Abstract
The human TGFB1 gene is polymorphic, and genetic variants are associated with altered cancer risk. However, human genetic association studies have had variable outcomes because TGFβ1 action is context-dependent. We used the murine skin model of chemical carcinogenesis in genetic linkage analysis of three independent Mus musculus NIH/Ola × (Mus spretus × M. musculus NIH/Ola)F1 backcrosses, to identify a skin tumor susceptibility locus, Skts14, on proximal chromosome 7. Tgfb1 maps at the peak of linkage. The mouse Tgfb1 gene is polymorphic, resulting in cis-regulated differential allelic mRNA expression between M. spretus and M. musculus in F1 mouse skin. This phenomenon is reflected in differential phospho-SMAD2 levels, downstream of TGFβ signaling, between these two mouse species. In normal F1 mouse skin, the Tgfb1SPR allele is expressed at higher levels than the Tgfb1NIH allele, and this differential is accentuated by phorbol 12-myristate 13-acetate treatment. In benign F1 papillomas, this imbalance is reversed, possibly by selection against expression of a hyperactive Tgfb1SPR allele in TGFβ growth-responsive tumors. We demonstrate that skin tumor susceptibility is altered by Tgfb1 gene dosage, but that manifestation of Tgfb1-linked skin tumor susceptibility in M. musculus NIH/Ola × (M. spretus × M. musculus NIH/Ola)F1 backcross mice depends on interactions with another unlinked tumor modifying locus, Skts15, that overlaps Tgfbm3 on chromosome 12. These findings illustrate the power of complex genetic interactions in determining disease outcome and have major implications to the assessment of disease risk in individuals harboring variant TGFB1 alleles.
Keywords: carcinoma, chemical skin carcinogenesis, genetic interaction, TGFβ
TGFβ acts as a negative growth regulator of normal and benign proliferative epithelial cells (1–3), but stimulates tumor progression once accumulation of oncogenic mutations dampens the tumor’s negative growth response to TGFβ (4–6). Additionally, TGFβ overproduced by both the malignant cell and tumor stroma may act on the tumor microenvironment to indirectly stimulate tumor progression (7, 8). TGFβ alters stromal cell characteristics, such as extracellular matrix deposition, secretion of proteases and other cytokines that enhance angiogenesis, and tumor growth and plasticity (9, 10). Moreover, it can act as a very potent local and systemic immunosuppressor (11–13).
Genes encoding components of the TGFβ1-signaling pathway, including TGFΒ1 (14) and TGFBRI (15, 16), have been shown to be functionally polymorphic in humans, and genetic associations have been found between carriers of specific TGFB1 and TGFBR1 polymorphic variants and cancer susceptibility (15–20). The TGFB1 gene harbors polymorphisms in its promoter, plus amino acid polymorphisms in its signal peptide that influence protein secretion and levels of freely circulating TGFβ1 (14, 21–23). Several independent groups have demonstrated a genetic association between variant TGFB1 alleles and altered risk for breast cancer (17, 19, 20, 24). The most extensive report was that of a case-control study of >3,900 early-onset (median age 50) invasive breast cancer patients and a similar number of controls (17). Dunning et al. (17) demonstrated that homozygosity for the high expressing TGFΒ1Pro-10 allele was associated with an increased invasive cancer risk (odds ratio 1.25), which would support a positive role of TGFΒ1 in tumor progression. Conversely, in a cohort study of >3,000 women >65 years old at recruitment, of which 146 developed breast cancer over the following 9 years, it was found that women homozygous for the high-expressing TGFB1Pro-10 allele were at a reduced risk of developing breast cancer, suggesting that TGFβ1 has breast tumor-suppressing activity in this cohort (19). One explanation for these seemingly discrepant findings is ascertainment bias in selecting only young women with invasive breast cancer (17) vs. women who had reached 65 years of age cancer-free (19). Taken together, these studies support the model of the dual role of TGFβ in tumorigenesis (4). They illustrate that the prediction of cancer risk associated with a particular TGFΒ1 allele depends on interacting genetic and environmental factors such that the hyperactive TGFΒ1 allele may confer either cancer protection or increased risk. This thesis was recently substantiated by the studies of Shin et al. (24) which emphasize the importance of understanding the context-dependent action of TGFβ1 and hint at the importance of genes that interact with TGFΒ1 in determining disease risk. Similar conclusions were recently made for risk of invasive prostate cancer (18).
In the current study, we demonstrate the existence of another skin tumor susceptibility locus, Skts14, containing the Tgfb1 gene on proximal chromosome 7. We show that cis-acting regulatory elements of the mouse Tgfb1 gene are polymorphic, leading to allelic mRNA expression, which could account for altered tumor susceptibility between mouse strains. Importantly, we demonstrate that the outcome of Tgfb1 allelic variation depends on genetic context, particularly with respect to another modifier locus, Skts15, on chromosome 12 with which Tgfb1 interacts.
Results
Skts14, a Skin Papilloma Susceptibility Locus on Proximal Mouse Chromosome 7.
Previous studies demonstrated the existence of a skin tumor susceptibility locus, Skts1, on proximal chromosome 7 that controls papilloma development (25). This linkage was originally mapped at a low resolution with a peak located at ≈50 Mb. In the current study, a panel of markers mapping at a higher resolution was used to regenotype the chromosome 7-linkage region in 306 (NIH/Ola × outbred Mus spretus)F1 × NIH/Ola (NSP) backcross animals, revealing two linkage peaks and suggesting two distinct loci, with a more proximal locus, Skts14, mapping at ≈14 Mb on chromosome 7 (Fig. 1).
