proteomic analysis has proven to be the most powerful method for identification of proteins and is suitable for studying how protein expression in an organism changes under various environmental conditions (6). Together with transcriptional analysis, proteomic analysis reveals features of posttranscriptional secretion regulation and also clarifies posttranscriptional modification of extracellular proteins. In addition, expressed sequence tags (ESTs) of A. oryzae have been reported by the A. oryzae genome consortium, and a draft genome sequencing project has been completed (23). This technology and valuable data allowed us to carry out the analysis of extracellular proteins of A. oryzae in solid-state culture.
In this study, we carried out a comparative proteome analysis of extracellular proteins under solid-state and submerged culture conditions and determined protein secretion profiles. We classified four groups of protein according to their secretion profiles. Northern analysis of seven genes from the four groups revealed posttranscriptional control for secretion of extracellular proteins under both growth conditions.
MATERIALS AND METHODS
Fungal strain and culture conditions.
Aspergillus oryzae strain RIB40 was employed for cultivation, protein extraction, and total RNA isolation. As a solid-state medium, autoclaved wheat bran with 100% water content was used. An A. oryzae RIB40 conidial suspension was inoculated onto 50 g of solid-state medium (giving 1.0 × 108 conidia/g wheat bran) and incubated at 30°C in 95% humidity. The culture was mixed every 12 h. Submerged culture was carried out as follows. A 6% (wt/vol) wheat bran suspension was autoclaved at 121°C for 15 min, cooled to room temperature, and then filtered through Miracloth (Calbiochem). Twofold-diluted filtrate containing 1% (NH4)2SO4 and 3% KH2PO4 was used as the medium for submerged culture. A conidial suspension was inoculated into 150 ml of this medium in a 500-ml baffle flask (giving 1 × 106 conidia/ml) and then cultured at 30°C with rotary shaking (120 rpm).
The mycelium content in the solid-state culture was determined by measuring the N-acetylglucosamine content by the method of Gomi et al. (10) with an Aspergillus assay kit (Kikkoman). The standard unit was determined with release of N-acetylglucosamine from dried mycelium of A. oryzae RIB40 from submerged culture by cell wall-degrading enzyme according to the manufacturer's instructions.
Preparation of proteins from A. oryzae culture. (i) Extracellular proteins.
The protein concentration in each step was determined by the Bradford protein assay (Bio-Rad) according to the manufacturer's instructions.
For extracellular protein preparation from solid-state culture, 10 g of cultured wheat bran was soaked for 8 h in 50 ml of 50 mM acetate buffer (pH 5.0) containing 90 mM NaCl, 1 mM phenylmethylsulfonyl fluoride (PMSF), and a protease inhibitor cocktail (Complete Mini; Roche). The suspension was centrifuged at 16,000 × g for 10 min twice to remove the large particles. The supernatant was then filtered through a 0.45-μm filter (Millipore).
To prepare extracellular protein from submerged culture, the culture broth was centrifuged at 12,000 × g for 20 min, and the supernatant was filtered through a 0.45-μm filter.
For two-dimensional (2-D) electrophoresis, protein samples were prepared as follows. For each sample from solid-state and submerged cultures, (NH4)2SO4 was added to the filtrate to a concentration of 95% saturation, and the solution was kept at 4°C overnight. The protein was precipitated by centrifugation at 12,000 × g for 30 min. The precipitate was dissolved in 2 ml of 50 mM acetate buffer (pH 5.0) containing 1 mM PMSF and Complete Mini. The protein solution was desalted with the same buffer and concentrated by ultrafiltration with an Amicon Ultra 10,000-molecular-weight-cutoff filter (Millipore). The protein concentration was determined by the Bio-Rad protein assay. The protein solution was used as the sample for 2-D electrophoresis.
(ii) Preparation of cell wall-bound protein from submerged culture.
Mycelia from the submerged culture were harvested with Miracloth, washed with ice-cold water, washed again with ice-cold 50 mM acetate buffer (pH 5.0) containing 90 mM NaCl, and dried completely with a paper towel. Three grams of mycelium was soaked in 10 ml of 50 mM acetate buffer containing 1 mM PMSF and Complete Mini (buffer A) and disrupted with a Multi beads shocker (Yasuikikai, Japan) (2,500 rpm; 1 min on, 1 min off) for 50 min at 4°C. The homogenate was centrifuged at 2,400 × g for 5 min, and the supernatant was discarded. The cell wall fraction was washed three times with 50 mM acetate buffer A containing 1 mM PMSF and then was washed three times in 30 ml of 4 M LiCl by centrifugation at 2,400 × g for 5 min. The mycelium debris was washed twice with buffer A containing 1% Tween 80, and then Tween 80 was washed out with buffer A three times. The cell wall fraction was resuspended in 1.5 ml of 1 M Tris-HCl (pH 8.0), 0.15 mM EDTA, 3% sodium dodecyl sulfate (SDS), 1.2 mM dithiothreitol (DTT) and boiled for 10 min. The supernatant was then obtained after centrifugation at 2,400 × g for 10 min at room temperature. The supernatant containing cell wall protein was desalted with Amicon Ultra 10,000-molecular-weight-cutoff ultrafiltration tubes. SDS in the concentrated cell wall protein was washed out with an SDS Out kit (Pierce). The protein solution obtained was used as the protein sample for 2-D electrophoresis.
2-D gel electrophoresis (differential display). (i) Isoelectric focusing and 2-D polyacrylamide gel electrophoresis (PAGE) conditions.
The protein solution (300 μg protein in a 350-μl solution containing 0.5% immobilized pH gradient [IPG] buffer, 8 M urea, 18 mM DTT, 2% Triton X-100, bromophenol blue) was loaded by rehydration onto an 18-cm-long, pH 3 to 10 or 4 to 7 IPG strip (Amersham Pharmacia) for 12 h at 20°C. Isoelectric focusing was carried out with Multiphor (Amersham Pharmacia) for a total of 37.5 kV · h (1 h at 500 V, 1 h at 1,000 V, 1 h at 4,000 V, and 4 h at 8,000 V). After focusing, the gels were reduced (2% DTT) for 30 min at room temperature and then alkylated (2.5% iodoacetamide) for 30 min at room temperature in equilibration buffer (6 M urea, 50 mM Tris-HCl [pH 6.8], 30% glycerol, 2% SDS). The gel strips were transferred onto a 12% Ettan DALT II precasting gel (Amersham Pharmacia), and the second-dimension separation was carried out on an Ettan DALT 6 electrophoresis system (Amersham Pharmacia) at constant current (24 mA/gel). The gels were fixed and stained with Coomassie brilliant blue (CBB) R-250.
(ii) Image analysis.
Two-dimensional images were captured by scanning CBB-stained gels using a GS-800 imaging densitometer (Bio-Rad) and were digitized with Multi Analyst software (Bio-Rad). Different two-dimensional images were processed, including detection and volumetric quantification using PDQuest version 7.1 software (Bio-Rad).
(iii) HPLC separation of solid-state extracellular protein.
