Abstract
The pathway of gene expression in higher eukaryotes involves a highly complex network of physical and functional interactions among the different machines involved in each step of the pathway. Here we established an efficient in vitro system to determine how RNA polymerase II (RNAP II) transcription is functionally coupled to pre-mRNA splicing. Strikingly, our data show that nascent premessenger RNA (pre-mRNA) synthesized by RNAP II is immediately and quantitatively directed into the spliceosome assembly pathway. In contrast, nascent pre-mRNA synthesized by T7 RNA polymerase is quantitatively assembled into the nonspecific H complex, which consists of heterogeneous nuclear ribonucleoprotein (hnRNP) proteins and is inhibitory for spliceosome assembly. Consequently, RNAP II transcription results in a dramatic increase in both the kinetics of splicing and overall yield of spliced mRNA relative to that observed for T7 transcription. We conclude that RNAP II mediates the functional coupling of transcription to splicing by directing the nascent pre-mRNA into spliceosome assembly, thereby bypassing interaction of the pre-mRNA with the inhibitory hnRNP proteins.
Keywords: RNAP II transcription, coupling transcription to splicing, pre-mRNA splicing, spliceosome assembly
In higher eukaryotes, pre-mRNAs are synthesized by RNA polymerase II (RNAP II) as large precursors containing multiple introns and exons. The premessenger RNAs (pre-mRNAs) undergo several processing steps, including capping at the 5′ end, splicing to remove introns, and polyadenylation at the 3′ end. All of these processing steps occur cotranscriptionally (Hirose and Manley 2000; Cramer et al. 2001b; Maniatis and Reed 2002; Orphanides and Reinberg 2002; Kornblihtt et al. 2004; Proudfoot 2004; Bentley 2005). In addition, the three processing steps appear to be functionally coupled to RNAP II transcription (Proudfoot 2004; Bentley 2005). Functional coupling of two processes has been defined as one process affecting the rate and/or efficiency of another (Moteki and Price 2002; Adamson et al. 2005). At present, the most is known about functional coupling of transcription to capping (Ho et al. 1998; Ho and Shuman 1999; McCracken et al. 1997a; Yue et al. 1997; Cho et al. 1997, 1998; Hirose and Manley 2000; Moteki and Price 2002). In this case, the capping complex interacts directly with the C-terminal domain (CTD) of RNAP II and caps the nascent pre-mRNA when only ∼25 nucleotides (nt) have emerged from the exit pore of the polymerase (for review, see Hirose and Manley 2000). This functional coupling is thought to serve as an important timing mechanism, which is required to prevent degradation of the nascent transcript. The recent development of efficient in vitro systems for functional coupling of RNAP II transcription to 3′-end cleavage/polyadenylation has also led to insights into the mechanisms involved in this coupling event (Yonaha and Proudfoot 1999, 2000; Adamson et al. 2005; Rigo et al. 2005). In particular, the nascent pre-mRNA itself is thought to play a key role in establishing an interaction between RNAP II and the 3′-end cleavage machinery (Rigo et al. 2005).
Cotranscriptional splicing in metazoans has been directly visualized in electron microscopic studies of the giant Balbiani ring of Chironomus tentans and of the Drosophila melanogaster chorion genes (Beyer and Osheim 1988; Bauren and Wieslander 1994; Kiseleva et al. 1994; Wetterberg et al. 2001). Several lines of evidence indicate that splicing in metazoans is also functionally coupled to transcription (Bentley 1999, 2002; Hirose and Manley 2000). For example, certain mutations in the CTD specifically affect splicing, but not transcription or other steps in RNA processing (McCracken et al. 1997b; Fong and Bentley 2001). In addition, different promoters, enhancers, and transcriptional activators can affect the choice of splice sites in vivo (Cramer et al. 1997, 1999, 2001a; Monsalve et al. 2000; Auboeuf et al. 2002; Kadener et al. 2002; Kwek et al. 2002; Nogues et al. 2002). Moreover, the rate of transcription elongation can also affect the choice of alternative splice sites (de la Mata et al. 2003; Nogues et al. 2003a, b). At present, however, it is not understood how transcription exerts its effects on splicing.
Advances in understanding the basic mechanism of splicing were made possible by the development of an in vitro splicing system in which pre-mRNAs synthesized with bacteriophage RNA polymerase (T7 or SP6) are spliced in HeLa cell nuclear extract (Krainer et al. 1984). These studies revealed that splicing takes place in two catalytic steps after the pre-mRNA assembles into the large and highly dynamic spliceosome (Burge et al. 1999; Black 2003). To determine how RNAP II transcription is functionally coupled to splicing, we developed an efficient in vitro system in which pre-mRNAs are synthesized by RNAP II and spliced in the same nuclear extract. Using this system, we show that RNAP II transcription is functionally coupled to spliceosome assembly, which in turn results in an increase in both the rate and efficiency of splicing. By coupling transcription to an early step in the splicing pathway (i.e., formation of the spliceosome), the nascent pre-mRNAs avoid improperly associating with highly abundant splicing inhibitors such as heterogeneous nuclear ribonucleoprotein (hnRNP) proteins, which can only access the nascent transcript by free diffusion.
Results
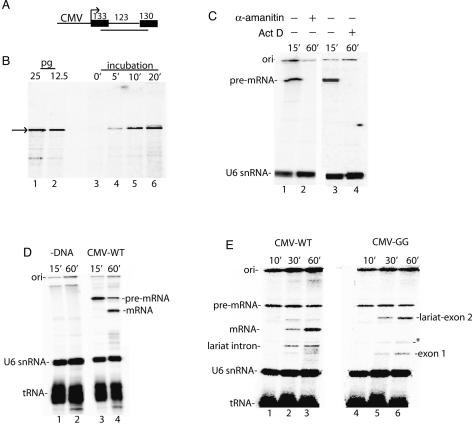
In previous work, significant progress was made in establishing an in vitro system for coupling RNAP II transcription to splicing, but the efficiency of this system was not sufficient to investigate the mechanisms of coupling (Ghosh and Garcia-Blanco 2000). To establish an efficient coupled system, we first fused the CMV promoter to a DNA template encoding AdML pre-mRNA (Fig. 1A). This template (designated Derivative of AdML, DoA1) was then used to optimize conditions for transcription and splicing in HeLa cell nuclear extracts (see Materials and Methods). Using the RNase protection assay, we found that a discrete band corresponding to unspliced pre-mRNA accumulates to picogram levels under optimized conditions (Fig. 1B). When the nuclear extract was incubated directly in 32P-UTP, this RNAP II transcript was detected as one main band (Fig. 1C, lanes 1,3). This band was not present when the general transcription inhibitor Actinomycin D or the RNAP inhibitor α-amanitin was added at the start of the reaction (Fig. 1C, lanes 2,4), or when DNA was omitted from the reaction (Fig. 1D, lanes 1,2) (as observed previously, the endogenous U6 snRNA and tRNA in the extract are labeled; see Reddy et al. [1987). Previous work has shown that the three RNAPs are differentially sensitive to α-amanitin with RNAP I being refractory, RNAP II sensitive to low concentrations (0.10–1 μg/mL), and RNAP III inhibited at higher levels. When we carried out a titration of α-amanitin, we found that transcription is sensitive in the range that inhibits RNAP II, as we observe strong inhibition at 40 ng/mL (data not shown) and complete inhibition at 400 ng/mL (e.g., Fig. 1C). We conclude that RNAP II is responsible for the observed transcription.
Figure 1.
RNAP II transcription and pre-mRNA splicing in vitro. (A) Structure of the DoA1 DNA template used to optimize transcription and splicing. The sizes of the exons and intron are indicated. The line below the DoA1 DNA template indicates the SP6 RNA probe, which protects 234 nt of T7-AdML and CMV-DoA1 (54 nt of exon 1, the entire intron, and 57 nt of exon 2). (B, lanes 1,2) RNase protection of known amounts of AdML T7 transcript. (Lanes 3–6) RNase protection of RNAP II transcript, which was generated by incubating the DoA1 DNA template in HeLa nuclear extract for the indicated times. The arrow indicates the protected transcript. (C, lanes 1,3) 32P-UTP and the CMV DNA template were incubated under transcription/splicing conditions for 15 min. α-Amanitin (lane 2) or Actinomycin D (lane 4) was added at the beginning of the transcription/splicing reaction and incubation was continued for 60 min. (D) 32P-UTP and the DoA1 DNA template were incubated under transcription/splicing conditions for 15 min (lane 3) or 60 min (lane 4). The DNA was omitted in the reactions shown in lanes 1 and 2. (E) Same as D, except that the CMV GG and wild-type (WT) DNA templates were compared. Asterisk indicates a nonspecific band. The endogenous U6 snRNA and tRNA are indicated. RNA was fractionated on an 8% denaturing polyacrylamide gel and detected by PhosphorImager.
When the reaction was incubated for 60 min, a band was detected that comigrates with spliced mRNA generated from the corresponding T7 pre-mRNA (Fig. 1D, lane 4; data not shown). In contrast, using a CMV DNA template containing a GG substitution for the AG at the 3′-splice site, the second step of splicing is blocked, and the lariat intermediate and exon 1 accumulate (Fig. 1E, lanes 1–6), as has been well established for T7 premRNAs. All RNA species in this study were identified based on comigration with known size markers and/or the corresponding T7 transcripts (data not shown). We conclude that pre-mRNA is synthesized by RNAP II in our system, as the transcription is both α-amanitin-sensitive and requires the RNAP II-dependent CMV promoter. The RNAP II transcripts are then spliced in this transcription/splicing system.
Transcription by RNAP II is functionally coupled to spliceosome assembly
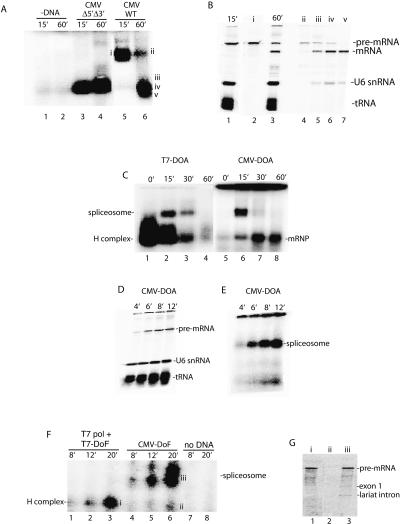
Cotranscriptional splicing of long pre-mRNAs has been well-documented in vivo (Beyer and Osheim 1988; Bauren and Wieslander 1994; Kiseleva et al. 1994; Wetterberg et al. 2001). We have not yet established conditions for detecting such cotranscriptional splicing in vitro. However, the goal of our present work is not to recapitulate cotranscriptional splicing, but rather to address the unanswered question of how RNAP II transcription is functionally coupled to splicing. To investigate this question, we first asked whether transcription by RNAP II has any effect on spliceosome assembly. Using optimized conditions (see Materials and Methods), no specific complexes were detected on a native agarose gel in the absence of the CMV-DoA1 DNA template (Fig. 2A, lanes 1,2) or in the presence of the DNA template and α-amanitin (data not shown). Using a CMV Δ5′Δ3′ DoA1 DNA template, which lacks functional splice sites, one small complex was detected after 15 or 60 min of incubation (Fig. 2A, lanes 3,4). This small complex contains only unspliced pre-mRNA and comigrates with the nonspecific H complex that assembles on the corresponding T7 Δ5′Δ3′ AdML pre-mRNA (Das and Reed 1999; data not shown).
Figure 2.
RNAP II transcription is functionally coupled to spliceosome assembly. (A) Nuclear extract was incubated under transcription/splicing conditions for the indicated times in the absence of DNA template (lanes 1,2) or presence of CMV-DoA1 Δ5′Δ3′ DNA template (lanes 3,4) or CMV-DoA1 DNA template (lanes 5,6). Heparin was added to each sample, which was then loaded on a 1.2% low-melting-point agarose mini-gel. Bands were detected by PhosphorImager. (B) RNA was extracted from the indicated regions (i–v) of the gel shown in A and fractionated on an 8% denaturing polyacrylamide gel (lanes 2,4–7); 15 and 60 min transcription/splicing reactions were run as markers (lanes 1,3). (C) Naked DoA T7 pre-mRNA or the corresponding CMV-DoA DNA template was incubated under transcription/splicing conditions for the times indicated. For the CMV-DoA DNA template, Actinomycin D was added at time 0 and incubation continued for 60 min (lane 5), or Actinomycin D was added at 15 min and incubation was continued for 0 min (lane 6), 15 min (lane 7), or 45 min (lane 8). Heparin was added to each sample, which were then fractionated side-by-side on a 1.2% low-melting-point agarose gel. The spliceosome, spliced mRNP, and nonspecific H complex are indicated. (D) DoA CMV DNA template was incubated under transcription/splicing conditions for the indicated times, and total RNA was fractionated on an 8% polyacrylamide gel. (E) DoA CMV DNA template was incubated under transcription/splicing conditions for the indicated times; heparin was added to each sample, which was then run on a 1.2% low-melting-point agarose gel. (F) T7 DoF DNA template with T7 polymerase or corresponding CMV-DoF DNA template was incubated under transcription/splicing conditions for the indicated times; heparin was added to each sample, which was then fractionated on a 1.2% low-melting point agarose gel. (G) RNA was extracted from the bands indicated (left of i–iii) of the gel shown in F and fractionated on an 8% denaturing polyacrylamide gel.
When the CMV DNA template containing wild-type splice sites is incubated in the RNAP II transcription/splicing system, a large complex is detected after 15 min of incubation and a smaller complex by 60 min (Fig. 2A, lanes 5,6). Analysis of the RNA in the large complex revealed that it contains unspliced pre-mRNA (Fig. 2B, lane 2), whereas the small complex mainly contains spliced mRNA (Fig. 2B, lanes 6,7). In addition, the large complex precisely comigrates with spliceosomes assembled on T7 transcripts (see below, Fig. 2C). We conclude that the large and small complexes assembled on the wild-type CMV DNA template correspond to the spliceosome and spliced mRNP, respectively.
To carry out a side-by-side comparison of a time course of spliceosome assembly on T7 versus RNAP II transcripts, we constructed PCR templates encoding the corresponding T7 and CMV pre-mRNAs (designated T7-DoA and CMV-DoA, respectively; see Materials and Methods). Virtually all of the CMV-DoA pre-mRNA generated in the RNAP II transcription/splicing system assembles into the spliceosome by 15 min of incubation, and little if any of the nonspecific H complex is detected (Fig. 2C; the smaller complex detected at the 15-min time point contains spliced mRNA). In contrast, the T7-DoA pre-mRNA is quantitatively assembled into the H complex, and then a portion of it subsequently assembles into the spliceosome (Fig. 2C). A striking feature of spliceosome assembly on the RNAP II transcript is the absence of the nonspecific H complex. To determine whether the H complex could be detected at all on the RNAP II transcript, we analyzed spliceosome assembly at earlier time points. Significantly, as early as the RNAP II transcript is synthesized (4 min) (Fig. 2D), the spliceosome is detected, and its levels increase as the level of transcript increases (Fig. 2E). Moreover, little if any of the H complex is detected on the RNAP II transcript at any time point (Fig. 2E).
To determine whether the immediate and quantitative assembly of the spliceosome on the RNAP II transcript is simply transcription-dependent or is instead directed by RNAP II, we added a DNA template containing the T7 promoter and encoding Ftz pre-mRNA (designated T7-DoF) to the extract together with T7 RNA polymerase or added the corresponding CMV-DoF DNA template to the extract. Significantly, only the H complex was detected on the T7 pre-mRNA at early times, whereas the spliceosome, but not the H complex, was detected on the RNAP II pre-mRNA (Fig. 2F,G). The same results were obtained comparing T7-DoA and CMV-DoA (data not shown). We conclude that the rapid and efficient spliceosome assembly on the RNAP II transcript occurs generally and is directed by RNAP II.
Yields of spliced mRNA are dramatically increased in the RNAP II transcription/splicing system
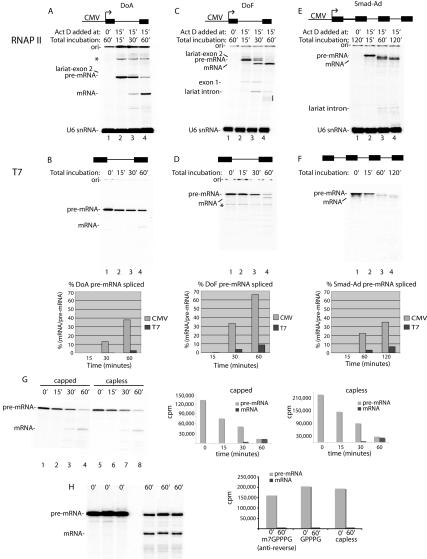
To determine whether there is a correlation between the efficient spliceosome assembly observed in our RNAP II transcription/splicing system and splicing of the nascent transcripts, we first compared splicing of a naked T7 pre-mRNA with the corresponding RNAP II transcript generated in the transcription/splicing system (Fig. 3). To establish a “zero” time point for the RNAP II transcription/splicing system, the DoA transcript was synthesized for 15 min and then Actinomycin D was added (Fig. 3A, lane 2). Incubation was then continued to allow splicing (Fig. 3A, lanes 3,4). For comparison, an equivalent amount of T7-DoA transcript (∼10 pg) was used in an identical side-by-side splicing reaction (Fig. 3B). As revealed by the quantitation, ∼38% of the RNAP II pre-mRNA was detected as spliced mRNA by 60 min. In contrast, only ∼3% of the T7-DoA pre-mRNA was detected as spliced mRNA by 60 min (Fig. 3B, lanes 1–4). Thus, the yield of mRNA is increased ∼14-fold in the RNAP II transcription/splicing system relative to the T7 system. The difference in mRNA yield is not due to the presence of Actinomycin D (data not shown).
Figure 3.
Yields of spliced mRNA are dramatically enhanced in the RNAP II transcription/splicing system. DoA (A), DoF (C), and Smad-Ad (E) CMV DNA templates or the corresponding DoA (B), DoF (D), and Smad-Ad (F) T7 transcripts were incubated side-by-side in nuclear extracts under the same reaction conditions (see Materials and Methods). For DoF and DoA CMV DNAs, Actinomycin D was added at time 0 and incubation continued for 60 min (lane 1), or Actinomycin D was added at 15 min and incubation was continued for 0 min (lane 2), 15 min (lane 3), or 45 min (lane 4). For Smad-Ad CMV DNA, Actinomycin D was added at time 0 and incubation was continued for 120 min (lane 1), or Actinomycin D was added at 15 min and incubation was continued for 0 min (lane 2), 45 min (lane 3), and 105 min (lane 4). For the T7 transcripts, DoA (10 pg), DoF (10 pg), and Smad-Ad (1 ng) were incubated for the times indicated, which correspond to the total incubation times used for the CMV DNAs. Total RNA was fractionated on an 8% denaturing polyacrylamide gel. The indicated RNA species were detected and quantitated by PhosphorImager. (*) Nonspecific band; (ori) gel origin. The line on the right side of the gel in C indicates breakdown of the lariat intron. The bands between the pre-mRNA and spliced mRNA in E are intermediate species containing one or two introns. These intermediates are readily resolved on a lower percentage gel (6.5%) (data not shown). The first-step products are not detected because they are so efficiently converted into second-step products. The yield of spliced mRNA was calculated as the percentage of spliced mRNA relative to pre-mRNA at the start of the reaction. (We note that T7-DoA splices approximately twofold less efficiently than the T7-AdML pre-mRNA used in our previous studies [e.g., Zhou et al. 2002]; see G [lanes 1–4] and B [lanes 1–4] for a comparison of T7-AdML and T7-DoA splicing efficiencies). (G) T7-AdML pre-mRNA either containing or lacking the GpppG cap (added during transcription) was incubated under transcription/splicing conditions for the times indicated and then fractionated on an 8% gel. The total counts per minute (cpm) for the pre-mRNA and mRNA are indicated at each time point for the capped and capless RNAs. (H) T7-AdML pre-mRNA containing the m7GpppG anti-reverse cap, GpppG, or no cap was incubated under standard splicing conditions for the times indicated and then fractionated on a 8% denaturing gel. The total counts per minute (cpm) for the pre-mRNA at 0 min and for mRNA at 60 min was quantitated by PhosporImager and is shown in graph.
A large increase in yield of mRNA was also observed when the corresponding DoF (derivative of Ftz) RNAP II and T7 transcripts were compared, with yields of ∼66% and ∼9%, respectively (Fig. 3C,D). Moreover, a Smad-Ad pre-mRNA containing four Smad exons and three AdML introns also generated more spliced mRNA when the transcript was produced by RNAP II than when the corresponding T7 transcript was spliced (Fig. 3E,F). We conclude that the yield of spliced mRNA is greatly increased with pre-mRNAs containing one or more introns in the RNAP II transcription/splicing system relative to the corresponding T7 transcripts. This difference in overall mRNA yield is general, as it has been observed with all transcripts compared in the two systems (this study; data not shown). The difference in mRNA yield does not appear to be due to 5′ capping of the T7 versus RNAP II transcript as T7-AdML transcripts that are capless or capped with GpppG or the anti-reverse m7GpppG (Stepinski et al. 2001) all have the same stability, and the yields of spliced mRNA are the same (Fig. 3G,H). Moreover, we excised from a gel DoA or DoF RNAP II transcripts, which should be capped during transcription in the nuclear extract, and used these transcripts for splicing. The data reveal that they are spliced with the same efficiency as their T7 counterparts, providing further evidence that the cap is not responsible for the enhanced splicing observed in the RNAP II transcription/splicing system (data not shown).
Increased yield of spliced mRNA is directed by RNAP II transcription
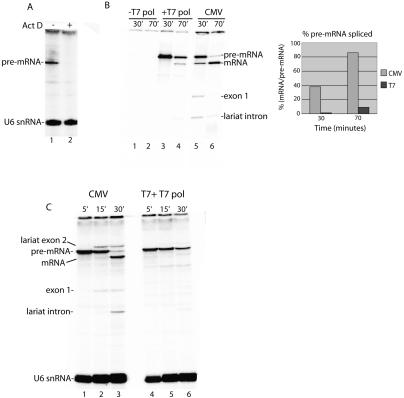
In the data presented above (Fig. 3) and in previous studies of splicing in vitro, T7 transcripts are added all at once to the nuclear extract. In contrast, RNAP II transcripts are synthesized over time in the extract. Thus, to determine whether the difference in yield of spliced mRNA is simply transcription-dependent or instead is directed by RNAP II, a DNA construct containing the T7 promoter was added to the extract together with T7 RNA polymerase. Actinomycin D, which blocks T7 transcription (Fig. 4A), was used so that splicing of T7 and RNAP II transcripts could be directly compared. Strikingly, although both DoF transcripts were efficiently synthesized (Fig. 4B, lanes 3,5), barely any spliced mRNA was generated from the T7 transcript, whereas high levels of mRNA were generated from the corresponding RNAP II transcript (Fig. 4B, cf. lanes 4 and 6). Thus, whether T7 transcripts are directly added to the extract (Fig. 3) or synthesized in the extract (Fig. 4), the yield of spliced mRNA is low relative to its RNAP II counterpart. The same results were obtained comparing DoA T7 versus CMV constructs (data not shown). In all cases, the same amount of T7 transcript and RNAP II transcript were synthesized and were assayed under the same conditions, side-by-side. On the basis of the α-amanitin sensitivity and RNAP II promoter specificity of the RNAP II transcription (Fig. 1) and the observation that transcription with T7 RNA polymerase does not result in increased yields of spliced mRNA, we conclude that these increased yields observed in the RNAP II transcription/splicing system are directed by RNAP II and not simply the process of transcription itself.
Figure 4.
Transcription by RNAP II results in an increase in mRNA yield and splicing kinetics. (A) T7 RNA polymerase and T7 DNA template were incubated in nuclear extract for 30 min in the absence (lane 1) or presence (lane 2) of Actinomycin D. (B) T7-DoF DNA template was incubated in nuclear extract in the absence (lane 1,2) or presence (lane 3,4) of T7 RNA polymerase. Actinomycin D was added at 30 min and incubation continued for 0 min (lanes 1,3) or 40 min (lanes 2,4). The corresponding CMV-DoF DNA template was incubated for 30 min; Actinomycin D was added and incubation continued for 0 min (lane 5) or 40 min (lane 6). Total RNA was fractionated on an 8% denaturing polyacrylamide gel. The percent pre-mRNA spliced (mRNA at each time point/pre-mRNA at 30 min) was quantitated by PhosphorImager and is shown in the graph. (C) Same as B except that transcriptions were carried out for 5 min, followed by the addition of Actinomycin D and continued incubation for 15 and 30 min.
Splicing kinetics are increased by RNAP II transcription
To determine whether the quantitative spliceosome assembly observed in the RNAP II transcription/splicing system enhances the kinetics of splicing, we incubated the CMV DNA for 5′ and then carried out a splicing time course. For comparison, we used an equal amount of T7 pre-mRNA generated by adding T7 RNA polymerase and T7 DNA template to the nuclear extract. As shown in Figure 4C, splicing of the RNAP II transcript begins at 15 min and is spliced by the 30-min time point. In contrast, the corresponding T7 transcript has barely started to splice by the 30-min time point (Fig. 4C, lane 6). The faster kinetics observed with the RNAP II transcript may occur because the spliceosome is recruited more quickly to RNAP II transcripts and/or because the T7 transcript assembles into the spliceosome after the H complex, which may require dissociation of inhibitory hnRNP proteins followed by recruitment of the spliceosome.
Discussion
In this study we investigated the basis for functional coupling of RNAP II transcription to pre-mRNA splicing using an efficient in vitro system for the coupling reaction. We found that the spliceosome assembles on RNAP II transcripts as rapidly as the transcripts are synthesized. The immediate spliceosome assembly, in turn, results in increased yields of spliced mRNA and a dramatic increase in the kinetics of splicing. Indeed, when the pre-mRNA is synthesized by RNAP II for 5 min, splicing is completed by 30 min. To our knowledge, there are no known examples of splicing kinetics even approaching these with the currently used T7 splicing system. The dramatic enhancement of splicing/spliceosome assembly is directed by RNAP II based on the observations that the transcription is specific to RNAP II promoters, is α-amanitin-sensitive, and is not a general property of transcription as it does not occur on nascent transcripts generated by adding a T7 DNA template and T7 RNA polymerase to the nuclear extract.
In contrast to the nascent RNAP II transcripts, naked T7 pre-mRNAs and nascent T7 transcripts assemble quantitatively into the nonspecific H complex, which results from the immediate binding of the highly abundant nuclear hnRNP proteins to naked RNA (Bennett et al. 1992). This complex is not splicing-specific, as it assembles not only on T7 pre-mRNAs but also on T7 RNAs lacking functional splice sites (Konarska and Sharp 1986; Michaud and Reed 1991; Bennett et al. 1992). It is well known that at least some hnRNP proteins inhibit splicing and that these proteins compete with essential splicing factors for binding to pre-mRNA in vitro (Caputi et al. 1999; Wagner and Garcia-Blanco 2001; Zhu et al. 2001; Black 2003; Rooke et al. 2003; Fu 2004; Shen et al. 2004; Wang et al. 2004; Han et al. 2005; Rothrock et al. 2005). Consistent with this conclusion, previous studies indicate that the H complex is not a functional precursor to the spliceosome (Michaud and Reed 1991, 1993). Specifically, when the H complex is isolated and added back to nuclear extract, only a small portion is capable of assembling into the spliceosome (Michaud and Reed 1991). The rest is either degraded or remains as H complex, depending on the transcript (Michaud and Reed 1991). It is not known whether the low level of spliceosomes that do assemble after H complex assembly results from the H complex falling apart and reassembling into the spliceosome, or whether a portion of the H complex contains hnRNP proteins that are bound to regions of the pre-mRNA that do not preclude spliceosome assembly. In either case, the net result of H complex assembly is an overall reduction in yields of spliced mRNA. The observation that transcription by RNAP II directs immediate spliceosome assembly and results in a bypass of the nonspecific H complex indicates that RNAP II transcription is functionally coupled to spliceosome assembly in metazoans. Recent studies show that spliceosome assembly occurs cotranscriptionally in Saccharomyces cerevisiae, but it is not yet known whether these two processes are functionally coupled (Gornemann et al. 2005; Lacadie and Rosbash 2005).
As previous studies in metazoans indicate that mutations in the RNAP II CTD affect splicing (Fong and Bentley 2001), it is possible that the effect of RNAP II transcription on spliceosome assembly results from association of specific spliceosome components with the CTD. These components, by associating with RNAP II, have immediate access to the nascent pre-mRNA as it emerges from the RNAP II exit pore and thereby readily out-compete inhibitory proteins such as hnRNP proteins, which can only access the pre-mRNA by free diffusion. As a consequence, the RNAP II transcripts are efficiently directed into the spliceosome assembly pathway rather than into the inhibitory H complex. A number of spliceosomal proteins that associate with RNAP II or the CTD in vitro have been identified, including Prp40, SCAFs, PSF, p54, SR proteins and all of the spliceosomal small nuclear RNPs (snRNPs) (Mortillaro et al. 1996; Yuryev et al. 1996; Corden and Patturajan 1997; Kim et al. 1997; Patturajan et al. 1998; Morris and Greenleaf 2000; Tian 2001; Emili et al. 2002; Robert et al. 2002; Kameoka et al. 2004; Phatnani and Greenleaf 2004), but a function for these proteins in coupling remains to be demonstrated.
Previous studies have provided detailed mechanistic insights into the coupling between transcription and capping, and significant advances have recently been made in understanding the mechanisms that couple transcription and polyadenylation (Yonaha and Proudfoot 1999, 2000; Adamson et al. 2005; Rigo et al. 2005). By establishing an efficient in vitro system for coupling transcription to splicing, and showing that functional coupling of these two steps in gene expression occurs during spliceosome assembly, it should now be possible to identify the factors involved.
Materials and methods
Plasmids and transcription DNA templates
Plasmid DoA1 was constructed by inserting a KpnI and XbaI fragment from AdML HMS81 (Michaud and Reed 1993) into pcDNA3.1, which contains the CMV and T7 promoters (Invitrogen). The DoA1 plasmid encodes a splicing substrate consisting of exon 1 (133 base pairs [bp]), intron 1 (123 bp), and exon 2 (130 bp). Plasmid DoF1 was constructed by inserting a HindIII and EcoRI fragment from Ftz HMS404 (Luo and Reed 1999) into pcDNA3.1. The DoF1 plasmid encodes a splicing substrate consisting of exon 1 (331 bp), intron 1 (147 bp), and exon 2 (277 bp). Plasmid Smad-Ad1 containing three introns was constructed by inserting the AdML intron (123 bp) into three exon junctions of an EcoRI and XbaI fragment containing Smad cDNA, which was then inserted into the same sites of pcDNA3.1. PCR products were generated from plasmids DoA1, DoF1, and Smad-Ad1 to yield T7 or CMV transcription templates designated DoA, DoF, and Smad-Ad, respectively. Exon 1, intron 1, and exon 2 generated by transcription of T7-DoA PCR product are 84, 123, and 130 bp, respectively, and for DoF are 281, 147, and 277 bp, respectively. The pre-mRNA generated using the CMV promoter putative start site corresponds to the pre-mRNA generated by the T7 promoter except for the presence of three additional nucleotides at the 5′ end of the T7 pre-mRNA. For Smad-Ad, the RNAP II transcript from the Smad-Ad PCR product contains exons 1, 2, 3, and 4 of 241, 89, 193, and 135 bp, respectively. The T7 transcript from this PCR product is identical except for 49 fewer nucleotides in the first exon of the T7 transcript. T7-AdML HMS 81 was used for the splicing assays with the capped and capless pre-mRNA (Fig. 3). The GG CMV-DoA1 plasmid was constructed by inserting a KpnI–XbaI fragment into the same site in pcDNA3.1. In vitro transcriptions with T7 polymerase were carried out using standard procedures in the presence of GpppG, anti-reverse m7GpppG (Stepinski et al. 2001), or no cap (Fig. 3G,H). For Figure 3H, the cap analogs were present at a 10:1 molar ratio over GTP and in Figure 3G, a 2.5:1 molar ratio.
RNAP II transcription and splicing
In vitro transcription/splicing reactions were performed at 30°C for the indicated times in 25 μL of reaction mixtures containing 200 ng DNA template, 1 μL 32P-UTP (800 Ci/mmol; Perkin Elmer Life Sciences catalog #BLU507x), 0.5 mM ATP, 3.2 mM MgCl2, 20 mM creatine phosphate (di-Tris salt), and 15 μL standard HeLa nuclear extract used for splicing (Krainer et al. 1984). The buffer conditions and extract used for the RNAP II transcription/splicing system are the same as those previously used for in vitro splicing of T7 transcripts. Omitting the final 20-min 10,000-rpm spin that follows the 4-h dialysis of the extract results in increased levels of transcription. The RNAP II-coupled transcription/splicing reactions work with similar efficiency in multiple preparations of nuclear extract. It is not necessary to add UTP, CTP, or GTP to the reaction mixture as they are not limiting for transcription in the extract. These nucleotides are not removed by dialysis due to binding to proteins present in the extract. The addition of extra NTPs does not increase the levels of transcription. The efficiency of transcription depends on the quality of the DNA template. Specifically, a PCR product without nicks and that has minimal sequences flanking the transcribed region generates the highest level of specific transcript. Nicks result in promoter-independent transcription at the site of the nick (Slattery et al. 1983). The amount of DNA used in the transcription/splicing system must be optimized for maximal transcription in different preparations of nuclear extract. Splicing of naked T7 transcripts was carried out under identical conditions except that the DNA template and 32P-UTP were omitted, and 10 pg (DoA and DoF) or 1 ng (Smad-Ad) 32P-UTP labeled pre-mRNA was added. T7 RNA polymerase transcription/splicing reactions were performed under identical conditions except that 0.5 μL T7 RNA polymerase (50, 000 U/mL, NEB) was added per 25 μL reaction mixtures. Actinomycin D (250 ng/25 μL reaction) or α-amanitin (400 ng/25 μL reaction) was used to block transcription. RNase protection assays were carried out using an RPA kit (Ambion). The probe for RNase protection was generated using SP6 RNA polymerase to transcribe the AdML HMS81 plasmid that was cut with EcoR1. The probe is 264 nt, and the protected region of the RNAP II transcript and T7 AdML is 234 nt. The protected region contains 54 nt of exon 1, the entire intron, and 57 nt of exon 2.
Spliceosome assembly
Spliceosomes were assembled on AdML pre-mRNAs by incubating the indicated T7 transcripts or the indicated CMV DNA templates under RNAP II transcription/splicing conditions. Spliceosomes were also assembled using T7-DoF template and T7 polymerase and the corresponding CMV-DoF template under transcription/splicing conditions. After the reactions, G-50 micro columns (Amersham Biosciences) were used to remove unincorporated 32P-UTP. One microliter of heparin (6.5 mg/mL) was added to 10 μL of G-50 column-purified reactions prior to loading on 1.2% low-melting-point agorose gels (Das and Reed 1999). The RNA species in the complexes were eluted and analyzed on an 8% denaturing polyacrylamide gel as described (Das and Reed 1999).
Acknowledgments
We are grateful to Chris Burge, Adrian Krainer, Tim Nilsen, Bob Roeder, Michael Green, Tom Maniatis, El Cherif Ibrahim, Claudine Christoforides, Patricia Valencia, Hong Cheng, Jeanne Hsu, Seiji Masuda, and Malcolm Whitman for useful discussions. We thank Dickie Hertel for communicating results prior to publication. HeLa cells were obtained from the National Cell Culture Center (Minneapolis, MN). This work was supported by an NIH grant to R.R.
Footnotes
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1397406
References
- Adamson T.E., Shutt D.C., Price D.H. Functional coupling of cleavage and polyadenylation with transcription of mRNA. J. Biol. Chem. 2005;280:32262–32271. doi: 10.1074/jbc.M505532200. [DOI] [PubMed] [Google Scholar]
- Auboeuf D., Honig A., Berget S.M., O’Malley B.W. Coordinate regulation of transcription and splicing by steroid receptor coregulators. Science. 2002;298:416–419. doi: 10.1126/science.1073734. [DOI] [PubMed] [Google Scholar]
- Bauren G., Wieslander L. Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell. 1994;76:183–192. doi: 10.1016/0092-8674(94)90182-1. [DOI] [PubMed] [Google Scholar]
- Bennett M., Pinol-Roma S., Staknis D., Dreyfuss G., Reed R. Differential binding of heterogeneous nuclear ribonucleoproteins to mRNA precursors prior to spliceosome assembly in vitro. Mol. Cell. Biol. 1992;12:3165–3175. doi: 10.1128/mcb.12.7.3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D. Coupling RNA polymerase II transcription with pre-mRNA processing. Curr. Opin. Cell Biol. 1999;11:347–351. doi: 10.1016/S0955-0674(99)80048-9. [DOI] [PubMed] [Google Scholar]
- Bentley D. The mRNA assembly line: Transcription and processing machines in the same factory. Curr. Opin. Cell Biol. 2002;14:336–342. doi: 10.1016/s0955-0674(02)00333-2. [DOI] [PubMed] [Google Scholar]
- Bentley D.L. Rules of engagement: Co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 2005;17:251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Beyer A.L., Osheim Y.N. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes & Dev. 1988;2:754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- Black D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Burge C.B., Tuschl T.H., Sharp P.A. Splicing of precursors to mRNAs by the spliceosomes. In: Gesteland R.F., et al., editors. The RNA world. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1999. pp. 525–560. [Google Scholar]
- Caputi M., Mayeda A., Krainer A.R., Zahler A.M. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 1999;18:4060–4067. doi: 10.1093/emboj/18.14.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho E.J., Takagi T., Moore C.R., Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes & Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho E.J., Rodriguez C.R., Takagi T., Buratowski S. Allosteric interactions between capping enzyme subunits and the RNA polymerase II carboxy-terminal domain. Genes & Dev. 1998;12:3482–3487. doi: 10.1101/gad.12.22.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corden J.L., Patturajan M. A CTD function linking transcription to splicing. Trends Biochem. Sci. 1997;22:413–416. doi: 10.1016/s0968-0004(97)01125-0. [DOI] [PubMed] [Google Scholar]
- Cramer P., Pesce C.G., Baralle F.E., Kornblihtt A.R. Functional association between promoter structure and transcript alternative splicing. Proc. Natl. Acad. Sci. 1997;94:11456–11460. doi: 10.1073/pnas.94.21.11456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer P., Caceres J.F., Cazalla D., Kadener S., Muro A.F., Baralle F.E., Kornblihtt A.R. Coupling of transcription with alternative splicing: RNA pol II promoters modulate SF2/ASF and 9G8 effects on an exonic splicing enhancer. Mol. Cell. 1999;4:251–258. doi: 10.1016/s1097-2765(00)80372-x. [DOI] [PubMed] [Google Scholar]
- Cramer P., Bushnell D.A., Kornberg R.D. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science. 2001a;292:1863–1876. [Google Scholar]
- Cramer P., Srebrow A., Kadener S., Werbajh S., de la Mata M., Melen G., Nogues G., Kornblihtt A.R. Coordination between transcription and pre-mRNA processing. FEBS Lett. 2001b;498:179–182. doi: 10.1016/s0014-5793(01)02485-1. [DOI] [PubMed] [Google Scholar]
- Das R., Reed R. Resolution of the mammalian E complex and the ATP-dependent spliceosomal complexes on native agarose mini-gels. RNA. 1999;5:1504–1508. doi: 10.1017/s1355838299991501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Mata M., Alonso C.R., Kadener S., Fededa J.P., Blaustein M., Pelisch F., Cramer P., Bentley D., Kornblihtt A.R. A slow RNA polymerase II affects alternative splicing in vivo. Mol. Cell. 2003;12:525–532. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Emili A., Shales M., McCracken S., Xie W., Tucker P.W., Kobayashi R., Blencowe B.J., Ingles C.J. Splicing and transcription-associated proteins PSF and p54nrb/nonO bind to the RNA polymerase II CTD. RNA. 2002;8:1102–1111. doi: 10.1017/s1355838202025037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong N., Bentley D.L. Capping, splicing, and 3′ processing are independently stimulated by RNA polymerase II: Different functions for different segments of the CTD. Genes & Dev. 2001;15:1783–1795. doi: 10.1101/gad.889101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X.D. Towards a splicing code. Cell. 2004;119:736–738. doi: 10.1016/j.cell.2004.11.039. [DOI] [PubMed] [Google Scholar]
- Ghosh S., Garcia-Blanco M.A. Coupled in vitro synthesis and splicing of RNA polymerase II transcripts. RNA. 2000;6:1325–1334. doi: 10.1017/s1355838200992537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gornemann J., Kotovic K.M., Hujer K., Neugebauer K.M. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol. Cell. 2005;19:53–63. doi: 10.1016/j.molcel.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Han K., Yeo G., An P., Burge C.B., Grabowski P.J. A combinatorial code for splicing silencing: UAGG and GGGG motifs. PLoS Biol. 2005;3:e158. doi: 10.1371/journal.pbio.0030158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y., Manley J.L. RNA polymerase II and the integration of nuclear events. Genes & Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- Ho C.K., Shuman S. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol. Cell. 1999;3:405–411. doi: 10.1016/s1097-2765(00)80468-2. [DOI] [PubMed] [Google Scholar]
- Ho C.K., Sriskanda V., McCracken S., Bentley D., Schwer B., Shuman S. The guanylyltransferase domain of mammalian mRNA capping enzyme binds to the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 1998;273:9577–9585. doi: 10.1074/jbc.273.16.9577. [DOI] [PubMed] [Google Scholar]
- Kadener S., Fededa J.P., Rosbash M., Kornblihtt A.R. Regulation of alternative splicing by a transcriptional enhancer through RNA pol II elongation. Proc. Natl. Acad. Sci. 2002;99:8185–8190. doi: 10.1073/pnas.122246099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameoka S., Duque P., Konarska M.M. p54(nrb) associates with the 5′ splice site within large transcription/splicing complexes. EMBO J. 2004;23:1782–1791. doi: 10.1038/sj.emboj.7600187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E., Du L., Bregman D.B., Warren S.L. Splicing factors associate with hyperphosphorylated RNA polymerase II in the absence of pre-mRNA. J. Cell Biol. 1997;136:19–28. doi: 10.1083/jcb.136.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiseleva E., Wurtz T., Visa N., Daneholt B. Assembly and disassembly of spliceosomes along a specific pre-messenger RNP fiber. EMBO J. 1994;13:6052–6061. doi: 10.1002/j.1460-2075.1994.tb06952.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska M.M., Sharp P.A. Electrophoretic separation of complexes involved in the splicing of precursors to mRNAs. Cell. 1986;46:845–855. doi: 10.1016/0092-8674(86)90066-8. [DOI] [PubMed] [Google Scholar]
- Kornblihtt A.R., de la Mata M., Fededa J.P., Munoz M.J., Nogues G. Multiple links between transcription and splicing. RNA. 2004;10:1489–1498. doi: 10.1261/rna.7100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krainer A.R., Maniatis T., Ruskin B., Green M.R. Normal and mutant human β-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell. 1984;36:993–1005. doi: 10.1016/0092-8674(84)90049-7. [DOI] [PubMed] [Google Scholar]
- Kwek K.Y., Murphy S., Furger A., Thomas B., O’Gorman W., Kimura H., Proudfoot N.J., Akoulitchev A. U1 snRNA associates with TFIIH and regulates transcriptional initiation. Nat. Struct. Biol. 2002;9:800–805. doi: 10.1038/nsb862. [DOI] [PubMed] [Google Scholar]
- Lacadie S.A., Rosbash M. Cotranscriptional spliceosome assembly dynamics and the role of U1 snRNA:5′ss base pairing in yeast. Mol. Cell. 2005;19:65–75. doi: 10.1016/j.molcel.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Luo M., Reed R. Splicing is required for rapid and efficient mRNA export in metazoans. Proc. Natl. Acad. Sci. 1999;96:14937–14942. doi: 10.1073/pnas.96.26.14937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- McCracken S., Fong N., Rosonina E., Yankulov K., Brothers G., Siderovski D., Hessel A., Foster S., Shuman S., Bentley D.L. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes & Dev. 1997a;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S., Fong N., Yankulov K., Ballantyne S., Pan G., Greenblatt J., Patterson S.D., Wickens M., Bentley D.L. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997b;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- Michaud S., Reed R. An ATP-independent complex commits pre-mRNA to the mammalian spliceosome assembly pathway. Genes & Dev. 1991;5:2534–2546. doi: 10.1101/gad.5.12b.2534. [DOI] [PubMed] [Google Scholar]
- Michaud S., Reed R. A functional association between the 5′ and 3′ splice site is established in the earliest prespliceosome complex (E) in mammals. Genes & Dev. 1993;7:1008–1020. doi: 10.1101/gad.7.6.1008. [DOI] [PubMed] [Google Scholar]
- Monsalve M., Wu Z., Adelmant G., Puigserver P., Fan M., Spiegelman B.M. Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Mol. Cell. 2000;6:307–316. doi: 10.1016/s1097-2765(00)00031-9. [DOI] [PubMed] [Google Scholar]
- Morris D.P., Greenleaf A.L. The splicing factor, Prp40, binds the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 2000;275:39935–39943. doi: 10.1074/jbc.M004118200. [DOI] [PubMed] [Google Scholar]
- Mortillaro M.J., Blencowe B.J., Wei X., Nakayasu H., Du L., Warren S.L., Sharp P.A., Berezney R. A hyperphosphorylated form of the large subunit of RNA polymerase II is associated with splicing complexes and the nuclear matrix. Proc. Natl. Acad. Sci. 1996;93:8253–8257. doi: 10.1073/pnas.93.16.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moteki S., Price D. Functional coupling of capping and transcription of mRNA. Mol. Cell. 2002;10:599–609. doi: 10.1016/s1097-2765(02)00660-3. [DOI] [PubMed] [Google Scholar]
- Nogues G., Kadener S., Cramer P., Bentley D., Kornblihtt A.R. Transcriptional activators differ in their abilities to control alternative splicing. J. Biol. Chem. 2002;277:43110–43114. doi: 10.1074/jbc.M208418200. [DOI] [PubMed] [Google Scholar]
- Nogues G., Kadener S., Cramer P., de la Mata M., Fededa J.P., Blaustein M., Srebrow A., Kornblihtt A.R. Control of alternative pre-mRNA splicing by RNA Pol II elongation: Faster is not always better. IUBMB Life. 2003a;55:235–241. doi: 10.1080/1521654031000119830. [DOI] [PubMed] [Google Scholar]
- Nogues G., Munoz M.J., Kornblihtt A.R. Influence of polymerase II processivity on alternative splicing depends on splice site strength. J. Biol. Chem. 2003b;278:52166–52171. doi: 10.1074/jbc.M309156200. [DOI] [PubMed] [Google Scholar]
- Orphanides G., Reinberg D. A unified theory of gene expression. Cell. 2002;108:439–451. doi: 10.1016/s0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- Patturajan M., Wei X., Berezney R., Corden J.L. A nuclear matrix protein interacts with the phosphorylated C-terminal domain of RNA polymerase II. Mol. Cell. Biol. 1998;18:2406–2415. doi: 10.1128/mcb.18.4.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phatnani H.P., Greenleaf A.L. Identifying phosphoCTD-associating proteins. Methods Mol. Biol. 2004;257:17–28. doi: 10.1385/1-59259-750-5:017. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr. Opin. Cell Biol. 2004;16:272–278. doi: 10.1016/j.ceb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Reddy R., Henning D., Das G., Harless M., Wright D. The capped U6 small nuclear RNA is transcribed by RNA polymerase III. J. Biol. Chem. 1987;262:75–81. [PubMed] [Google Scholar]
- Rigo F., Kazerouninia A., Nag A., Martinson H.G. The RNA tether from the poly(A) signal to the polymerase mediates coupling of transcription to cleavage and polyadenylation. Mol. