Abstract
Cdc7 kinase, conserved through evolution, is known to be essential for mitotic DNA replication. The role of Cdc7 in meiotic recombination was suggested in Saccharomyces cerevisiae, but its precise role has not been addressed. Here, we report that Hsk1, the Cdc7-related kinase in Schizosaccharomyces pombe, plays a crucial role during meiosis. In a hsk1 temperature-sensitive strain (hsk1-89), meiosis is arrested with one nucleus state before meiosis I in most of the cells and meiotic recombination frequency is reduced by one order of magnitude, whereas premeiotic DNA replication is delayed but is apparently completed. Strikingly, formation of meiotic dsDNA breaks (DSBs) are largely impaired in the mutant, and Hsk1 kinase activity is essential for these processes. Deletion of all three checkpoint kinases, namely Cds1, Chk1, and Mek1, does not restore DSB formation, meiosis, or Cdc2 activation, which is suppressed in hsk1-89, suggesting that these aberrations are not caused by known checkpoint pathways but that Hsk1 may regulate DSB formation and meiosis. Whereas transcriptional induction of some rec genes and horsetail movement are normal, chromatin remodeling at ade6-M26, a recombination hotspot, which is prerequisite for subsequent DSB formation at this locus, is not observed in hsk1-89. These results indicate unique and essential roles of Hsk1 kinase in the initiation of meiotic recombination and meiosis.
Keywords: Cdc7-related kinase, checkpoint control, chromatin remodeling, meiosis, meiotic recombination
Although recent studies revealed the presence of a number of conserved factors involved in mitotic DNA replication (1), it is largely unknown whether they play roles in meiotic replication and recombination (2, 3). Studies in fission and budding yeasts indicated essential roles of Cdc2 kinase (1) and Clb5- and Clb6-dependent Cdc28 kinase (4), respectively, in premeiotic DNA replication. Orc, Cdc18, and MCM are also required for premeiotic DNA synthesis (3, 5), although their requirement may not be identical to that during mitotic replication (2). It is not known whether initiation is regulated by a common mechanism or the same set of replication origins is used during premeiotic S phase. In budding yeast, strong coupling of DNA replication and meiotic recombination has been shown (6–9).
Cdc7 kinase plays an essential role in firing replication origins during mitotic growth (10, 11). It was reported previously that meiotic recombination frequency is reduced, whereas premeiotic DNA replication proceeds, in the budding yeast cdc7ts mutants and that Cdc7 may play a role in synaptonemal complex formation during meiosis, a step stabilizing homologous pairing during meiotic recombination (12, 13), but the precise meiotic roles of Cdc7 have remained elusive. We report here that Hsk1, the fission yeast homologue of Cdc7 kinase, is required for efficient meiotic recombination and meiotic nuclear division. During meiosis, Hsk1 is required specifically for formation of dsDNA breaks (DSBs), an early event for meiotic recombination. The loss of DSB and meiosis are not corrected by loss of three major checkpoint kinases of fission yeast, suggesting that known checkpoint pathways are not involved. Our results also indicate requirement of hsk1+ for chromatin remodeling at the site of meiosis-specific DSBs. These results show unique and essential roles for Hsk1 kinase during meiotic processes.
Results
Defective Meiosis in hsk1-89 Mutant.
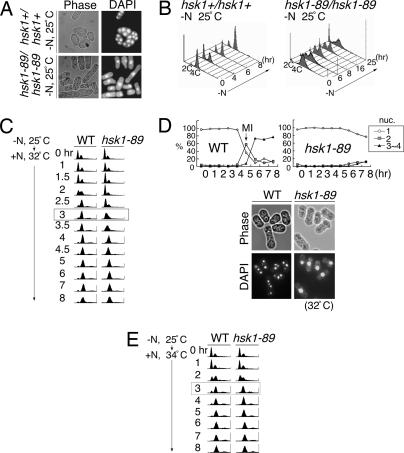
We previously reported hsk1-89, a temperature-sensitive allele of hsk1+ encoding the fission yeast Cdc7 homologue, and showed that hsk1+ function is essential for mitotic S phase (14). We noticed that the hsk1-89/hsk1-89 homozygous diploid cells frequently failed to complete meiosis, and most cells were arrested with one nucleus (Fig. 1A), indicative of the requirement of Hsk1 kinase for early stage(s) of meiosis, including premeiotic DNA synthesis, meiotic recombination, or meiosis I. When vegetatively growing diploid cells were shifted to nitrogen-free media at 25°C, DNA content in the majority of the WT cells initially shifted to 2C (C = haploid genome equivalent) (4 h after shift), and then resifted to 4C by 8 h, consistent with successful progression of premeiotic DNA replication (Fig. 1B Left). In contrast, the hsk1-89 diploid cells exhibited slow transition to 2C DNA content and 40–50% population kept being arrested in G1 phase until 16 h after nitrogen deprivation. They shifted to 4C DNA content by 25 h (Fig. 1B Right). These results show that initial transfer to 2C DNA content after nitrogen starvation and subsequent premeiotic DNA replication are delayed in the hsk1-89/hsk1-89 homozygous diploid.
Fig. 1.
Meiotic defect in hsk1-89 cells. (A and B) Diploid cells, NI298 (hsk1+/hsk1+) or NI325 (hsk1-89/hsk1-89), grown in minimal medium to 5 × 106 cells per ml, were starved for nitrogen at 25°C for 24 h and examined under a microscope after staining with DAPI (A) or analyzed by FACS at the indicated times after starvation (B). (C) JZ767 (pat1-114, WT) and NI394 (pat1-114 hsk1-89) cells were starved for nitrogen sources for 16 h at 25°C and released into media containing nitrogen at 32°C, nonpermissive for pat1-114 (0 h), to permit induction of meiosis. Cells were collected at various times after release, and DNA contents were analyzed by FACS. The difference of timing for premeiotic DNA replication among the two strains is most obvious at 3 h after induction (highlighted by a box). (D) Morphology of JZ767 (pat1-114, WT) and NI394 (pat1-114 hsk1-89) cells induced into meiosis. The photos were taken at 8 h after release at 32°C. Spore-like compartments containing three to four nuclei were observed in pat1-114, whereas most cells were arrested with one nucleus in pat1-114 hsk1-89. The graphs indicate the fractions of cells with one, two, or three to four nuclei at various time points after release. (E) JZ767 (pat1-114, WT) or NI394 (pat1-114 hsk1-89) cells were induced into meiosis, as described in Materials and Methods, except that release from nitrogen starvation was conducted at 34°C, semipermissive for hsk1-89. DNA content was analyzed by FACS at each time point after release. At this temperature, DNA replication proceeds in a kinetics similar to that of hsk1+ cells, apparently completing the process at 3 h after induction (highlighted by a box). (Magnifications: ×250, A; ×290, D.)
pat1+ encodes a protein kinase that phosphorylates Mei2 protein and negatively regulates meiosis at both the stage of premeiotic DNA replication and that of meiosis I (15, 16). In pat1-114 cells, meiosis can be induced in a haploid state at a nonpermissive temperature in a more synchronous manner. After nitrogen starvation for 16 h at 25°C, pat1-114 was shifted to 32°C (nonpermissive for mitotic growth) in medium containing nitrogen sources. Analyses of DNA content at various times after release indicate that premeiotic DNA replication occurs at 1.5–3 h after shift to a nonpermissive temperature in pat1-114 (Fig. 1C Left). By 8 h after shift, four nuclei compartments similar to spores were observed (Fig. 1D Lower). In the hsk1-89 background, transition to 2C DNA, detectable on FACS, did not start until 2.5 h after temperature shift, and DNA content slowly increased until it became a 2C amount by 4–5 h after the induction (Fig. 1C Right). Most cells were with one nucleus even at 8 h after shift (Fig. 1D). Therefore, meiosis induced in haploid cells by inactivation of Pat1 is also arrested in hsk1-89 cells before meiosis I, and premeiotic DNA replication is delayed.
hsk1-89 cells do not form colonies at 30–32°C, but can grow at 37°C, albeit at a reduced rate (17). This growth property may be caused by induction of chaperone protein(s) at 37°C, which may partially reactivate the kinase-compromised Hsk1-89 protein. When meiosis was induced in pat1-114 hsk1-89 at 34°C, at which hsk1-89 cells could form small colonies after prolonged incubation, very little delay of premeiotic S phase was observed, and DNA replication was mostly completed within 3 h after temperature shift, suggesting that the partially restored mutant Hsk1-89 protein could support premeiotic DNA replication in nearly normal timing (Fig. 1E). However, most cells still arrested with one nucleus under this condition (data not shown), indicating that meiosis was blocked.
Meiosis-Induced DSBs Are Not Generated in hsk1-89 Cells.
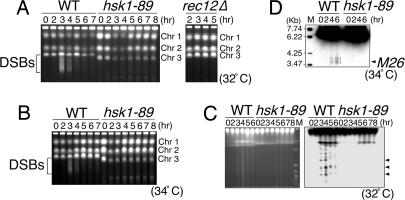
Initiation of meiotic recombination is marked by induction of multiple DSBs on the chromosomes (7, 18). DSBs generate broken chromosomes that can be detected in pulsed-field gel electrophoresis as fragments migrating faster in the gel. In hsk1+ cells, DSBs appear between 2 and 4 h after temperature shift of pat1-114 cells, and their intensity reaches maximum at 3 h (Fig. 2A), roughly correlating with the completion of premeiotic DNA synthesis. These broken chromosomes are not detected in a mutant lacking rec12+ encoding the protein that makes DSBs (ref. 19 and Fig. 2A Right), suggesting that they are indeed intermediates of meiotic recombination. In contrast, broken chromosomes are not detected in hsk1-89 even at later times when the bulk of DNA has been synthesized (5–8 h after temperature shift; Fig. 2A Left). DSBs were not detected in hsk1-89 cells at 34°C either, where premeiotic DNA replication proceeded in normal timing (Figs. 1E and 2B).
Fig. 2.
Induction of meiotic DSBs in hsk1-89 cells. (A and B) JZ767 (pat1-114, WT), NI394 (pat1-114 hsk1-89), and KO272 (pat1-114 rec12Δ) cells were induced into meiosis by temperature shift to 32°C (A) or 34°C (B). At the times indicated, DNAs were analyzed on pulsed-field gel electrophoresis. DNA fragments derived from DSBs induced during meiosis are indicated by brackets. Chr 1, Chr 2, and Chr 3 indicate the positions of chromosomes 1, 2, and 3, respectively. DNA plugs were prepared as described (3). The size of chromosome 3 in hsk1-89 cells is shorter than that in hsk1+ cells. A similar observation was also made in another hsk1ts cell (hsk1-1312; ref. 44) and other replication mutant cells. The alteration of the size of chromosome 3 is most likely to be caused by the expansion or shrinkage of the rRNA-encoding DNA repeats on this chromosome (data not shown). (C) DNA from hsk1+ (WT) or hsk1-89 cells was analyzed on a pulsed-field gel under an altered electrophoresis condition. (Left) Ethidium bromide staining of the gel. (Right) Southern blot hybridization of the same gel with a radioactive probe containing the ura1 gene located ≈0.75 Mb from the left end of the chromosome I. Arrowheads indicate three major fragments generated during meiotic DSB, which were detected by this probe (19). M indicates Saccharomyces cerevisiae chromosome DNA markers (BioWhittaker). (D) DSBs induced at the ade6-M26 locus during pat1-induced meiosis were examined under K342 (WT) and KO162 (hsk1-89) backgrounds. DNA plugs were prepared as described (25). Digestion of DNA plugs and preparation of the probe for Southern blotting were conducted as described (20). M indicates EcoT14I-digested DNA size markers (TAKARA). The arrowhead indicates the position of M26. The bracket indicates the position of the breaks.
DSBs can be detected in a more sensitive manner by analyzing the broken chromosomes near one end of chromosome I by hybridization using a radioactive ura1 probe (19). Chromosome DNAs were separated on pulsed-field gel electrophoresis under a different condition. Broken chromosomes were visible in the ethidium bromide-stained gel in hsk1+ cells at 3–6 h after induction, whereas they were not obvious in hsk1-89 cells (Fig. 2C Left). When this gel was hybridized with the ura1 probe, multiple bands, including three major bands previously identified (19), were detected in the hsk1+ cells, whereas the intensities of these bands were significantly reduced in the hsk1-89 background (Fig. 2C Right). We next examined DSBs at ade6–M26, a recombination hot spot, where specific meiotic DSBs have been detected (20). These DSBs can be detected at 4–6 h after induction of meiosis in pat1-114 cells with the rad50S background that accumulates unprocessed meiotic DSBs (21, 22). However, these cleavages were not detected in the hsk1-89 background (Fig. 2D). These data indicate an essential role of the hsk1+ function in the induction of meiotic DSBs.
Premeiotic DNA Replication Is Likely to Be Completed in hsk1-89 Cells Under a Nonpermissive Condition for Mitotic DNA Replication.
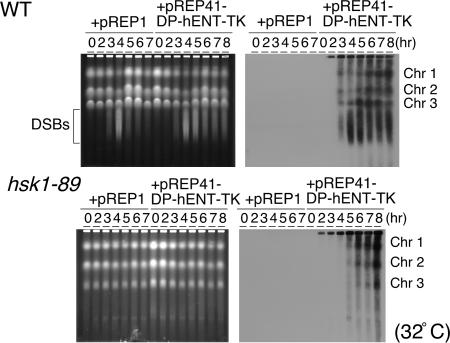
Strong coupling of premeiotic DNA replication and recombination in budding yeast (6–9) suggests a possibility that DNA replication may not be completed in hsk1-89 cells, which may be affecting the initiation of meiotic recombination. Therefore, we have examined the extent of premeiotic DNA replication in hsk1-89 cells. It is known that the replicating DNA does not enter agarose in pulsed-field gel electrophoresis. Only after completion and decatenation of the replicated chromosomes can the chromosomes enter agarose and become separated as distinct chromosome bands. We labeled the DNA with BrdU and monitored the progression of premeiotic DNA replication by Western blotting of the chromosomes fractionated by pulsed-field gel electrophoresis using anti-BrdU antibody (23). For this purpose, we have introduced a thymidine kinase (TK)-expressing plasmid in yeast cells. Under this condition, the progression of pat1-induced meiosis at 32°C was delayed for some unknown reason. Complete transition to 2C DNA content required 4 h in WT cells and 7 h in hsk1-89 cells (Fig. 7, which is published as supporting information on the PNAS web site). In WT cells, the chromosome bands incorporating BrdU were visible at 3 h after the induction of meiosis and increased at later hours (Fig. 3Upper), whereas they were detectable at 5 h after the induction and reached maximum at 8 h in hsk1-89 cells (Fig. 3 Lower). Delayed appearance of the replicated chromosomes in hsk1-89 cells is consistent with the results of FACS analyses of DNA contents. Nevertheless, this result strongly indicates that the three chromosomes are replicated to completion during the meiotic process in the mutant cells. The DSBs were not observed even at 12 h after induction, the timing long after completion of the premeiotic DNA replication in hsk1-89 cells (Fig. 8, which is published as supporting information on the PNAS web site), indicating that the formation of DSBs is not simply delayed but is indeed impaired. We also confirmed the replication of the specific segments of the chromosome by directly analyzing the replicated molecules (Fig. 9, which is published as supporting information on the PNAS web site). These results mostly rule out the possibility that loss of DSB is caused by the incomplete replication of the genome. However, we cannot completely rule out the possibility that minor defects in DNA replication that cannot be detected by the methods used in this study are responsible for the loss of DSBs. In budding yeast, it was previously reported that the overall extent of chromosome replication is not significantly affected in cdc7ts mutants during the course of meiosis (12, 13), and our results in fission yeast are consistent with these reports.
Fig. 3.
BrdU labeling of the replicating chromosomes and analyses on pulsed-field gel electrophoresis. JZ767 (pat1-114, WT) or NI394 (pat1-114 hsk1-89) cells harboring the plasmid indicated were grown in the absence of thiamine, and meiosis was induced as described in Materials and Methods. At the times indicated, genomic DNAs were analyzed on pulsed-field gel electrophoresis. (Right) After the run, the gel was blotted with anti-BrdU antibody. (Left) Ethidium bromide staining of the same gel. The bracket indicates the positions of meiotic DSBs.
Hsk1 Kinase Activity Is Required for Timely Premeiotic DNA Synthesis and DSB Formation.
Both premeiotic DNA synthesis and DSBs were restored in the mutant by introduction of a plasmid expressing WT Hsk1, whereas kinase-negative (K129D) or kinase-attenuated (K129R,K130S) forms of Hsk1 (24) did not restore them under the same conditions (Fig. 10 A and B, which is published as supporting information on the PNAS web site, and data not shown), indicating that the kinase activity of Hsk1 is required for timely DNA synthesis and induction of DSBs during meiosis. The K129R,K130S mutant can complement the temperature-sensitive growth of hsk1-89 when overexpressed (24). However, this mutant did not restore defective meiosis, including timing of premeiotic DNA synthesis (Fig. 10A) and DSB formation even after overproduction (Fig. 10B). These results show that the requirement of Hsk1 kinase activity during meiosis may be more stringent than that for mitotic growth. The size of chromosome 3 is shorter in hsk1-89 than in WT cells, but is partially corrected by introduction of a plasmid expressing the WT Hsk1 but not the kinase-attenuated one. The shortening of chromosome 3 is observed not only in nitrogen-starved cells but also in vegetatively growing cells, and thus is not caused by the problems in the process of meiosis. We have shown that it is caused by reduction of the copy numbers of the rRNA-encoding DNA on chromosome 3 (data not shown).
Transcriptional Induction of Some rec Genes and Horsetail Movement in hsk1-89 Are Normal.
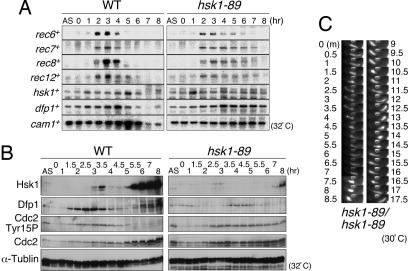
A series of rec genes are transcriptionally activated during meiosis. The induction of these rec genes, which depends on the Cdc10–Rep1 transcriptional complex (7, 25), is needed for meiotic recombination. We have examined whether this transcriptional activation is affected in hsk1-89 cells. Under the hsk1+ background, rec6+, rec7+, rec8+, and rec12+ genes were induced at 2 h after temperature shift in pat1-114 cells and disappeared by 5–6 h. Under the hsk1-89 background, all of these genes were induced at identical timings. The transcripts were detected until 5–7 h after induction and disappeared by 8 h (Fig. 4A and Fig. 11A, which is published as supporting information on the PNAS web site). The hsk1+ transcript, on the other hand, displayed biphasic induction, peaking at 1 and 5 h after induction of meiosis, whereas the dfp1+/him1+ transcript, encoding an activation subunit for Hsk1 (10, 24, 26, 27), increased during the first 4 h and decreased after that in hsk1+ cells, suggesting that transcriptions of hsk1+ and dfp1+/him1+ may be under regulation distinct from that for rec genes. They were induced with similar timing in hsk1-89 cells, and their levels were maintained until later hours. On the protein level, Dfp1/Him1 protein starts to increase at 1.5 h after temperature shift, around the time of premeiotic DNA synthesis, whereas the Hsk1 protein level transiently increased at 3–3.5 h after the shift, roughly the time of DSB formation, and diminished again until it started to increase again at 5.5 h after induction. In hsk1-89 cells, Hsk1 protein slightly increased at 2.5–3 h after the shift and increased again only at 7–8 h, whereas Dfp1/Him1 protein showed delayed induction at 3.5–5.0 h after the shift (Figs. 4B and 11B). These results indicate that at least the transcriptional induction of some meiotic recombination genes and replication factors are intact in hsk1-89 cells. Similarly, telomere clustering and horsetail movement, required for efficient homologue pairing and meiotic recombination (28), were detected in the hsk1-89/hsk1-89 diploid cells as actively as in the WT diploid cells (Fig. 4C).
Fig. 4.
Expression of various proteins and horsetail movement during meiosis in hsk1-89 cells. (A and B) JZ767 (pat1-114, WT) and NI394 (pat1-114 hsk1-89) cells were induced into meiosis, and RNA and whole-cell extracts were prepared at the times indicated after temperature shift. They were analyzed, respectively, by Northern analyses using the probes for the genes indicated (A) or by Western analyses with the various antibodies indicated (B). AS, culture growing asynchronously at 25°C. The band intensities were quantified, and relative intensities are presented in Fig. 11. (C). Time-lapse observation of horsetail movement of hsk1-89/hsk1-89. To examine the effect of hsk1-89 mutation on horsetail movement, NI322 (hsk1-89 h+) and NI324 (hsk1-89 h−) cells were conjugated on a sporulation agar (SPA) plate. After incubation at 30°C for 17 h, cells were stained with Hoechst 33342 (4 μg/ml in SPA) and mounted on a thin SPA layer on a glass slide, and pictures were taken every 30 s with a Zeiss Axiophoto equipped with an AQUACOSMOS imaging system (Hamamatsu Photonics, Hamamatsu City, Japan). Photos of representative cells at selected time points are shown.
Known Checkpoint Kinases Are Not Involved in Suppression of DSB and Meiosis in hsk1-89.
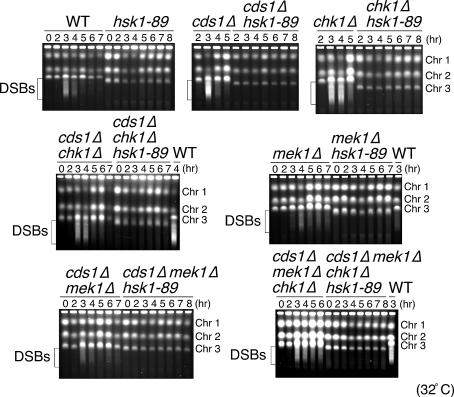
We then examined whether the meiotic defects in hsk1-89 result from checkpoint-mediated inhibition caused by perturbation of the preceding events. We therefore investigated the effects of mutations in checkpoint kinases known to be involved in DNA damage and replication checkpoint responses. Delay of premeitoic DNA synthesis and generation of one nucleus cells were not suppressed by either chk1 or cds1 mutations (Fig. 12, which is published as supporting information on the PNAS web site, and data not shown). Similarly, DSBs were not generated even in the chk1 or cds1 null background (Fig. 5). Furthermore, neither replication timing nor DSB formation was restored in cds1Δ chk1Δ double mutant. We also examined the effect of mek1 mutation, which encodes a meiosis-specific checkpoint kinase related to Cds1 (29, 30). Introduction of mek1Δ in hsk1-89 did not restore timely premeiotic DNA replication, DSBs, or meiosis. Furthermore, they were not restored even in the presence of cds1Δ mek1Δ double or cds1Δ mek1Δ chk1Δ triple mutations. The inability of checkpoint mutations to rescue the defects of hsk1-89 is in a sharp contrast to hydroxyurea-induced inhibition of DSB formation, which is restored by mutations in the Rad3–Cds1 checkpoint pathway (31, 32).
Fig. 5.
Effect of disruption of Cds1, Chk1, Mek1, and combinations of these checkpoint kinase mutations on DSB formation. pat1-114 (WT) or pat1-114 hsk1-89 cells in combination with checkpoint mutations indicated were induced into meiosis, and DSB formation was analyzed by pulsed-field gel electrophoresis. The brackets indicate the positions of the meiotic breaks. Chr 1, Chr 2, and Chr 3 indicate the positions of chromosomes 1, 2, and 3, respectively.
Cdc2 kinase needs to be activated before cell division, and this activation is partly regulated by phosphorylation of Tyr-15 residue (1, 33). Cdc2 protein was present at more or less constant levels throughout meiosis, except for later stages when it significantly increased in its abundance (Fig. 4B). In contrast, phosphorylation of Tyr-15 of Cdc2 increased at 2–3.5 h after the shift and then disappeared in hsk1+ cells, whereas this phosphorylation was maintained until later stages in hsk1-89 cells (Figs. 4B and 11). This sustained phosphorylation of Tyr-15 of Cdc2 may be at least partially responsible for inhibition of meiosis I and II in the hsk1 mutant. We examined whether the Tyr-15 phosphorylation in hsk1-89 is regulated by checkpoint kinases. Under cds1Δ, chk1Δ, mek1Δ, cds1Δ mek1Δ, cdslΔ chk1Δ, or cdslΔ chk1Δ mek1Δ backgrounds, sustained phosphorylation of Cdc2 Tyr-15 was still observed (Fig. 13, which is published as supporting information on the PNAS web site), suggesting that these checkpoint kinases are not involved in inhibition of dephosphorylation of this tyrosine residue.
These results show that delay of premeiotic DNA replication, inhibition of meiotic DSB, or meiotic divisions in hsk1-89 is not under the regulation of known checkpoint kinases.
Meiotic Recombination Is Impaired in hsk1-89 Cells.
Reduced levels of DSBs in hsk-89 predict the reduced recombination frequency. We have therefore measured recombination frequency in spores generated in hsk1-89 cells. Approximately 10% of the hsk1-89 cells undergo sporulation and generate four-spored asci. We measured the intragenic recombination frequency between ade6-469 and ade6-M26 and intergenic recombination frequency in the leu1-mat1 and leu1-his5 loci (Table 1). We found that the intragenic recombination frequencies decreased by ≈7-fold in hsk1-89 cells (0.78% and 0.11% in WT and hsk1-89, respectively). The intergenic recombination frequencies in the WT cells were 12.0% for leu1/mat1 and 13.5% (leu+ his+)/12.5% (leu− his−) for leu1/his5, respectively, which were roughly consistent with the values reported (29). In contrast, they were 0.92% and 2.08% (leu+ his+)/1.56% (leu− his−), respectively, in the hsk1-89 cells, which are about the same as the values observed in some rec mutants (34). These results show that recombination is indeed impaired in hsk1-89 cells, as was reported in cdc7ts cells in budding yeast (12, 13).
Table 1.
hsk1-89 mutant is defective in meiotic recombination
| Cell type | Intragenic recombination frequencies (Ade+ recombinants/total colonies counted), M26 × 469 | Intergenic recombinantion frequencies, % |
||
|---|---|---|---|---|
| Leu+ Mat1-M* | Leu+ His+† | Leu− His−† | ||
| WT | 82/10572 (0.78%) | 12 | 13.5 | 12.5 |
| hsk1-89 | 8/7080 (0.11%) | 0.92 | 2.08 | 1.56 |
The frequencies of meiotic intragenic recombination were measured in the WT (K37 × K41, WT) and hsk1-89 (KO185 × KO461) backgrounds by random spore analyses. Ade+ frequencies were determined by red (Ade−) or white (Ade+) colony color selection on yeast extract plates. The frequencies of meiotic intergenic recombination was measured on the leu1-mat1 and leu1-his5 intervals on the chromosome 2 by random spore analyses of crosses in the following:
*, WT (NT145 × K27, WT) and hsk1-89 (KO147 × KO159) backgrounds or
†, diploid WT (D20) and hsk1-89 (D21) cells. Glusulase-treated cells were further treated by 30% ethanol for 5 min to kill remaining diploid cells. The surviving colonies were examined for the markers indicated. All the Leu+ His+ colonies were confirmed to be haploids.
Chromatin Remodeling at the Site of DSB Is Impaired in hsk1-89 Cells.
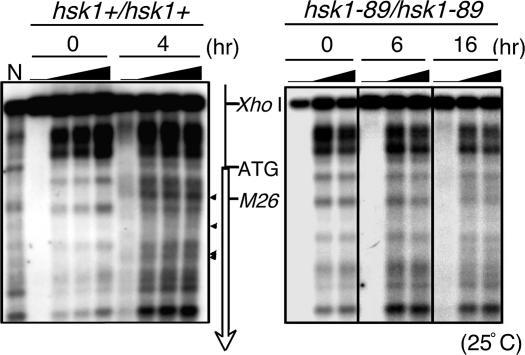
Chromatin remodeling is known to play an important role in the generation of DSBs at ade6-M26 (20, 35, 36). We have examined whether the hsk1-89 mutation affects chromatin remodeling at this locus. In the hsk1+ background, the alteration of micrococcal nuclease cleavage patterns started to appear at 3–4 h after induction of meiosis in diploid cells and continued to increase until 6 h (Fig. 6Left and data not shown), indicative of alteration of chromatin structures. In contrast, in hsk1-89 cells, very little change in the cleavage pattern was detected even at 16 h after induction of meiosis (Fig. 6 Right). Strong chromatin remodeling observed in the WT cells was not obvious in hsk1-89 cells during pat1-induced meiosis (Fig. 14, which is published as supporting information on the PNAS web site). These results show that the hsk1 mutation abrogates chromatin remodeling at the site of meiotic DSBs, which may cause the loss of DSBs.
Fig. 6.
Chromatin remodeling during meiosis in hsk1-89 cells. Chromatin remodeling was examined in the ade6-M26 region during meiosis induced in diploid cells D20 (M26 hsk1+/M26 hsk1+, Left) and D21 (M26 hsk1-89/M26 hsk1-89, Right). The arrowheads indicate the bands derived from the chromatin remodeling observed in the WT cells. Micrococcal nuclease was titrated (0, 5, 10, or 20 units per ml for hsk1+/hsk1+ and 0, 10, or 20 units per ml for hsk1-89/hsk1-89) at each time point, as indicated by the filled triangles. The open arrow indicates the coding region and direction of the ade6 gene. The positions of the XhoI site, ATG initiation codon, and M26 DSB sites are also indicated. N, MNase digestion of naked DNA.
Discussion
The data presented in this study demonstrate essential roles of hsk1+, the fission yeast homologue of Cdc7 kinase known to be essential for mitotic S phase (10, 26, 27), during meiosis. Our results indicate that hsk1+ is required for both meiotic recombination and meiotic nuclear and cell divisions. Most strikingly, our data show that hsk1+ is required for induction of DSBs, one of the initial events for meiotic recombination. These meiotic defects in the hsk1 mutant are not caused by known checkpoint pathways.
Strong coupling between premiotic DNA replication and meiotic recombination has been reported in budding yeast (6–9). Although premeiotic DNA replication is slightly delayed in hsk1-89 cells, evidence indicates that recombination and meiotic defects may not be consequences of incomplete DNA replication. (i) Analyses of replicated chromosomes on pulsed-field gel electrophoresis strongly indicate the completion and proper decatenation of all of the chromosomes (Figs. 3 and 7). (ii) Analyses of BrdU-labeled replicated molecules strongly suggest the replication of the entire genome in hsk1-89 cells (Fig. 9). (iii) At 34°C, a temperature that permits a limited growth of hsk1-89, premeiotic DNA replication is induced apparently in normal timing, and yet no DSB is induced and cells arrest with one nucleus (Figs. 1 and 2). (iv) Both intragenic and intergenic recombination frequency is significantly reduced in the mutant spores, which may have completed meiotic processes, including premeiotic DNA replication and meiotic division (Table 1). Although the deteriorative effect of minor defects in premeiotic DNA replication on DSB formation cannot be formally ruled out, these findings strongly indicate that Hsk1 either directly or indirectly regulates meiotic recombination. They are in keeping with previous reports showing that the bulk amount of DNA synthesis during meiosis is not affected in cdc7ts mutants (12, 13). Apparent completion of premeiotic DNA replication in hsk1-89 under the condition nonpermissive for mitotic DNA replication suggests differential roles of Cdc7 kinase in DNA replication during mitotic and meiotic phases.
Chromatin remodeling at a recombination hot spot is not induced in the hsk1-89 cells, suggesting a possibility that Hsk1 may regulate the processes of chromatin remodeling. We previously reported that sister chromatid cohesion during mitosis is partially impaired in hsk1-89 cells (14). It is an intriguing possibility that defective meiotic sister chromatid cohesion in hsk1-89 may lead to impaired chromatin remodeling, a prerequisite for meiotic recombination.
The requirement of Hsk1 for meiotic nuclear division was unexpected. It was reported that meiosis I can proceed in mutants lacking recombination functions, including rec12+, essential for DSB induction (37). Therefore, the block of meiosis I in hsk1-89 cells may not be simply caused by the absence of recombination. In mammalian cells, Cdc7 has been implicated in mitosis as well (38). Therefore, we speculate that Hsk1 may play some unknown essential functions during meiotic nuclear and cell divisions independent from its S-phase functions.
We recently reported severe defects in the development of testes and ovaries in genetically engineered mice with an attenuated Cdc7 kinase level (39). Analyses of defective testes indicate arrest of spermatogenesis at premeiotic phases or early prophase I of meiosis I, which is roughly consistent with premeiotic arrest in hsk1-89 cells. Thus, the essential roles of Cdc7 kinase during the processes of meiosis may be conserved through evolution.
Materials and Methods
Yeast Strains, Media, and Genetics.
Schizosaccharomyces pombe strains used in this study are listed in Table 2, which is published as supporting information on the PNAS web site. Cells were grown in rich medium or minimal medium (MM) containing the required supplements. Sporulation agar medium containing 10 g/liter of glucose, 1 g/liter of KH2PO4, 1 ml of ×1,000 vitamin stock solution per liter−1, or malt extract agar medium was used for genetic crosses and sporulation. Nitrogen-free derivative MM-N was used for nitrogen starvation experiments as described (40). General genetic methods (41) and the procedure for transformation (42) were as described.
Induction of Meiosis in pat1-114 Cells.
Cells, grown at 25°C in MM (1% glucose) supplemented with 100 μg/ml of leucine and 100 μg/ml of adenine per liter−1 to 5 × 106 cells/ml, were washed with MM-NH4Cl (1% glucose) four times and resuspended in MM-NH4Cl (1% glucose) supplemented with 25 μg/ml of leucine, followed by incubation for 16 h at 25°C. After addition of an equal volume of prewarmed MM (containing 1 g/liter of NH4Cl and 1% glucose) supplemented with 50 μg/ml of leucine and 70 μg/ml of adenine, meiosis was induced by shifting the temperature to 32°C or 34°C. Equal volumes of samples were withdrawn at each time point from each culture for FACS analyses and DNA analyses on pulsed-field gel electrophoresis.
Detection of Meiotic DSBs by Pulsed-Field Gel Electrophoresis.
pat1-114 cells released from nitrogen starvation were harvested every hour and treated as described (3). Pulsed-field gel electrophoresis was carried out in 0.6% chromosomal grade agarose gel (Bio-Rad) on a Bio-Rad CHEF-DRIII apparatus and recirculated at 14°C. Electorophoresis was performed for 48–72 h at 1.5 V/cm in 0.5× TAE buffer (Tris-acetate-EDTA), with a switch time of 30 min at an included angle of 106°. For analyses by Southern hybridization, electrophoresis was conducted in 0.8% chromosomal grade agarose gel (Bio-Rad) for 24 h at 6 V/cm in 0.5× TBE buffer (89 mM Tris/89 mM boric acid/2 mM EDTA, pH 8.0), with a linearly ramped 60- to 120-s switch time at an included angle of 120°, as described (19).
Incorporation of BrdU.
JZ767 and NI394 harboring pREP41-DP-hENT-TK (expressing human TK gene; a gift from Hisao Masukata, Osaka University, Osaka, Japan) or pREP1, grown in the absence of thiamine, were starved for nitrogen source for 16 h and transferred to 32°C to induce meiosis. BrdU (400 μg/ml) was added to the culture 30 min before temperature shift, and cells were harvested every hour (23). Genomic DNA was prepared, and pulsed-field gel electrophoresis was carried out as described above. After electrophoresis, the gel was stained with ethidium bromide and soaked in 0.25 M HCl for 20 min, followed by incubation in a solution of 0.5 M NaOH and 1.5 M NaCl for 30 min and transfer to nylon membrane (Hybond N+, Amersham Pharmacia Biosciences). The membrane was washed in 2× SSC briefly and dried. Immunodetection was conducted with anti-BrdU antibody (a gift from Katsuyuki Tamai, Cyclex, Nagano, Japan).
Antibodies.
Anti-Hsk1 and anti-Dfp1/Him1 antibodies were affinity-purified and used as described (26, 43). B-5-1-2 (Sigma), anti-Cdc2 (sc-53, PSTAIRE; Santa Cruz Biotechnology), and anti-phospho-Cdc2 (Tyr-15; Cell Signaling Technology, Beverly, MA) antibodies were used to detect α-tubulin, Cdc2, and tyrosine-phosphorylated Cdc2, respectively.
Analyses of Chromatin Remodeling at the ade6–M26 Locus.
Analyses of chromatin structures by indirect end labeling were done as reported (35). To analyze chromatin structures around the ade6+ promoter, genomic DNA was digested with XhoI and separated by electrophoresis in 1.5% agarose gel (40 cm long) in TAE buffer (Tris-acetate-EDTA). The separated DNA fragments were alkali-transferred to charged nylon membranes (Biodyne B membrane, Pall). The probe used for detection of the ade6+ locus was as described (35).
See Supporting Text, which is published as supporting information on the PNAS web site, for more details.
Supplementary Material
Acknowledgments
We thank Dr. Yuichi Iino (University of Tokyo, Tokyo) for the pat1-114 strain; Dr. Yoshinori Watanabe (University of Tokyo, Tokyo) for various strains, valuable suggestions for many aspects of this work, and advice for recombination frequency measurement; Dr. Takehiko Shibata (RIKEN, Saitama, Japan) for valuable suggestions; Dr. Hisao Masukata for the gift of pREP41-DP-hENT-TK; Dr. Pedro San-Segundo (Universidad de Salamanca, Salamanca, Spain) for the mek1Δ strain; Dr. Katsuyuki Tamai for anti-BrdU antibody; and Ai Ishii (Tokyo Metropolitan Institute of Medical Science) for suggestions in conducting FACS analyses. This work was supported in part by grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to H.M.) and grants for basic research from Bio-oriented Technology Research Advancement Institution (to K. Ohta and T. Shibata).
Abbreviations
- DSB
dsDNA break
- TK
thymidine kinase
- MM
minimal medium.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Iino Y., Hiramine Y., Yamamoto M. Genetics. 1995;140:1235–1245. doi: 10.1093/genetics/140.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Forsburg S. L., Hodson J. A. Nat. Genet. 2000;25:263–268. doi: 10.1038/77015. [DOI] [PubMed] [Google Scholar]
- 3.Murakami H., Nurse P. Nat. Genet. 2001;28:290–293. doi: 10.1038/90142. [DOI] [PubMed] [Google Scholar]
- 4.Stuart D., Wittenberg C. Genes Dev. 1998;12:2698–2710. doi: 10.1101/gad.12.17.2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindoner K., Gregan J., Montgomery S., Kearsey S. E. Mol. Biol. Cell. 2002;13:435–444. doi: 10.1091/mbc.01-11-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borde V., Goldman A. S. H., Lichten M. Science. 2000;290:806–809. doi: 10.1126/science.290.5492.806. [DOI] [PubMed] [Google Scholar]
- 7.Davis L., Smith G. R. Proc. Natl. Acad. Sci. USA. 2001;98:8395–8402. doi: 10.1073/pnas.121005598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith K. N., Penkner A., Ohta K., Klein F., Nicolas A. Curr. Biol. 2001;11:88–97. doi: 10.1016/s0960-9822(01)00026-4. [DOI] [PubMed] [Google Scholar]
- 9.Murakami H., Botde V., Shibata T., Lichten M., Ohta K. Nucleic Acids Res. 2003;31:4085–4090. doi: 10.1093/nar/gkg441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masai H., Arai K. J. Cell. Physiol. 2002;190:287–296. doi: 10.1002/jcp.10070. [DOI] [PubMed] [Google Scholar]
- 11.Sclafani R. A. J. Cell Sci. 2000;113:2111–2117. doi: 10.1242/jcs.113.12.2111. [DOI] [PubMed] [Google Scholar]
- 12.Schild D., Byers D. Chromosoma. 1978;70:109–130. doi: 10.1007/BF00292220. [DOI] [PubMed] [Google Scholar]
- 13.Hollingsworth E. R., Sclafani A. R. Chromosoma. 1993;102:415–420. doi: 10.1007/BF00360406. [DOI] [PubMed] [Google Scholar]
- 14.Takeda T., Ogino K., Tatebayashi K., Ikeda H., Arai K., Masai H. Mol. Biol. Cell. 2001;12:1257–1274. doi: 10.1091/mbc.12.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iino Y., Yamamoto M. Proc. Natl. Acad. Sci. USA. 1985;82:2447–2451. doi: 10.1073/pnas.82.8.2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watanabe Y., Yamamoto M. Cell. 1994;78:487–498. doi: 10.1016/0092-8674(94)90426-x. [DOI] [PubMed] [Google Scholar]
- 17.Matsumoto S., Ogino K., Noguchi E., Russell P., Masai H. J. Biol. Chem. 2005;280:42536–42542. doi: 10.1074/jbc.M510575200. [DOI] [PubMed] [Google Scholar]
- 18.Fox M. E., Smith G. R. Prog. Nucleic Acid Res. Mol. Biol. 1998;61:345–378. doi: 10.1016/s0079-6603(08)60831-4. [DOI] [PubMed] [Google Scholar]
- 19.Cervantes M. D., Farah J. A., Smith G. R. Mol. Cell. 2000;5:883–888. doi: 10.1016/s1097-2765(00)80328-7. [DOI] [PubMed] [Google Scholar]
- 20.Steiner W. W., Schreckhise R. W., Smith G. R. Mol. Cell. 2002;9:847–855. doi: 10.1016/s1097-2765(02)00489-6. [DOI] [PubMed] [Google Scholar]
- 21.Cao L., Alani E., Kleckner N. Cell. 1990;61:1089–1101. doi: 10.1016/0092-8674(90)90072-m. [DOI] [PubMed] [Google Scholar]
- 22.Young J. A., Schreckhise R. W., Steiner W. W., Smith G. R. Mol. Cell. 2002;9:253–263. doi: 10.1016/s1097-2765(02)00452-5. [DOI] [PubMed] [Google Scholar]
- 23.Lengronne A., Pasero P., Bensimon A., Schwob E. Nucleic Acid Res. 2001;29:1433–1442. doi: 10.1093/nar/29.7.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogino K., Takeda T., Matsui E., Iiyama H., Taniyama C., Arai K., Masai H. J. Biol. Chem. 2001;276:31376–31387. doi: 10.1074/jbc.M102197200. [DOI] [PubMed] [Google Scholar]
- 25.Lin Y., Smith G. R. Genetics. 1994;136:769–779. doi: 10.1093/genetics/136.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masai H., Miyake T., Arai K. EMBO J. 1995;14:3094–3104. doi: 10.1002/j.1460-2075.1995.tb07312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown G. W., Kelly T. J. J. Biol. Chem. 1998;273:22083–22090. doi: 10.1074/jbc.273.34.22083. [DOI] [PubMed] [Google Scholar]
- 28.Chikashige Y., Ding D. Q., Funabiki H., Haraguchi T., Mashiko S., Yanagida M., Hiraoka Y. Science. 1994;264:270–273. doi: 10.1126/science.8146661. [DOI] [PubMed] [Google Scholar]
- 29.Pérez-Hidalgo L., Moreno S., San-Segundo P. A. J. Cell Sci. 2003;116:259–271. doi: 10.1242/jcs.00232. [DOI] [PubMed] [Google Scholar]
- 30.Shimada M., Nabeshima K., Tougan T., Nojima H. EMBO J. 2002;21:2807–2818. doi: 10.1093/emboj/21.11.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tonami Y., Murakami H., Shirahige K., Nakanishi M. Proc. Natl. Acad. Sci. USA. 2005;19:5797–5801. doi: 10.1073/pnas.0407236102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogino K., Masai H. J. Biol. Chem. 2006;281:1338–1344. doi: 10.1074/jbc.M505767200. [DOI] [PubMed] [Google Scholar]
- 33.Gold K. L., Nurse P. Nature. 1989;342:39–45. doi: 10.1038/342039a0. [DOI] [PubMed] [Google Scholar]
- 34.DeVeaux L. C., Hoagland N. A., Smith G. R. Genetics. 1992;130:251–262. doi: 10.1093/genetics/130.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mizuno K., Emura Y., Baur M., Kohli J., Ohta K., Shibata T. Genes Dev. 1997;11:876–886. doi: 10.1101/gad.11.7.876. [DOI] [PubMed] [Google Scholar]
- 36.Yamada T., Mizuno K., Hirota K., Kon N., Wahls W. P., Hartsuiker E., Murofushi H., Shibata T., Ohta K. EMBO J. 2004;23:1792–1803. doi: 10.1038/sj.emboj.7600138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis L., Smith G. R. Genetics. 2003;163:857–874. doi: 10.1093/genetics/163.3.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoshizawa-Sugata N., Ishii A., Taniyama C., Matsui E., Arai K., Masai H. J. Biol. Chem. 2004;280:13062–13070. doi: 10.1074/jbc.M411653200. [DOI] [PubMed] [Google Scholar]
- 39.Kim J. M., Takemoto N., Arai K., Masai H. EMBO J. 2003;22:5260–5272. doi: 10.1093/emboj/cdg497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Isshiki T., Mochizuki N., Maeda T., Yamamoto M. Gen. Dev. 1992;6:2455–2462. doi: 10.1101/gad.6.12b.2455. [DOI] [PubMed] [Google Scholar]
- 41.Moreno S., Klar A., Nurse P. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 42.Okazaki K., Okazaki N., Kume K., Jinno S., Tanaka K., Okayama H. Nucleic Acids Res. 1990;18:6485–6489. doi: 10.1093/nar/18.22.6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takeda T., Ogino K., Matsui E., Cho M. K., Kumagai H., Miyake T., Arai K., Masai H. Mol. Cell. Biol. 1999;19:5535–5547. doi: 10.1128/mcb.19.8.5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Snaith H. A., Brown G. W., Forsburg S. L. Mol. Cell. Biol. 2000;21:7922–7932. doi: 10.1128/mcb.20.21.7922-7932.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.