Fig. 1.
Skts14, a skin tumor susceptibility locus on proximal mouse chromosome 7. LOD scores for genetic linkage on chromosome 7 in the F1 NSP backcross (n = 206; ♦), and in that subset of the NSP backcross sharing Haplotype 4 (n = 44; ○). Also shown is a linkage analysis in the N4 backcross (n = 76; ▵).
Information on shared haplotypes in outbred populations can be used to refine the locations of potential disease susceptibility genes (26). A haplotype map was previously constructed for chromosome 7 by using variation in microsatellite lengths between the M. spretus alleles in the outbred colony (26). Interestingly, when the linkage data were stratified according to M. spretus haplotype, it was found that mice from one haplotype, namely “Haplotype 4,” showed linkage only to the Skts14 locus, in the absence of linkage to the original, more distal, Skts1 locus, thus demonstrating that the two chromosome 7 loci act independently (Fig. 1). Skts14 was further validated by linkage analysis on N4 backcross mice that had been backcrossed four more generations to the NIH strain while selecting for those with papilloma resistance at each generation (N4 mice). The LODMAX at Skts1 was less than LODMAX − 1 at Skts14, again suggesting independence of Skts14 from Skts1 (Fig. 1).
Skts14NIH-Linked Tumor Susceptibility Depends on Genetic Interaction with Skts15NIH on Chromosome 12.
Genetic interaction between Skts1 and Skts5 on chromosomes 7 and 12, respectively, has been shown (27). This interaction analysis was repeated by using the higher density marker set over chromosome 7 and three markers located at 17.7, 34.1, and 60.7 Mb on chromosome 12. Interestingly, two peaks of significant interaction were found (see Table 2, which is published as supporting information on the PNAS web site). As seen in ref. 27, Skts1 on chromosome 7 at 39.3 Mb shows genetic interaction with Skts5 on chromosome 12 at 34.1 Mb. Independently, Skts14 on chromosome 7 at 14 Mb interacts strongly with another tumor susceptibility locus, Skts15, on chromosome 12 at 17.7 Mb. Conversely, interactions between Skts14 and Skts5 and between Skts1 and Skts15 were both insignificant (see Table 2), illustrating the independence of the two genetic interactions between chromosomes 7 and 12.
Table 1 shows that homozygosity for NIH alleles at Skts14 and Skts15 confers higher tumor susceptibility in two independent M. spretus × NIH backcrosses (Table 1). The presence of a single M. spretus allele at either of these interacting loci results in relative resistance to papilloma development, with additional M. spretus alleles (Tgfb1Spr/NIH;D12Nds11Spr/NIH) having little additional effect (Table 1). The Skts1/Skts5 interaction also shows this effect, but only in the NSP cross (Table 1), emphasizing the independence of the two interactions and the importance of the Skts14/Skts15 interaction.
Table 1.
Homozygosity for NIH alleles at both Tgfb1 and D12Nds11 associates with increased papilloma yield per mouse
| Experiment | Tgfb1 |
D12Nds11 |
P value | Experiment | D7Mit18 |
D12Mit154 |
P value | ||
|---|---|---|---|---|---|---|---|---|---|
| NS | NN | NS | NN | ||||||
| NSP | NS | 2.52 (n = 59) | 2.31 (n = 81) | 2.2 × 10−4 | NSP | NS | 1.87 (n = 91) | 2.68 (n = 79) | <10−16 |
| NN | 2.48 (n = 90) | 5.03 (n = 96) | NN | 2.07 (n = 69) | 5.94 (n = 87) | ||||
| NSE | NS | 3.14 (n = 22) | 3.84 (n = 32) | 3.1 × 10−10 | NSE | NS | 2.12 (n = 25) | 4.32 (n = 19) | 0.006 |
| NN | 3.61 (n = 28) | 10.83 (n = 24) | NN | 5.09 (n = 22) | 10.56 (n = 27) | ||||
P values were determined by the Kruskal–Wallis test. N, NIH allele; S, spretus allele; NSP, F1 (NIH/O1a × outbred M. spretus) × NIH/Ola; NSE, F1 (NIH/Ola × inbred SEG/Pas) × NIH/Ola.
Haploinsufficiency for Tgfb1, a Candidate Gene at Skts14, Increases Papilloma Incidence.
Tgfb1 is an excellent candidate as a tumor susceptibility gene at Skts14, especially in light of the evidence that human TGFB1 is functionally polymorphic and alters risks for breast and prostate cancer (17–19). We previously showed that keratinocyte-targeted TGFβ1 gene expression in the chemically induced model of mouse skin carcinogenesis reduces papilloma outgrowth (4). Fig. 2 demonstrates that 7,12-dimethylbenz[a]anthracene (DMBA)/phorbol 12-myristate 13-acetate (PMA) treatment of Tgfb1+/− mice leads to a significant increase in papilloma incidence compared with control Tgfb1+/+ mice, demonstrating that global alterations in TGFβ1 levels also influence mouse skin tumor susceptibility.
Fig. 2.
Haploinsufficiency for Tgfb1 increases papilloma incidence. Twenty-three WT and 30 Tgfb1+/− age-matched adult female mice were subjected to a standard chemical carcinogenesis protocol, and papilloma numbers were counted weekly. Tgfb1+/− developed significantly more papillomas than Tgfb1+/+ mice (P = 0.025 at 20 weeks; Student’s t test).
Basal and PMA-Inducible Gene Expression Levels of Tgfb1Spr in Skin Are Higher than Those of Tgfb1NIH.
The coding region of Tgfb1 was sequenced in its entirety in M. spretus and in three strains of Mus musculus (NIH, 129, and C57). No amino acid differences were found between any of the strains, although several silent SNPs were detected.
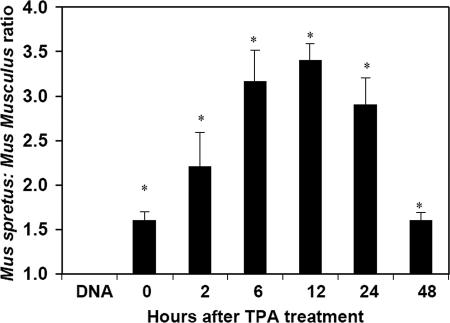
In the absence of TGFβ1 amino acid polymorphisms, functional polymorphism between the two mouse species could occur at the RNA level due to differential transcription, splicing, or message stability. To investigate this possibility, allele-specific TaqMan probes that distinguish between NIH/Ola and SPRET/Ei Tgfb1 transcripts were designed on the basis of a silent SNP in the cDNA. Allele-specific expression patterns of Tgfb1 were quantified within the skin of normal F1 mice [(NIH/Ola × SPRET/Ei)F1] at various times after topical treatment with PMA. TaqMan analyses of cDNA transcribed from RNA of normal mouse skin showed a significant and reproducible difference in basal Tgfb1 gene expression of ≈1.7-fold in favor of the M. spretus allele (Fig. 3). This expression difference between the two alleles was enhanced after PMA treatment, reaching a maximum of a 3.5-fold differential at 12 h after PMA treatment (Fig. 3). In F1 mice, both alleles of the Tgfb1 gene are present within the same cell; thus, regulation of their differential expression must occur in cis. As a control, similar experiments using genomic DNA from the same F1 hybrid mice invariably showed a 1:1 ratio by TaqMan analysis.
Fig. 3.
Differential allelic expression of Tgfb1 in normal mouse skin. RNA was prepared from normal skins of (NIH/Ola × SPRET/Gla)F1 mice, untreated (0) or at various times (2, 6, 12, 24, or 48 h) after topical application of PMA. The M. spretus to M. musculus Tgfb1 transcript ratio was determined by TaqMan analysis of cDNA by using allele-specific probes. All samples overexpressed the M. spretus Tgfb1 allele, and this effect was accentuated 12 h after PMA treatment. For comparison, we also determined the M. spretus/M. musculus ratio in genomic DNA. As expected, this ratio is close to 1 for normal F1 mice. Asterisks indicate significant differences between genomic DNA and cDNA samples. The means and standard deviations of three independent experiments are shown.
Sequencing of 5 kb of genomic DNA upstream from the translation initiation site, and the evolutionarily conserved first intron, revealed complete intraspecific conservation of Tgfb1 within the M. musculus strains, but considerable interspecific polymorphism between M. spretus and M. musculus. Polymorphisms occur within the gene promoter, 5′ untranslated region of the mRNA and within the first intron, with some sequence variants altering potential transcription factor-binding sites, thus introducing the possibility of both transcriptional and posttranscriptional regulation of differential Tgfb1 message levels between the two species (see Fig. 6, which is published as supporting information on the PNAS web site).
Reduced Basal Phospho-SMAD2 Levels in M. musculus Compared with M. spretus Epidermis.
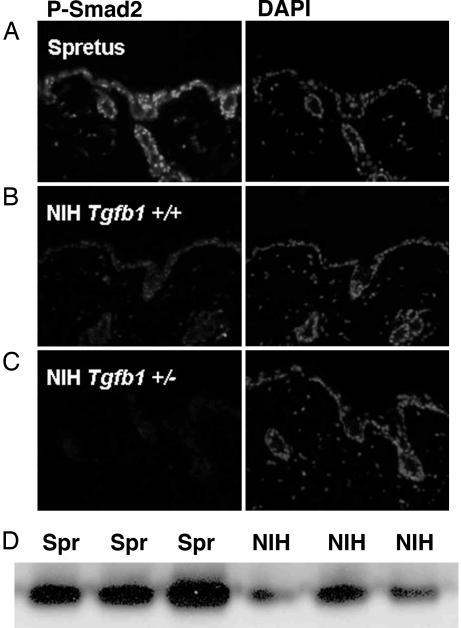
We investigated additional components of the TGFβ-signaling pathway downstream of TGFβ1 itself. Phosphorylation and nuclear translocation of the receptor-associated SMADs, SMAD2 and SMAD3, are early events in signal transduction downstream of the TGFβ receptor complex (5). To determine whether differential expression of the Tgfb1 gene results in altered SMAD signaling between M. spretus and M. musculus, immunohistochemistry and Western blot analysis was performed to assess levels and localization of this marker of TGFβ signaling in WT M. spretus skin vs. WT and heterozygous Tgfb1+/− M. musculus skin. As predicted, heterozygous Tgb1+/− M. musculus epidermis had reduced levels of phospho-SMAD2 compared with M. musculus WT mice. Importantly, phospho-SMAD2 levels were greatly elevated in M. spretus compared with M. musculus epidermis (Fig. 4). Taken together, these results indicate that the TGFβ-signaling pathway is up-regulated substantially in cells and tissues from M. spretus mice.
Fig. 4.
Basal phospho-Smad2 levels are elevated in the skin of M. spretus compared with M. musculus. (A–C) Immunohistochemistry using an anti-phospho-SMAD2 antibody (that recognizes both phospho-Smad2 and phospho-Smad3) on untreated skins from M. spretus (A), M. musculus Tgfb1+/+ (B), and Tgfb1+/− (C) mice. (D) Different levels of phospho-SMAD2 were also observed by Western blot analysis. Cell lysates from three different M. spretus and three different M. musculus mice were immunoprecipitated with an anti-phospho-SMAD2-specific antibody, followed by Western blot analysis using the same antibody.
Tgfb1Spr and Tgfb1NIH Are Differentially Expressed in Tumor Cell Lines and Primary Papilloma.
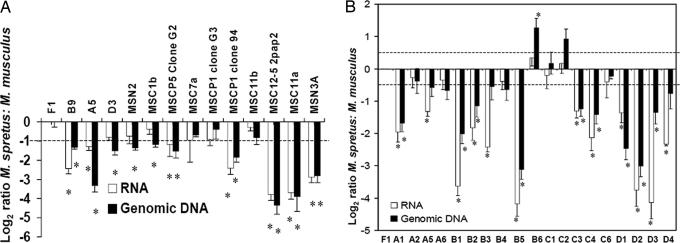
It is widely accepted that mouse and human tumors have elevated expression of TGFβ1 (7, 28) that may ultimately favor malignant progression (9). The Tgfb1 gene is autoinductive and up-regulated by ras via its AP-1-binding site (29, 30). Moreover, during multistage carcinogenesis in the mouse skin model, cytogenetic abnormalities accumulate and, at an early stage, tumor cells frequently show trisomy of chromosome 7 due to duplication of the chromosome harboring mutant H-ras. Subsequently, this chromosome may be further amplified and the chromosome bearing normal H-ras may also be lost (31, 32). The chromosomal imbalance involving chromosome 7 will also lead to duplication of Tgfb1 in the proximal part of the chromosome. Because duplication of the M. spretus allele would lead to very high levels of TGFβ signaling due to the endogenously high levels of the Tgfb1 M. spretus transcript (Fig. 3), it would be anticipated that there may be selection for duplication of the M. musculus chromosome during acquisition of trisomy. This selective trisomy would allow duplication of the mutant H-ras allele on distal chromosome 7 without causing major increases in expression levels of TGFβ1 in the proximal region. This hypothesis was verified by allele-specific TaqMan analysis of genomic DNA samples from 13 tumor cell lines derived from F1 mice. Of these cell lines, 11 showed a genomic DNA imbalance in favor of the M. musculus chromosome, and TaqMan analysis demonstrated relatively reduced expression of the M. spretus Tgfb1SPR allele (Fig. 5A), despite the fact that the M. spretus allele is more highly expressed in normal skin (Fig. 3). We conclude that this reflects selection against the high level-expression of the M. spretus Tgfb1 allele, which would tend to reduce growth of early stage primary tumors.
Fig. 5.
Genomic imbalance and differential allelic expression of Tgfb1 in tumors. (A) cDNA was prepared from skin tumor-derived M. spretus/M. musculus F1 hybrid cell lines. Seven of these lines showed significant overexpression of the M. musculus allele, which is indicated by asterisks. Genomic DNA was isolated from the same samples, and TaqMan analysis was carried out to measure the relative copy number of different Tgfb1 alleles. Ten of the samples (denoted by asterisks) showed significant amplification of the M. musculus allele of Tgfb1. (B) cDNA and genomic DNA was generated from primary papillomas of M. musculus NIH mice, congenic for M. spretus on proximal chromosome 7. TaqMan analysis was carried out as above. Nine tumors showed significant genomic imbalance at Tgfb1 in favor of the M. musculus allele. These nine, together with an additional three had significant overexpression of Tgfb1NIH, indicated by asterisks. Two tumors had genomic imbalance in favor of the M. spretus allele, but in both cases the Tgfb1SPR/Tgfb1NIH expression ratio was not significantly different from 1.0. The dashed line in A and B denote the significance level for differential expression/genomic imbalance determined by Student’s t test.
To exclude the possibility that preferential duplication of the M. musculus chromosome 7 is driven by an alternative tumor susceptibility locus, Skts2, which lies near the H-ras gene on distal chromosome 7 (25), we generated congenic mice containing only the proximal region (0–19 cM) of chromosome 7 from M. spretus on the NIH background. These animals are homozygous M. musculus in the distal portion of the chromosome containing the H-ras gene, and any preferential duplication of parental alleles cannot be driven by genes in the region of Skts2. As shown in Fig. 5B, 13 of 19 primary tumors examined had duplicated the M. musculus rather than the M. spretus chromosome 7 region containing Tgfb1, and these papillomas showed relative overexpression of the M. musculus compared with the M. spretus Tgfb1 allele. Only two papillomas showed genomic overrepresentation of the M. spretus Tgfb1 allele. In these two exceptional cases, the effects of genomic imbalance in favor of Tgfb1Spr were minimized at the RNA expression level of Tgfb1Spr compared with Tgfb1NIH. This finding again supports the hypothesis of selection against Tgfb1Spr expression even in papillomas where the Tgfb1Spr allele is in excess (Fig. 5B; samples B6 and C2).
Discussion
It has been demonstrated that, genomewide, the strongest pairwise interaction between two genetic loci that modifies tumor susceptibility in the mouse skin model is between proximal chromosome 7 (Skts1) and proximal chromosome 12 (Skts5) (27). Fine genetic mapping now shows that Skts1 and Skts5 can each be dissected into two independently interacting regions of the genome. Skts14 at 14.4 Mb on chromosome 7 interacts with Skts15 at 17.7 Mb on chromosome 12 to drive papilloma susceptibility, and an equally strong interaction occurs between NIH alleles at Skts1 at 39.3 Mb on chromosome 7 and Skts5 at 34.1 Mb on chromosome 12. The LODMAX for Skts14 is at Tgfb1, a very strong candidate gene for influencing cancer risk. Intriguingly, LODMAX for Skts15 maps precisely at Tgfbm3, a locus previously identified by its ability to modify the phenotypic outcome of Tgfb1 nullizygosity (33), giving indirect support to the concept that Tgfb1 is the tumor modifier at 14.4 Mb at Skts14.
Unlike the situation for humans, there are no TGFβ1 amino acid polymorphisms between the different mouse strains examined. All strains encode the equivalent of the hyperactive signal peptide isoform observed in humans. Nevertheless, like the human gene, the mouse Tgfb1 gene has a polymorphic gene promoter and 5′ noncoding region and drives differential expression of the two alleles in a cis-acting manner. In F1 mouse skin, the M. spretus Tgfb1 allele is expressed 1.7-fold higher than its M. musculus partner, and this differential is enhanced to nearly 4-fold after induction of the gene by PMA. Elevated ligand expression in M. spretus results in generalized up-regulation of basal TGFβ-signaling activity, as demonstrated by enhanced nuclear staining with a phospho-SMAD2 antibody in the epidermis. It would therefore appear that the hyperactive M. spretus Tgfb1 allele is protective against tumor outgrowth due to the tumor-suppressing (growth inhibitory/differentiation-inducing) activity of TGFβ1 at early stages of tumor outgrowth (4, 34). It is therefore not surprising that this hyperactive M. spretus allele is selectively down-regulated either genetically and/or epigenetically in papillomas that are still growth-responsive to TGFβ. Indeed, selective loss of the M. spretus chromosome 7 in papillomas, which is frequently observed in F1 and F1 backcross mice (32), may indeed be driven, at least in part, by selection against the M. spretus hyperactive Tgfb1 allele.
The tumor-protective role of the M. spretus Tgfb1 allele is redundant if the animal possesses a M. spretus allele at Skts15. High tumor susceptibility is only seen in animals of the F1 backcross that are homozygous NIH at both loci. A similar genetic interaction is seen regulating developmental angiogenesis in the mouse in which possession of NIH alleles at Tgfbm3NIH reduces the developmental dependence on TGFβ1 for normal angiogenesis (33). It is interesting that the same interacting loci, namely Tgfb1 and Skts15/Tgfbm3, appear to alter susceptibility for both cancer and defective developmental angiogenesis. This phenomenon may be because the altered cancer risk seen here is related to alterations in angiogenic capacity or because this chromosome 7/12 interaction modifies fundamental processes in cell biology common to both tumorigenesis and angiogenesis and affecting several cell types (e.g., cell proliferation, survival, and migration). Indeed, the Skts15/Tgfbm3 locus on chromosome 12 encompasses a small interval enriched in genes involved in regulation of cell proliferation, survival, and plasticity, including several genes known to be directly on the TGFβ-signaling pathway (33). Tieg2b/Tieg3 is a TGFβ-inducible transcriptional repressor with antiproliferative and antiapoptotic functions (35). Idb2 encodes Id2 (inhibitor of differentiation), which is also involved in transcriptional inhibitory responses to TGFβ (36–38) in cell growth control and angiogenesis (36, 37, 39, 40). Other genes involved in cell proliferation include Ornithine decarboxylase (Odc) and Ribonucleotide reductase 2 (Rrm2), and those involved in modifying cellular plasticity and migration encode Rho kinase (Rock2), integrin β1-binding protein 1 (Itgb1bp1/LCAP1), and TNFα-converting enzyme (Adam17). It is possible that the gene, or combination of genes, at this locus responsible for Tgfb1 developmental redundancy vs. Tgfb1spr redundancy for tumor resistance are different from each other. This theory remains to be tested. From the cancer perspective, it should be noted that a 2-Mb interval of the human genome syntenic to chromosome 12 at D12Nds11 is amplified in >60% of human prostate tumors (41), a particularly provocative finding because the homozygous TGFB1Pro-10 genotype in humans is associated with a 2.5-fold elevated risk of invasive prostate cancer (18).
The current study illustrates the importance of considering genetic context when undertaking human genetic association studies, particularly for genes that can have either positive or negative effects on disease progression. Indeed, despite highly significant genetic interaction between Tgfb1 and Skts15 (P = 1.1 × 10−8) in the M. spretus (SEG/Pas) × NIH/Ola backcross, neither of these loci reached significance when scored as independent quantitative trait loci (P = 4.5 × 10−3 for Skts14 and P = 3.4 × 10−3 for Skts15). Thus, association of a single genetic polymorphism with potent phenotypic effects on disease risk may be masked by ignoring the contributions of interacting loci that either synergize or neutralize this genetic effect. The small 1.25-fold increase in relative risk for invasive breast cancer observed for homozygous TGFB1Pro-10 individuals (17) may be an under estimate of TGFB1-associated cancer risk. The homozygous TGFB1Pro-10 cohort may in fact be made up of a subpopulation of individuals in which TGFB1Pro-10 elicits very high cancer risk (i.e., their genetic context favors tumor promoting activity of TGFβ1), and another subpopulation in which the risk is much smaller or even negative (19, 20) due to the protective effects of TGFβ1 in tumorigenesis. Indeed, this phenomenon could explain the discrepant TGFB1 genetic association results seen in breast cancer for high-risk patients, namely those that develop breast cancer early in life (median age 50) (17) compared with those that have not developed cancer by age 65 (19). The discrepancy could also be due to definition of phenotypes. The studies of Dunning et al. (17) and Ewart-Toland et al. (18) focused on invasive cancer, whereas those of Ziv et al. (19) and Hishida et al. (20) did not distinguish between different cancer grades. Shin et al. (24) recently showed that high TGFβ1 levels may protect from low-grade cancer but predispose to invasive tumors. This type of effect has also been seen in cardiovascular disease whereby the hyperactive TGFB1 allele has been associated with an increased risk of hypertension (14, 21) but, counterintuitively, with protection from myocardial infarction (14, 22). Such an effect could be because the prevalent TGFβ1 target cell differs for the two conditions. A similar complex interaction may well occur in cancer in which TGFβ has potent effects on both the tumor cell and on all cell types of the tumor microenvironment.
Many proteins have biphasic actions in tumorigenesis, such as c-myc and p53, which can act as either oncogenes or tumor suppressors, dependent on cellular context (42). The issue of epistasis is therefore very important in the design of human genetic association studies for estimating contributions to cancer risk. The elucidation of genetic networks of interacting genes will provide tools for a more holistic and combinatorial approach to accurate determination of disease risks in humans.
Materials and Methods
Animals and Tumor Induction.
The mice used for these studies were described in refs. 25 and 27. Briefly, inbred NIH/Ola mice were purchased from Harlan Olac (Bicester, U.K). Outbred M. spretus and inbred SEG/Pas mice (derived from M. spretus) were obtained from S. Brown (Medical Research Council, Harwell, United Kingdom) and J.-L. Guenet (Institut Pasteur, Paris), respectively. SPRET/Ei mice were obtained from The Jackson Laboratory. M. spretus × NIH backcrosses were generated by breeding a male M. spretus to a female NIH mouse, and the female F1 mice were backcrossed to male NIH. In the backcrosses, NSP, NSE [(NIH/Ola × inbred SEG/Pas) × NIH/Ola], and NSJ [(NIH/Ola × inbred SPRET/Ei) × NIH/Ola] breeding and tumor induction protocols were used as described in ref. 25, and papilloma susceptibility was estimated by the number of papillomas at 20 weeks after initiation. The phenotype data of 106 NSE, 162 NSJ, and 326 NSP animals were reported in refs. 25 and 27.
N4 mice were generated by selecting a single mouse from the NSP backcross that developed no papillomas after chemical carcinogenesis treatment. The mouse was backcrossed through three generations onto the NIH strain, selecting another tumor-resistant mouse at each generation. At the N3 generation, the “line” was expanded to generate 76 N4 mice by backcrossing once more to the NIH strain. These mice were subjected to chemical carcinogenesis and genotyped across chromosome 7. Genetic linkage to tumor resistance was assessed by linear regression analysis using the spss statistics program (SPSS, Chicago).
Tgfb1+/− mice were on an NIH/Ola genetic background after backcrossing >10 generations These mice were subjected to the standard 7,12-dimethylbenz[a]anthracene/PMA chemical carcinogenesis protocol (25).
DNA Preparation, Genotyping, Linkage, and Haplotype Analysis.
Genomic DNAs were prepared from tails and amplified by standard methods by using microsatellites. Negative binomial regression analysis was used to screen for predisposition loci and to identify interacting loci, as reported in ref. 27. For fine mapping of quantitative trait loci, we constructed haplotypes in outbred M. spretus mice for association studies by using the variation in length of microsatellites between the different NSP alleles.
Allele-Specific TaqMan Analysis.
We measured allele-specific expression of Tgfb1 using the ABI Prism 7700 Sequence Detection System (Applied Biosystems). PCR for allele-specific expression (50 μl) contained 50 ng of reverse-transcribed RNA or 50 ng of genomic DNA, 1 × TaqMan universal PCR master mix, forward and reverse primers (900 nM), 200 nM VIC-labeled probe, and 200 nM FAM-labeled probe. Amplification conditions were as follows: 1 cycle of 50°C for 2 min, followed by 1 cycle of 95°C for 10 min, 40 cycles of 95°C for 15 s, and 60–64°C for 1 min. Completed PCRs were read on an ABI Prism 7700 Sequence Detector. PCR was done in triplicate for each sample, and experiments were repeated at least three times. ΔCT values were normalized to the average normal genomic ΔCT difference in each experiment. The ΔCT values between the two probes for the triplicates were then averaged. Probe specificity was assessed by analysis of pure M. spretus and pure M. musculus genomic DNAs as controls. ΔCT (CT difference between M. spretus and M. musculus probe) for pure M. spretus DNA was 16.1, and for pure M. musculus DNA it was −9.6.
Western Blotting Analysis.
Total protein extracts were prepared from mouse skin with STEN lysis buffer (50 mM Tris, pH 7,4/2 mM EDTA/150 mM NaCl/1% Nonidet P-40/0.1% SDS/0.5% Triton X-100) containing Complete Protease Inhibitor mixture (CPI; Roche). Phospho-Smad2 protein was immunoprecipitated from 500 μg of total protein by using a rabbit anti-phospho-Smad2 antibody (a gift from J. Yingling, Eli Lilly) with 20 μl of protein G-Sepharose beads (GE Healthcare, Piscataway, NJ) at room temperature for 3–4 h. The antibody complexes were denaturated and separated by NuPAGE Novex PAGE (Invitrogen), transferred onto a poly(vinylidene difluoride) membrane (Millipore), and immunoblotted with the rabbit anti-phospho-Smad2 antibody. The membranes were washed and incubated with secondary horseradish peroxidase-conjugated antibody (Sigma). The antigen–antibody reactions were detected by enhanced chemiluminescence (ECL; Amersham Pharmacia) and exposed to autoradiographic film.
Immunohistochemistry.
Freshly harvested mouse skin was fixed in 4% PFA and embedded in paraffin by the Comprehensive Cancer Center Immunohistochemistry Core (University of California, San Francisco). Immunofluorescence staining was performed after deparaffinization of 5-μm sections in xylene, rehydration, and antigen retrieval in 10 mM Na Citrate. The sections were washed in PBS and nonspecific antigens blocked for 30 min with 10% FCS in PBS. Proteins were bound with the rabbit anti-phospho-Smad2/3 antibody (Santa Cruz Biotechnology) and detected by using a secondary Alexa Fluor 488 anti-rabbit antibody (Molecular Probes). Slides were washed and mounted in Vectashield hard set mounting medium with DAPI (Vector Laboratories).
Supplementary Material
Acknowledgments
We thank Jonathan Yingling (Eli Lilly) for the gift of anti-phospho-SMAD2 antibody. This work was supported by National Institutes of Health Grants P01 AR050440 (to R.J.A. and A.B.), R01 GM60514 (to R.J.A.), and U01 CA84244 (to A.B.), and U.S. Department of Energy Grant DE-FG02-03ER63630 (to A.B. and J.-H.M.).
Abbreviations
- NSP
(NIH/Ola × outbred M. spretus) × NIH/Ola backcross
- PMA
phorbol 12-myristate 13-acetate
- LOD
logarithm of the odds
- ΔCT
differential between threshold cycles.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Akhurst R. J. J. Clin. Invest. 2002;109:1533–1536. doi: 10.1172/JCI15970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Muraoka R. S., Dumont N., Ritter C. A., Dugger T. C., Brantley D. M., Chen J., Easterly E., Roebuck L. R., Ryan S., Gotwals P. J., et al. J. Clin. Invest. 2002;109:1551–1559. doi: 10.1172/JCI15234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Y. A., Dukhanina O., Tang B., Mamura M., Letterio J. J., MacGregor J., Patel S. C., Khozin S., Liu Z. Y., Green J., et al. J. Clin. Invest. 2002;109:1607–1615. doi: 10.1172/JCI15333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui W., Fowlis D. J., Bryson S., Duffie E., Ireland H., Balmain A., Akhurst R. J. Cell. 1996;86:531–542. doi: 10.1016/s0092-8674(00)80127-0. [DOI] [PubMed] [Google Scholar]
- 5.Derynck R., Akhurst R. J., Balmain A. Nat. Genet. 2001;29:117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- 6.Roberts A. B., Wakefield L. M. Proc. Natl. Acad. Sci. USA. 2003;100:8621–8623. doi: 10.1073/pnas.1633291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derynck R., Goeddel D. V., Ullrich A., Gutterman J. U., Williams R. D., Bringman T. S., Berger W. H. Cancer Res. 1987;47:707–712. [PubMed] [Google Scholar]
- 8.Sieweke M. H., Thompson N. L., Sporn M. B., Bissell M. J. Science. 1990;248:1656–1660. doi: 10.1126/science.2163544. [DOI] [PubMed] [Google Scholar]
- 9.Akhurst R. J., Derynck R. Trends Cell Biol. 2001;11:S44–S51. doi: 10.1016/s0962-8924(01)02130-4. [DOI] [PubMed] [Google Scholar]
- 10.Bhowmick N. A., Chytil A., Plieth D., Govska A. E., Dumont N., Shappell S., Washington M. K., Neilson E. G., Moses H. L. Science. 2004;303:848–851. doi: 10.1126/science.1090922. [DOI] [PubMed] [Google Scholar]
- 11.Welch D. R., Fabra A., Nakajima M. Proc. Natl. Acad. Sci. USA. 1990;87:7678–7682. doi: 10.1073/pnas.87.19.7678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Torre-Amione G., Beauchamp R. D., Koeppen H., Park B. H., Schreiber H., Moses H. L., Rowley D. A. Proc. Natl. Acad. Sci. USA. 1990;87:1486–1490. doi: 10.1073/pnas.87.4.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arteaga C. L., Hurd S. D., Winnier A. R., Johnson M. D., Fendly B. M., Forbes J. T. J. Clin. Invest. 1993;92:2569–2576. doi: 10.1172/JCI116871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cambien F., Ricard S., Troesch A., Mallet C., Generenaz L., Evans A., Arveiler D., Luc G., Ruidavets J. B., Poirier O. Hypertension. 1996;28:881–887. doi: 10.1161/01.hyp.28.5.881. [DOI] [PubMed] [Google Scholar]
- 15.Chen T., Triplett J., Dehner B., Hurst B., Colligan B., Pemberton J., Graff J. R., Carter J. H. Cancer Res. 2001;61:4679–4682. [PubMed] [Google Scholar]
- 16.Pasche B., Kolachana P., Nafa K., Satagopan J., Chen Y. G., Lo R. S., Brener D., Yang D., Kirstein L., Oddoux C., et al. Cancer Res. 1999;59:5678–5682. [PubMed] [Google Scholar]
- 17.Dunning A. M., Ellis P. D., McBride S., Kirschenlohr H. L., Healey C. S., Kemp P. R., Luben R. N., Chang-Claude J., Mannermaa A., Kataja V., et al. Cancer Res. 2003;63:2610–2615. [PubMed] [Google Scholar]
- 18.Ewart-Toland A., Chan J. M., Yuan J., Balmain A., Ma J. Cancer Epidemiol. Biomarkers Prev. 2004;13:759–764. [PubMed] [Google Scholar]
- 19.Ziv E., Cauley J., Morin P. A., Saiz R., Browner W. S. J. Am. Med. Assoc. 2001;285:2859–2863. doi: 10.1001/jama.285.22.2859. [DOI] [PubMed] [Google Scholar]
- 20.Hishida A., Iwata H., Hamajima N., Matsuo K., Mizutani M., Iwase T., Miura S., Emi N., Hirose K., Tajima K. Breast Cancer. 2003;10:63–69. doi: 10.1007/BF02967627. [DOI] [PubMed] [Google Scholar]
- 21.Yamada Y., Fujisawa M., Ando F., Niino N., Tanaka M., Shimokata H. J. Hum. Genet. 2002;47:243–248. doi: 10.1007/s100380200033. [DOI] [PubMed] [Google Scholar]
- 22.Yokota M., Ichihara S., Lin T. L., Nakashima N., Yamada Y. Circulation. 2000;101:2783–2787. doi: 10.1161/01.cir.101.24.2783. [DOI] [PubMed] [Google Scholar]
- 23.Grainger D. J., Heathcote K., Chiano M., Snieder H., Kemp P. R., Metcalfe J. C., Carter N. D., Spector T. D. Hum. Mol. Genet. 1999;8:93–97. doi: 10.1093/hmg/8.1.93. [DOI] [PubMed] [Google Scholar]
- 24.Shin A., Shu X. O., Cai Q., Gao Y. T., Zheng W. Cancer Epidemiol. Biomarkers Prev. 2005;14:1567–1570. doi: 10.1158/1055-9965.EPI-05-0078. [DOI] [PubMed] [Google Scholar]
- 25.Nagase H., Bryson S., Cordell H., Kemp C. J., Fee F., Balmain A. Nat. Genet. 1995;10:424–429. doi: 10.1038/ng0895-424. [DOI] [PubMed] [Google Scholar]
- 26.Ewart-Toland A., Briassouli P., de Koning J. P., Mao J. H., Yuan J., Chan F., MacCarthy-Morrogh L., Ponder B. A., Nagase H., Burn J., et al. Nat. Genet. 2003;34:403–412. doi: 10.1038/ng1220. [DOI] [PubMed] [Google Scholar]
- 27.Nagase H., Mao J. H., de Koning J. P., Minami T., Balmain A. Cancer Res. 2001;61:1305–1308. [PubMed] [Google Scholar]
- 28.Fowlis D. J., Flanders K. C., Duffie E., Balmain A., Akhurst R. J. Cell Growth Differ. 1992;3:81–91. [PubMed] [Google Scholar]
- 29.Yue J., Mulder K. M. J. Biol. Chem. 2000;275:35656. [PubMed] [Google Scholar]
- 30.Van Obberghen-Schilling E., Roche N. S., Flanders K. C., Sporn M. B., Roberts A. B. J. Biol. Chem. 1988;263:7741–7746. [PubMed] [Google Scholar]
- 31.Bremner R., Balmain A. Cell. 1990;61:407–417. doi: 10.1016/0092-8674(90)90523-h. [DOI] [PubMed] [Google Scholar]
- 32.Kemp C. J., Fee F., Balmain A. Cancer Res. 1993;53:6022–6027. [PubMed] [Google Scholar]
- 33.Tang Y., Sook Lee K., Yang H., Logan D. W., Wang S., McKinnon M. L., Holt L. J., Condie A., Luu M. T., Akhurst R. J. Genomics. 2005;85:60–70. doi: 10.1016/j.ygeno.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 34.Akhurst R. J., Fee F., Balmain A. Nature. 1988;331:363–365. doi: 10.1038/331363a0. [DOI] [PubMed] [Google Scholar]
- 35.Cook T., Gebelein B., Mesa K., Mladek A., Urrutia R. T. J. Biol. Chem. 1998;273:25929–25936. doi: 10.1074/jbc.273.40.25929. [DOI] [PubMed] [Google Scholar]
- 36.Kowanetz M., Valcourt U., Bergström R., Heldin C.-H., Moustakas A. Mol. Biol. Cell. 2004;24:4241–4254. doi: 10.1128/MCB.24.10.4241-4254.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ota T., Fujii M., Sugizaki T., Ishii M., Miyazawa K., Aburatani H., Miyazono K. J. Cell Physiol. 2002;193:299–318. doi: 10.1002/jcp.10170. [DOI] [PubMed] [Google Scholar]
- 38.Siegel P. M., Sku W., Massague M. J. Biol. Chem. 2003;12:35444–35450. doi: 10.1074/jbc.M301413200. [DOI] [PubMed] [Google Scholar]
- 39.Lasorella A., Rothschild G., Yokota Y., Russell R. G., Iavarone A. Nature. 2000;407:592–598. doi: 10.1038/35036504. [DOI] [PubMed] [Google Scholar]
- 40.Benezra R., Rafii S., Lyden D. Oncogene. 2001;20:8334–8341. doi: 10.1038/sj.onc.1205160. [DOI] [PubMed] [Google Scholar]
- 41.Paris P. L., Andaya A., Fridlyand J., Jain A. N., Weinberg V., Kowbel D., Brebner J. H., Simko J., Watson J. E., Volik S., et al. Hum. Mol. Genet. 2004;13:1303–1313. doi: 10.1093/hmg/ddh155. [DOI] [PubMed] [Google Scholar]
- 42.Dang C. V., O’Donnell K. A., Juopperi T. Cancer Cell. 2005;8:177–178. doi: 10.1016/j.ccr.2005.08.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.