Solid-state extracellular protein (10 mg) in 20 mM acetate buffer (pH 5.0) was applied to a TSK gel Phenyl 5-PW column (21.5-mm [inner diameter] by 15 cm) (Tosoh) in a high-pressure liquid chromatography (HPLC) system (Shimadzu). The sample was washed with the same buffer containing 2.0 M NH4(SO4)2 for 20 min at a flow rate of 3.0 ml/min and then eluted with a linear gradient of NH4(SO4)2 (2.0 to 0 M) in 20 mM acetate buffer (pH 5.0) at a flow rate of 3.0 ml/min for 60 min. Protein was monitored at 280 nm. α-Amylase activity was assayed by monitoring the amount of reducing sugars released from starch by using an α-amylase activity assay kit (Kikkoman). Each fraction was concentrated with Biomax (UFV2BCC40) ultrafiltration tubes (Millipore) and subjected to SDS-PAGE on a 10 to 20% gradient gel. The gel was stained with CBB. Precision Plus protein standards (Bio-Rad) were used for molecular mass determination. Each band was excised and subjected to matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS) for identification.
MALDI-TOF mass spectrometry. (i) Sample preparation.
Spots of interest were manually excised from Coomassie blue-stained 2-D electrophoresis gels. The gel pieces were destained with 50% acetonitrile in 25 mM ammonium bicarbonate (Ambic) twice, washed with 100% acetonitrile, and vacuum dried. The gel pieces were reduced with 10 mM dithiothreitol in 25 mM Ambic for 1 h at 55°C, washed with 25 mM Ambic, and then alkylated with 55 mM iodoacetamide in 25 mM Ambic at room temperature for 45 min in the dark. Gel pieces were washed with 25 mM Ambic twice, followed by acetonitrile, and dried under vacuum. After that, the excised gel pieces were in-gel deglycosylated by treatment with 0.5 mU glycopeptidase F (PNGase F; TaKaRa) in 25 mM Ambic overnight at 37°C. Glycans were removed from the gel pieces by two changes of 25 mM Ambic with sonication for 10 min. We call this deglycosylation procedure the “in-gel deglycosylation method.” Gel pieces were dried by shrinking in acetonitrile followed by vacuum drying. All gel pieces were incubated with 12.5 ng/μl sequencing-grade modified trypsin (Promega) in 25 mM Ambic for 12 h at 37°C. After digestion, the supernatants were recovered. Peptides were extracted from the gel pieces twice with a 0.1% trifluoroacetic acid-50% acetonitrile solution with 10 min each of sonication and vortexing. All extracts were pooled, and the volume was reduced with a Speed Vac. Extracted peptides were purified with a Zip tip (C18; Millipore) as described in the instruction manual.
(ii) MALDI-TOF MS.
A 2-μl drop of sample was spotted onto a MALDI target, and then 1 μl of matrix solution (0.33 mg/ml α-cyano-4-hydroxycinnapic acid matrix [Bruker] in acetone-ethanol [1:2]) was added to the drop. MALDI-TOF MS analyses were performed on an Autoflex mass spectrometer (Bruker Daltonics). Peptides were selected in the mass range of 800 to 3,500 Da. All mass spectra were calibrated externally with a peptide mass standard kit (Bruker) and internally with trypsin autolysis peaks. Tandem MS (MS/MS) sequencing analyses were carried out with an Ultraflex spectrometer (Bruker Daltonics).
(iii) Database search.
The peptide mass fingerprinting and peptide fragment ion data obtained from MALDI-TOF and MS/MS analyses were used to search for protein candidates in the NCBI database and the A. oryzae EST and genome databases by using Mascot (Matrix Science) software programs. Information about coding sequences and proteins was obtained from the A. oryzae EST database (http://www.nrib.go.jp/ken/EST/db/index.html) and genome database (http://www.bio.nite.go.jp/dogan/Top). The genome database was developed by the Aspergillus oryzae genomic consortium (23). Initial search parameters were set as follows: Cys as the S-carbamidomethyl derivative and Met in oxidized form, one missed cleavage site, peptide mass tolerance of 100 ppm and MS/MS tolerance of ±0.5 Da.
RNA extraction and Northern analysis.
Total RNA from submerged cultures was extracted with Isogen (Nippon Gene) according to the manufacturer's instructions. Total RNA extraction from solid-state culture was done as described by Akao et al. (2). Denatured total RNAs (10 μg/lane) were electrophoresed on a formaldehyde-agarose gel and transferred in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) onto a Hybond N+ membrane (Amersham Pharmacia). Northern analysis hybridization was performed with a PCR digoxigenin probe synthesis kit (Roche) and detection starter kit II (Roche) according to the manufacturer's instructions. Forward and reverse primers, each 30 bp long, corresponding to the first and the last 30 bp of the open reading frame were used to synthesize the digoxigenin-labeled probe.
RESULTS
Differential displays of extracellular proteins from solid-state and submerged cultures.
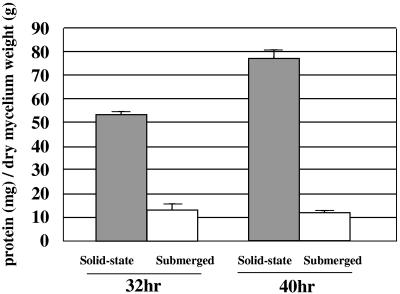
Many fungi secrete more proteins under solid-state culture conditions than under submerged culture conditions (1, 3, 8, 9, 25). To clarify this point, we examined extracellular proteins produced by A. oryzae during 32 h and 40 h of growth under both solid-state and submerged culture conditions. After the 32 h and 40 h of solid-state culture, A. oryzae secreted 53.7 ± 1.36 and 77.3 ± 2.6 mg protein per g dry mycelium, respectively. For the submerged culture, the values were 13.2 ± 2.60 and 11.9 ± 0.89 mg protein per g dry mycelium, respectively. The respective concentrations of mycelia at 32 and 40 h of culture were 6.26 ± 0.67 and 6.57 ± 0.51 mg/ml in submerged culture and 75.4 ± 2.78 and 70.9 ± 4.19 mg/g in solid-state culture. A. oryzae secreted about 4.0- to 6.4-fold more protein per mg mycelium under the solid-state culture conditions than under submerged culture conditions (Fig. 1).
FIG. 1.
Comparison of the amounts of secreted protein under solid-state and submerged culture conditions. Secreted proteins were prepared from 100 ml of submerged culture and 10 g of wheat bran as solid-state culture at 32 and 40 h, and the total protein content was measured after extraction. Dry mycelium weight was determined by N-acetylglucosamine content. Data are mean values and standard deviations from three independent experiments. In the solid state, the amount of protein secreted per gram of dry mycelium was 4- to 6.4-fold greater than the amount secreted in submerged culture.
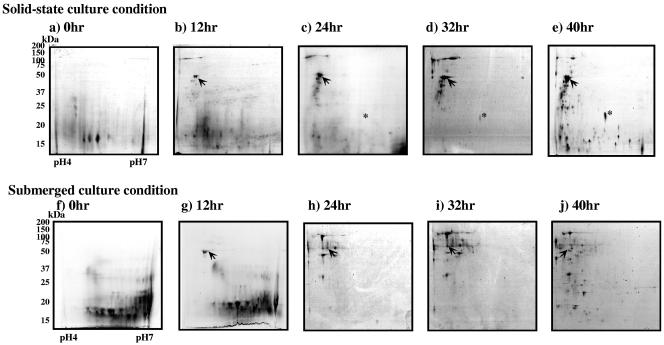
Profiles of the extracellular proteins in the solid-state condition were obtained by 2-D polyacrylamide gel electrophoresis and differential display. At first we used a pH range of from 3 to 10 for electrophoresis in the first dimension. In this case, protein spots on the 2-D gels had pI values of between 4 and 7. Most proteins produced in both cultures had pIs in the range of 4.5 to 5.5. Therefore, we used pH 4 to 7 IPG strips in subsequent analyses. In 2-D electrophoresis, the spot patterns depended on the culture conditions and the age of the culture (Fig. 2). Some protein spots on 2-D electrophoresis were rather diffuse because of the heterogeneity of the secreted proteins due to glycosylation. Wheat bran proteins were degraded and vanished at 24 h, and some extracellular proteins appeared after 12 h. Some smaller proteins detected at 32 h of culture increased greatly by 40 h under both culture conditions.
FIG. 2.
Sequential differential display of secreted proteins from A. oryzae cultured under solid-state and submerged conditions. Secreted proteins were prepared from solid-state and submerged cultures from 0 to 40 h. The protein extract was electrophoresed in an IPG of pH 4 to 7 (18 cm) in the first dimension and a 10 to 20% SDS-polyacrylamide gel in the second dimension. Each gel was loaded with 300 μg of secreted protein and stained with CBB. Gel images from solid-state and submerged conditions at 0, 12, 24, 32, and 40 h are aligned. Arrows indicate deduced α-amylase spots. Asterisks indicate oryzin spots.
The intensity of time-dependent deduced α-amylase spots increased in the solid-state culture but not in the submerged culture (Fig. 2). The appearance of protein spots originating from wheat bran after 12 h suggests that the cell walls of wheat bran were digested by proteins secreted from A. oryzae. The submerged culture condition produced many more spots of over 50 kDa than did the solid-state culture condition. Protein differential display revealed that A. oryzae secreted various kinds of proteins in a different manner in each culture condition.
Identification of extracellular proteins produced during 40 h of solid-state and submerged culture.
The majority of proteins secreted from filamentous fungi are highly glycosylated (30). N-linked oligosaccharides can be difficult to see by MALDI-TOF MS because they tend to make the masses of the typically digested peptides very large (above 3,500 kDa), which appear as broad-band signals in the mass spectra (29). This problem could be solved by deglycosylation of the N-linked oligosaccharides with PNGase F. Deglycosylation of proteins before 2-D PAGE caused the proteins to become insoluble during desalting. Therefore, we used an “in-gel deglycosylation” process after 2-D PAGE. We cut out six spots from the 40-h gel and subjected one half of each spot to in-gel deglycosylation (data not shown). Deglycosylation increased the number of peaks in the mass spectrum of each spot. Deglycosylation increased matched peaks and scores in four spots but had no effect in the other two spots. Thus, in-gel deglycosylation increased the number of A. oryzae proteins that could be identified by MALDI-TOF MS.
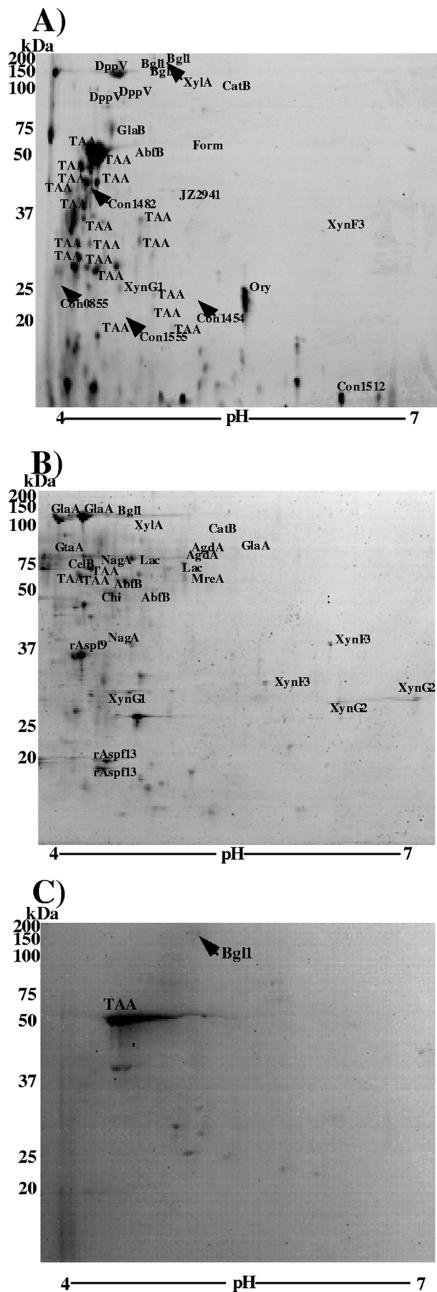
Considering the greater number of protein spots in the 40-h gel and growth phase of mycelium, we chose this gel to identify the extracellular protein under both culture conditions (Fig. 2). Under both culture conditions, all spots visualized by CBB staining were excised, in-gel deglycosylated with PNGase F, digested with trypsin, and analyzed by MS. Eighty-five spots from the solid-state condition (Fig. 3A) and 110 spots from the submerged condition (Fig. 3B) were detected reproducibly and analyzed by MS. Of the 85 spots obtained from the solid-state culture, 43 (50.5%) were identified and assigned to 21 different genes. Of all 110 spots from the submerged culture, 37 spots were identified (33.6%) and assigned to 27 different genes. An analysis of the solid-state and submerged culture 2-D gel images by PDQuest indicated that the identified proteins account for 60.9% and 60.4%, respectively, of the visible proteins in the gels. Eleven protein species were found only in the solid-state condition, 11 were found only in the submerged condition, and 8 were found in both culture conditions (Table 1). Proteins specifically identified in the solid-state condition included glucoamylase B (which is known as a solid-state-specific expression marker), formamidase, and alanyl dipeptidyl peptidase (Fig. 3A). Some hypothetical proteins having low homology to known proteins were found, indicating that A. oryzae secreted new proteins of unknown function in the solid-state condition. In contrast, many well-known proteins, such as glucoamylase A, α-glucosidase, and β-galactosidase, were identified in the submerged culture (Fig. 3B).
FIG. 3.
Comparative proteome analysis of secreted proteins under solid-state (A) and submerged (B) culture conditions and cell wall-bound protein under submerged culture conditions (C). The protein extract was electrophoresed in an IPG of pH 4 to 7 (18 cm) in the first dimension and a 10 to 20% SDS-polyacrylamide gel in the second dimension. Each gel was loaded with 300 μg of secreted protein and stained with CBB. Gel spots of interest in each culture condition were excised and in-gel deglycosylated with PNGase F prior to trypsin digestion and were identified by peptide mass fingerprinting using MALDI-TOF MS. The protein spots identified are labeled by the abbreviations of the proteins as described in Table 1. The spots that had scores of greater than 60 and were reproducibly identified were defined to be matched.
TABLE 1.
Summary of proteins identified from solid-state and submerged cultures of A. oryzae by peptide mass fingerprinting
| Culture condition and spota | Matched contig
|
Annotation
|
Scored | Coverage (%) | Matching peptides | ||||
|---|---|---|---|---|---|---|---|---|---|
| A. oryzae genome database | EST database | NCBI database | Protein | Species | E valueb | ||||
| Solid state | |||||||||
| GlaB | Con1580_homo100-Con1580-4329 | gil 2980896 | GlaB (solid-specific glucoamyl) | A. oryzae | 0 | 101 | 26 | 8/25 | |
| DppV | gil 5305698 | Alanyl dipeptidyl peptidase | A. oryzae | 0 | 167 | 29 | 16/40 | ||
| Form | Con1345_gdraw-f6 | Formamidase | Emericella nidulans | 0 | 75 | 39 | 13/63 | ||
| Man | Con0944_h_218 | α-Mannosidase | A. phoenisis | 0 | 65 | 28 | |||
| JZ2941 | JZ002941 | Transaldolase | Schizosaccharomyces pombe | 0 | 137 | 53 | 16/62 | ||
| con1555 | Con1555_h_1073 | Probable d-amino acid oxidase | S. pombe | 1.00E−26 | 62 | 19 | 7/15 | ||
| con1454 | Con1454_gdraw-f12 | Hypothetical protein T04F3.1 | Caenorhabditis elegans | 2 | 61 | 61 | 7/36 | ||
| con1393 | Con1393_gdraw-r7 | JZ004859 | Unknown | Pseudomonas syringae | 5 | 62 | 39 | 6/30 | |
| con1482 | Con1482_gdraw_f6 | Hypothetical ORF Yil105cp | Saccharomyces cerevisiae | 1E−52 | 61 | 11 | 7/20 | ||
| con1512 | Con1512_gdraw-f3 | Putative protein | Arabidopsis thaliana | 3.00E−03 | 61 | 36 | 8/15 | ||
| con0855 | Con0855_gdraw-f1 | JZ000734 | Hypothetical protein PAB05188E-08 | Pyrococcus abyssi | 0.42 | 60 | 15 | 7/36 | |
| Submerged | |||||||||
| XynG2 | gil 12082170 | Xylanase G2 | A. oryzae | 0 | 62 | 34 | 6/15 | ||
| GlaA | Con1350_e_3 | gil 543806 | GlaA (glucoamylase precursor) | A. oryzae | 0 | 225 | 35 | 16/32 | |
| AgdA | Con1713_h_1547 | JZ000452 | gil 3023272 | α-Glucosidase (AgdA) | A. oryzae | 0 | 62 | 7 | 7/17 |
| GtaA | Con0816_e_0 | gil 6456478 | Glutaminase A | A. oryzae | 0 | 61 | 13 | 7/24 | |
| Lac | Con0970_h_248 | β-Galactosidase precursor | A. niger | 0 | 69 | 8 | 9/28 | ||
| CelB | Con1438_headtail-Con1438-3307-r1 | 1,4-β-d-Glucanhydrolase B | A. niger | 0 | 103 | 32 | 10/20 | ||
| Chi | Con1459_gdraw-f6 | Chitinase | E. nidulans | E−167 | 80 | 15 | 10/24 | ||
| NagA | Con1525_headtail-Con1525-3904-f1 | N-Acetylglucosaminidase | E. nidulans | 0 | 60 | 18 | 8/30 | ||
| rAspf9 | Con1553_gdraw-r4 | rAsp f9 | A. fumigatus | E−104 | 64 | 11 | 7/27 | ||
| MreA | gil 10119790 | Isoamyl alcohol oxidase | A. oryzae | 0 | 64 | 8 | 5/7 | ||
| Aspf13 | Con1184_h_480 | Allergen Aspf15 (Aspf13) | A. fumigatus | 1.00E−37 | 63 | 18 | 3/8 | ||
| Both | |||||||||
| TAA | Con1713_h_1548 | gil 4139860 | α-Amylasec | A. oryzae | 0 | 108 | 25 | 11/83 | |
| Bgl | Con0854_h_130 | β-Glucosidasec | A. aculeatus | 0 | 87 | 26 | 15/47 | ||
| Ory | Con1418_h_836 | gil 129234 | Oryzin alkaline protease precursor | A. oryzae | 0 | 66 | 24 | 6/40 | |
| XynF3 | Con1587_e_6 | gil 15823785 | Xylanase F3 | A. oryzae | E−130 | 105 | 57 | 12/80 | |
| AbfB | Con0723_h_62 | gil 16754882 | α-l-Arabinofuranosidase | A. oryzae | 0 | 135 | 27 | 11/28 | |
| XynG1 | gil 1945601 | Xylanase G1 | A. oryzae | 0 | 61 | 29 | 5/32 | ||
| CatB | Con1475_h_928 | JZ002907 | Catalase B | A. oryzae | 0 | 149 | 23 | 12/25 | |
| XylA | Con1679_gdraw-r16 | Xylosidase/arabinosidase | Thermanaerobacter etha | E−128 | 111 | 25 | 13/38 | ||
E values were obtained from BLAST searches against the NCBI protein database for each gene contig.
Protein found in cell wall fraction.
Only results for proteins that had scores of greater than 60 and were reproducibly identified are shown.
In the solid-state condition, half of the identified spots were α-amylases and their proteolytic products. An analysis of the gel image by PDQuest revealed that α-amylases and their proteolytic products accounted for over 50% of the amount of visible proteins in the gel.
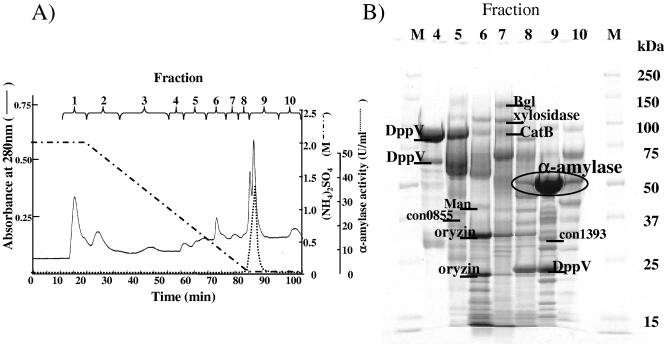
To exclude the effect of a large amount of α-amylase in the solid-state culture extract, we separated the extracellular proteins by hydrophobic chromatography using HPLC. Each fraction was subjected to SDS-PAGE, and a total of 110 bands were analyzed by MS (Fig. 4). Of the 110 bands, 24 bands were identified. Almost all of the identified bands were the same proteins shown in Table 1, and as a result, three new species of proteins were identified. One protein is α-mannosidase, and the other two were annotated as proteins of unknown function. Many different bands were identified as the same protein species, indicating that many proteins had been digested by protease during the 40 h of incubation. Some of the protein degradation may have also occurred during sample preparation in spite of the presence of protease inhibitors.
FIG. 4.
Fractionation of secreted proteins in solid-state culture by hydrophobic chromatography of Phenyl 5-PW (A) and SDS-PAGE analysis of Phenyl 5-PW fractions and identification of secreted proteins (B). (A) Proteins from solid-state culture at 40 h were applied to a Phenyl 5-PW high-performance liquid chromatography column and eluted as described in Materials and Methods. The absorbance at 280 nm, α-amylase activity, and concentration of ammonium sulfate in the elution buffer are shown. (B) Proteins of fractions 4 to 10 were concentrated and subjected to separation by SDS-PAGE. Fractions 1 to 3 contained only a small amount of protein and gave no apparent band on SDS-PAGE. Prominent bands were excised and identified by MALDI-TOF MS. Only the main identified bands are labeled, by the abbreviations of the proteins as described in Table 1. Lane numbers correspond to the fraction numbers in panel A. Lanes M, molecular mass markers.
Trapping of proteins in the cell wall.
Some proteins secreted in the solid-state culture were found to be trapped in the cell wall in the submerged condition (27). Such proteins were identified by 2-D electrophoresis and MS (Fig. 3C). Our preparation of cell wall-trapped protein did not include proteins that were covalently bound to the cell wall through a glycosylphosphatidylinositol anchor or other means. 2-D electrophoresis revealed two major spots, identified as α-amylase and β-glucosidase, and other faint spots. Based on the spot sizes, most of the secreted α-amylase and β-glucosidase was trapped in the cell wall in submerged culture.
Classification of extracellular protein from A. oryzae.
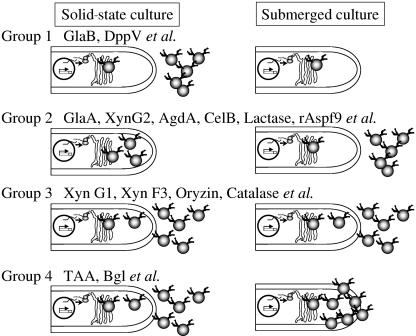
The extracellular proteins secreted under solid-state and submerged culture conditions are compared in Table 1. These results and the results of cell wall-bound protein analysis were used to divide the extracellular proteins into four groups (Fig. 5).
FIG. 5.
Classification of secreted proteins. Secreted proteins are classified into four groups by their secretion profiles. Group 1 proteins are secreted specifically in solid-state culture. Group 2 proteins are secreted specifically in submerged culture. Group 3 proteins are secreted under both submerged and solid-state conditions. Group 4 proteins are trapped as cell wall-bound protein in submerged culture but are secreted in solid-state culture. Abbreviations of the proteins are as described in Table 1.
Group 1 consisted of proteins found specifically in the solid-state condition, including glucoamylase B and alanyl dipeptidyl peptidase. Group 2 consisted of the proteins specifically found in the submerged condition, including glucoamylase A, xylanase G2, α-glucosidase A, cellulase B, lactase, and rAspf9. Group 3 consisted of the proteins found under both conditions, including oryzin, xylanase F3, catalase B, and xylanase G1. Group 4 consisted of the proteins found in the solid-state condition and trapped in the cell wall in the submerged condition, including α-amylase and β-glucosidase.
Gene expression analysis of the four groups of proteins under both conditions.
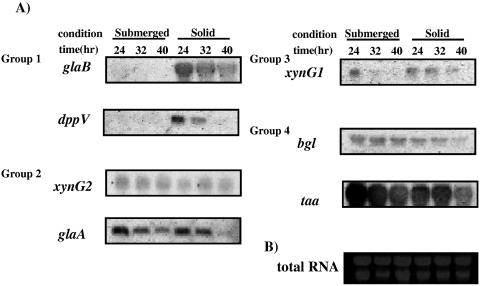
In fungi, secretion of proteins is controlled at various levels, including gene expression, gene translation, quality control in the endoplasmic reticulum, and vesicle transport in the secretion pathway. To determine whether proteins secreted in the solid-state condition were controlled at the translational or posttranslational level, the expression of seven genes (glaB, dppV, glaA, xynG2, xynG1, TAA, and bgl) that encode proteins in the four groups was assayed under both culture conditions by Northern analysis (Fig. 6). glaB and dppV (group 1) were found to be expressed only in the solid-state culture condition (Fig. 6). GlaB and DppV were secreted only in the solid-state condition according to the comparative proteome analysis. Thus, secretion of these proteins appears to be controlled at the transcriptional level.
FIG. 6.
Northern analysis of seven genes in the solid-state and submerged cultures. (A) A. oryzae RIB40 mycelia were harvested from submerged or solid-state culture at 24, 32, and 40 h. Following isolation of the total RNAs, Northern analysis was done as described in Materials and Methods, using PCR fragments of each gene open reading frame as probes. (B) An electrophoresed gel was stained with ethidium bromide as a quantitative control.
The proteome analysis showed that GlaA and XynG2 (group 2) were not secreted in the solid-state culture, but their gene expression was confirmed. This suggests that a specific posttranscriptional mechanism controls secretion of these enzymes in the solid-state condition.
xynG1 (group 3) was expressed under both conditions, although the gene expression was weak in submerged culture. Group 4 genes, including the TAA and bgl genes, were expressed at approximately the same levels under both conditions. TAA and Bgl appear to be controlled posttranscriptionally by trapping in the cell wall in the submerged condition.
DISCUSSION
In this report, we have described the proteins produced by A. oryzae under both solid-state and submerged culture conditions. Despite the importance of solid-state culture and submerged culture in traditional food and commercial enzyme industries, little is known about the extracellular proteins produced in solid-state culture. This is the first comparative proteome analysis of solid-state and submerged cultures of a filamentous fungus. Moreover, our comparative proteome analysis revealed that the cultivation environment greatly affects the types of secretion proteins. Our comparative proteome analysis and Northern analysis of some identified proteins revealed that not only transcriptional regulation but also posttranscriptional regulation plays important roles in protein production in the solid-state culture of A. oryzae. Our in-gel deglycosylation made it possible to identify more proteins by MS analysis.
Effect of deglycosylation on identification of extracellular proteins by MALDI-TOF MS.
The majority of proteins secreted from filamentous fungi are highly glycosylated. Glycosylation makes extracellular proteins stable, resistant to proteolysis, and soluble in the culture medium (30). Because extracellular fungal proteins are resistant to proteolysis, they are often difficult to identify by peptide mass fingerprinting. Eliminating the sugar chains of fungal proteins removes their protease resistance. In-gel deglycosylation made it possible to identify such proteins. Other Aspergillus spp. and other filamentous fungi also secrete such highly glycosylated proteins. In-gel deglycosylation might be used as a common protocol for identifying the extracellular proteins of many other fungi.
The rates of identification of extracellular proteins from solid-state and submerged cultures of A. oryzae by mass fingerprinting (50.5% and 33.6%, respectively) were still low, even though in-gel deglycosylation was used. One possible reason is that many of the proteins in the gel may be resistant to trypsin digestion so that a mass fingerprint could not be obtained. Most extracellular proteins from A. oryzae have acidic isoelectric points, indicating the presence of few basic lysine and arginine residues, which are specific sites of trypsin digestion. Other peptidases, such as asparaginylendopeptidase, might be more successful at digesting these proteins, which would then allow identification by mass fingerprinting. Another method for identifying these proteins is ion trap tandem mass spectrometry. This method is useful for those microorganisms with undetermined complete genome sequences.
Another reason for the low identification rate is that only 10 of 47 spots corresponding to peptides under 25 kDa in the solid-state gel at 40 h were identified, because the number of peptides obtained for the search was extremely low. The same thing was also observed in the proteome analysis of yeast protein using peptide mass fingerprinting, in which the rate of identification of proteins under 30 kDa by mass fingerprinting was extremely low (32, 33). Moreover, some extracellular proteins also contain O-linked oligosaccharides which result in a diversity of peptide masses, as occurs with N glycosylation. To overcome these problems, it is necessary to obtain peptide amino acid sequences. Tandem mass spectrometry (e.g., MALDI-TOF MS/MS) is one way of obtaining these sequences.
Amounts of extracellular proteins under both culture conditions.
Many studies have reported that protein production differs under solid-state and submerged culture conditions (1, 3, 8, 9, 12, 25). In many cases, the production of these proteins in solid-state culture frequently exceeds that in submerged culture. However, no reports have compared total protein production under the solid-state and submerged culture conditions.
In the present study, the total amounts of protein secreted at 32 and 40 h in the solid-state culture were about fourfold and sixfold greater, respectively, than those secreted in the submerged culture (Fig. 1). Data were not obtained for 24 h because a faint stained region, apparently from a contaminant from wheat bran, was observed in the 2-D electrophoresis gel of the sample (Fig. 2c and h). The amount of extracellular proteins in the solid-state culture at 24 h, like amounts at 32 and 40 h, seemed to be larger than that in submerged culture, based on appearance of new spots in the 2-D electrophoresis gel (Fig. 2). Still, many spots remained unidentified; some of them might be derived from wheat bran protein, but these should be few based on the image analysis of 2-D electrophoresis (Fig. 3). These results suggested that A. oryzae generally produced a much larger amount of total proteins in the solid-state culture than in submerged culture. This might be because A. oryzae has to secrete a large amount of enzymes to obtain essential nutrients under the solid-state conditions, since the water activity of solid-state culture is low and differentiation of nutrient components is restricted.
Identification of extracellular proteins.
We clarified the profiles of extracellular proteins under both culture conditions at the proteome level and classified the proteins into four groups according to the profiles. In group 1, we identified glucoamylase B (GlaB) and α-mannosidase. The glaB gene is specifically expressed in solid-state culture (13). Furthermore, GlaB and α-mannosidase were previously identified as AOS1 (A. oryzae solid-state culture-specific gene 1) and AOS22 in a subtraction analysis (2). In this respect, the results of our proteome analysis agreed well with previous results, and the production of these proteins will be controlled at the transcription level. However, some proteins of group 1 were not found in the AOS genes. As Akao et al. (2) reported, not all the AOS genes have been identified. In this context, we suggest that most group 1 proteins are controlled at the transcriptional level. However, proteins might also be regulated at the posttranscriptional level.
Some novel proteins of group 1 were annotated as functionally unknown, while most proteins of group 2 were shown to be functional. Some AOS genes were also annotated as functionally unknown. The functions of these proteins and genes should play important roles in the behavior of A. oryzae in solid-state culture. Thus, it is significant to know the molecular functions of these genes to understand the nature of A. oryzae, especially in solid-state culture. To elucidate the functions of these proteins, genetic and biological analysis on the molecular level will be required, such as overexpression or deletion of the genes.
Among group 3, the major proteins were carbohydrate hydrolases, such as xylanase G1, xylanase F1, xylosidase, and α-l-arabinofuranosidase, which were needed to degrade hemicellulose of wheat bran. Secretion of these enzymes may be controlled not by environmental factors which come from solid-state recognition but by nutrient factors in the culture. The huge amount of hemicellulose from the wheat cell wall is contained in both media, and these enzymes are required to digest it. For A. niger, it was reported that the production and gene expression of xylanolytic enzymes, such as xylanase, xylosidase, and α-l-arabinofuranosidase, were regulated by XlnR, which responds to xylose, xylan, and arabinose induction (35). In A. oryzae, the expression of xylanase G1, xylanase G2, xylosidase A, and cellulase B genes was also controlled by AoXlnR, which is a homolog of XlnR, under xylan induction (26). In fact, the levels of xylanase G1 gene expression were the same under both culture conditions, and AoXlnR will play an important role in wheat bran cultivation.
Regulation of protein secretion.
In fungi, protein secretion is regulated at various steps, including gene expression, mRNA modification and turnover, translation, protein folding, protein modification, vesicle transport, and cell wall passage (7, 11, 18). To understand the novel character of A. oryzae in solid-state culture, it is important to analyze the protein secretion mechanisms at the molecular level. Proteome and Northern analyses help to identify the most significant steps in protein secretion.
Northern analysis showed that solid-state-specific genes, such as glaB and dppV, were not transcribed in submerged culture (Fig. 6). GlaB and DppV are thought to be regulated at the transcription level. Therefore, in the submerged culture condition, overexpression of GlaB protein will be achieved by conversion of the promoter to that which is highly expressed under submerged conditions. Ishida et al. showed that the amount of GlaB protein production reaches approximately 1.0 g/liter of broth even in submerged culture using the melO promoter (17). Their results indicated that transcriptional regulation is significant for the production of GlaB protein. Thus, the regulation of gene expression is thought to be critical in the production of proteins belonging to group 1. However, the role of posttranscriptional regulation is still not clear.
In contrast, secretion of GlaA protein might be controlled at the posttranscriptional level. The results of Northern analysis and comparative proteome analysis showed that the glaA gene was transcribed under both conditions, but production of the protein was found only in the submerged culture. Moreover, Hata and Ishida indicated that GlaA protein production by transformants which possess several glaA genes with their 5′ untranslated regions increased five- to eightfold under submerged culture conditions but was not altered under solid-state culture conditions (14). These results indicated that secretion of GlaA in the solid-state culture was controlled at the posttranscriptional level. However, the process that controls the protein production remains to be clarified. Analysis of localization of a forced-expressed GlaA-enhanced green fluorescent protein fusion might clarify the bottleneck of protein production.
TAA and Bgl proteins were trapped in the cell wall of mycelia in the submerged culture condition, while they were released into the medium in the solid-state culture condition. Consequently, the production of these proteins also could be controlled posttranscriptionally, depending on the culture conditions. A. oryzae might alter the structure of the cell wall under both conditions, and consequently posttranscriptional secretion control would occur. The cell wall is not a static shield in yeast, but its highly dynamic structure can be changed according to the physiological needs of the cell (31). A. oryzae needs a static cell wall in submerged culture to cope with low external osmotic pressure. On the other hand, in solid-state culture, there is no need for a static cell wall, and the cell wall might be loose. As a result, secretion of the enzymes becomes easier.
In Aspergillus kawachii, about 80% of the β-glucosidase was localized in the cell wall in submerged culture (19), while about 80% of it was secreted in solid-state culture. β-Glucosidase and acid-stable α-amylase are secreted under solid-state conditions with the support of extracellular polysaccharide (EPS) (20, 28). A. kawachii produces EPS under the solid-state culture condition. EPS stabilized β-glucosidase and liberated β-glucosidase from the cell wall in the solid-state culture. A. oryzae might also possess the same mechanism for controlling β-glucosidase and α-amylase localization.
Together, these data indicate that A. oryzae drastically alters the manner of protein secretion in response to the culture conditions. Both transcriptional regulation and posttranscriptional regulation were involved and correlated in this manner. Several environmental factors affect gene expression in A. oryzae and also affect the posttranscriptional protein production (16). Considering this, it is essential to understand the effects of both transcriptional and posttranscriptional regulation to clarify the behavior of A. oryzae under solid-state culture conditions.
In this study, we determined the major proteins that are affected by culture conditions. These proteins are excellent markers to analyze posttranscriptional regulation. Using these marker proteins, it will be possible to evaluate how the dynamics of posttranscriptional regulation are affected by each environmental factor. If we could clarify and control the mechanisms of secretion control under both conditions, we would be able to produce the protein more efficiently under both culture conditions.
Acknowledgments
We thank the Aspergillus genome sequence consortium for kindly providing Aspergillus oryzae genome data.
This work was supported by the Bio-Oriented Technology Research Advancement Institution (BRAIN) of Japan.
REFERENCES
- 1.Acuna-Arguelles, M. E., M. Gutierrez-Rojas, G. Viniegra-Gonzalez, and E. Favela-Torres. 1995. Production and properties of three pectinolytic activities produced by Aspergillus niger in submerged and solid-state fermentation. Appl. Microbiol. Biotechnol. 43:808-814. [DOI] [PubMed] [Google Scholar]
- 2.Akao, T., K. Gomi, K. Goto, N. Okazaki, and O. Akita. 2002. Subtractive cloning of cDNA from Aspergillus oryzae differentially regulated between solid-state culture and liquid (submerged) culture. Curr. Genet. 41:275-281. [DOI] [PubMed] [Google Scholar]
- 3.Ashokkumar, B., N. Kayalvizhi, and P. Gunasekaran. 2001. Optimization of media for β-fructofuranosidase production by Aspergillus niger in submerged and solid state fermentation. Process Biochem. 37:331-338. [PubMed] [Google Scholar]
- 4.Bennett, J. W. 2001. Aspergillus and koji: history, practice and molecular biology. SIM News 51:65-71. [Google Scholar]
- 5.Christensen, T., H. Woeldike, E. Boel, S. B. Mortensen, K. Hjortshoej, L. Thim, and M. T. Hansen. 1988. High level expression of recombinant genes in Aspergillus oryzae. Bio/Technology 6:1419-1422. [Google Scholar]
- 6.Chu, P. W., M. N. Yap, C. Y. Wu, C. M. Huang, F. M. Pan, M. J. Tseng, and S. T. Chen. 2000. A proteomic analysis of secreted proteins from xylan-induced Bacillus sp. strain K-1. Electrophoresis 21:1740-1745. [DOI] [PubMed] [Google Scholar]
- 7.Conesa, A. P. J. Punt, N. van Luijk, and C. A. van den Hondel. 2001. The secretion pathway in filamentous fungi: a biotechnological view. Fungal Genet. Biol. 33:155-171. [DOI] [PubMed] [Google Scholar]
- 8.Diaz-Godinez, G., J. Soriano-Santos, C. Augur, and G. Viniegra-Gonzalez. 2001. Exopectinases produced by Aspergillus niger in solid-state and submerged fermentation: a comparative study. J. Ind. Microbiol. Biotechnol. 26:271-275. [DOI] [PubMed] [Google Scholar]
- 9.Elinbaum, S., H. Ferreyra, G. Ellenrieder, and C. Cuevas. 2002. Production of Aspergillus terreus alpha-l-rhamnosidase by solid state fermentation. Lett. Appl. Microbiol. 34:67-71. [DOI] [PubMed] [Google Scholar]
- 10.Gomi, K., N. Okazaki, T. Tanaka, C. Kumagai, H. Inoue, Y. Iimura, and S. Hara. 1987. Estimation of mycelial weight in rece-koji with use of fungal cell wall lytic enzyme. J. Brew. Soc. Jpn. 82:130-133. [Google Scholar]
- 11.Gouka, R. J., P. J. Punt, and C. A. van den Hondel. 1997. Efficient production of secreted proteins by Aspergillus: progress, limitations and prospects. Appl. Microbiol. Biotechnol. 47:1-11. [DOI] [PubMed] [Google Scholar]
- 12.Hashimoto, T., M. Morishita, K. Iwashita, H. Shimoi, Y. Nakata, Y. Tsuji, and K. Ito. 1999. Production and some properties of salt-tolerant β-xylosidases from a shoyu koji mold, Aspergillus oryzae in solid and liquid cultures. J. Biosci. Bioeng. 88:479-483. [DOI] [PubMed] [Google Scholar]
- 13.Hata, Y., H. Ishida, E. Ichikawa, A. Kawato, K. Suginami, and S. Imayasu. 1998. Nucleotide sequence of an alternative glucoamylase-encoding gene (glaB) expressed in solid-state culture of Aspergillus oryzae. Gene 207:127-134. [DOI] [PubMed] [Google Scholar]
- 14.Hata, Y., and H. Ishida. 2000. Glucoamylase-encoding genes of Aspergillus oryzae. Seibustu-kogaku 78:120-127. (In Japanese.) [Google Scholar]
- 15.Hatamoto, O., H. Sekine, E. Nakano, and K. Abe. 1999. Cloning and expression of a cDNA encoding the laccase from Schizophyllum commune. Biosci. Biotechnol. Biochem. 63:58-64. [DOI] [PubMed] [Google Scholar]
- 16.Ishida, H., Y. Hata, A. Kawato, Y. Abe, K. Suginami, and S. Imayasu. 2000. Identification of functional elements that regulate the glucoamylase-encoding gene (glaB) expressed in solid-state culture of Aspergillus oryzae. Curr. Genet. 37:373-379. [DOI] [PubMed] [Google Scholar]
- 17.Ishida, H., K. Matsumura, Y. Hata, A. Kawato, K. Suginami, Y. Abe, S. Imayasu, and E. Ichishima. 2001. Establishment of a hyper-protein production system in submerged Aspergillus oryzae culture under tyrosinase-encoding gene (melO) promoter control. Appl. Microbiol. Biotechnol. 57:131-137. [DOI] [PubMed] [Google Scholar]
- 18.Iwashita, K. 2002. Recent studies of protein secretion by filametous fungi. J. Biosci. Bioeng. 94:530-535. [DOI] [PubMed] [Google Scholar]
- 19.Iwashita, K., T. Nagahara, H. Kimura, M. Takano, H. Shimoi, and K. Ito. 1999. The bglA gene of Aspergillus kawachii encodes both extracellular and cell wall-bound beta-glucosidases. Appl. Environ. Microbiol. 65:5546-5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iwashita, K., H. Shimoi, and K. Ito. 2001. Extracellular soluble polysaccharide (EPS) from Aspergillus kawachii improves the stability of extracellular β-glucosidase (Ex-1 and Ex-2) and is improved in their localization. J. Biosci. Bioeng. 91:134-140. [DOI] [PubMed] [Google Scholar]
- 21.Kitano, H., Kataoka, K. Furukawa, and S. Hara. 2002. Specific expression and temperature-dependent expression of the acid protease-encoding gene (pepA) in Aspergillus oryzae in solid-state culture (rice-koji). J. Biosci. Bioeng. 93:563-567. [DOI] [PubMed] [Google Scholar]
- 22.Machida, M. 2002. Progress of Aspergillus oryzae genomics. Adv. Appl. Microbiol. 51:81-106. [DOI] [PubMed] [Google Scholar]
- 23.Machida, M., K. Asai, M. Sano, T. Tanaka, T. Kumagai, G. Terai, K.-I. Kusumoto, T. Arima, O. Akita, Y. Kashiwagi, K. Abe, K. Gomi, H. Horiuchi, K. Kitamoto, T. Kobayashi, M. Takeuchi, D. W. Denning, J. E. Galagan, W. C. Nierman, J. Yu, D. B. Archer, J. W. Bennett, D. Bhatnagar, T. E. Cleveland, N. D. Fedorova, O. Gotoh, H. Horikawa, A. Hosoyama, M. Ichinomiya, R. Igarashi, K. Iwashita, P. Rao Juvvadi, M. Kato, Y. Kato, T. Kin, A. Kokubun, H. Maeda, N. Maeyama, J.-I. Maruyama, H. Nagasaki, T. Nakajima, K. Oda, K. Okada, I. Paulsen, K. Sakamoto, T. Sawano, M. Takahashi, K. Takase, Y. Terabayashi, J. R. Wortman, O. Yamada, Y. Yamagata, H. Anazawa, Y. Hata, Y. Koide, T. Komori, Y. Koyama, T. Minetoki, S. Suharnan, A. Tanaka, K. Isono, S. Kuhara, N. Ogasawara, and H. Kikuchi. 2005. Genome sequencing and analysis of Aspergillus oryzae. Nature 438:1157-1161. [DOI] [PubMed] [Google Scholar]
- 24.Maeda, H., M. Sano, Y. Maruyama, T. Tanno, T. Akao, Y. Totsuka, M. Endo, R. Sakurada, Y. Yamagata, M. Machida, O. Akita, F. Hasegawa, K. Abe, K. Gomi, T. Nakajima, and Y. Iguchi. 2004. Transcriptional analysis of genes for energy catabolism and hydrolytic enzymes in the filamentous fungus Aspergillus oryzae using cDNA microarrays and expressed sequence tags. Appl. Microbiol. Biotechnol. 65:74-83. [DOI] [PubMed] [Google Scholar]
- 25.Maldonado, M. C., and A. M. Strasser de Saad. 1998. Production of pectinesterase and polygalacturonase by Aspergillus niger in submerged and solid state systems. J. Ind. Microbiol. Biotechnol. 20:34-38. [DOI] [PubMed] [Google Scholar]
- 26.Marui, J., A. Tanaka, S. Mimura, L. H. de Graaff, J. Visser, N. Kitamoto, M. Kato, T. Kobayashi, and N. Tsukagoshi. 2002. A transcriptional activator, AoXlnR, controls the expression of genes encoding xylanolytic enzymes in Aspergillus oryzae. Fungal Genet. Biol. 35:157-169. [DOI] [PubMed] [Google Scholar]
- 27.Messner, R., K. Hagspiel, and C. P. Kubicek. 1990. Isolation of a β-glucosidase binding and activating polysaccharide from cell wall of Trichoderma reesei. Arch. Microbiol. 154:150-155. [Google Scholar]
- 28.Nagamine, K., K. Murashima, T. Kato, H. Shimoi, and K. Ito. 2003. Mode of alpha-amylase production by the shochu koji mold Aspergillus kawachii. Biosci. Biotechnol. Biochem. 67:2194-2202. [DOI] [PubMed] [Google Scholar]
- 29.Packer, N. H., and L. Keatinge. 2001. Glycobiology and proteomics, p. 257-275. In S. R. Pennington and M. J. Dunn (ed.), Proteomics from protein sequence to function. BIOS Scientific Publishers, Oxford, United Kingdom.
- 30.Peberdy, J. F. 1994. Protein secretion in filamentous fungi—trying to understand a highly productive black box. Trends Biotechnol. 12:50-57. [DOI] [PubMed] [Google Scholar]
- 31.Popolo, L., T. Gualtieri, and E. Ragni. 2001. The yeast cell-wall salvage pathway. Med. Mycol. 39(Suppl. 1):111-121. [PubMed] [Google Scholar]
- 32.Sagliocco, F., J. C. Guillemot, C. Monribot, J. Capdevielle, M. Perrot, E. Ferran, P. Ferrara, and H. Boucherie. 1996. Identification of proteins of the yeast protein map using genetically manipulated strains and peptide-mass fingerprinting. Yeast 12:1519-1533. [DOI] [PubMed] [Google Scholar]
- 33.Shevchenko, A., O. N. Jensen, A. V. Podtelejnikov, F. Sagliocco, M. Wilm, O. Vorm, P. Mortensen, H. Boucherie, and M. Mann. 1996. Linking genome and proteome by mass spectrometry: large-scale identification of yeast proteins from two dimensional gels. Proc. Natl. Acad. Sci. USA 93:14440-14445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsuchiya, K., T. Nagashima, Y. Yamamoto, K. Gomi, K. Kitamoto, C. Kumagai, and G. Tamura. 1994. High level secretion of calf chymosin using a glucoamylase-prochymosin fusion gene in Aspergillus oryzae. Biosci. Biotechnol. Biochem. 58:895-899. [DOI] [PubMed] [Google Scholar]
- 35.van Peij, N. N., M. M. Gielkens, R. P. de Vries, J. Visser, and L. H. de Graaff. 1998. The transcriptional activator XlnR regulates both xylanolytic and endoglucanase gene expression in Aspergillus niger. Appl. Environ. Microbiol. 64:3615-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]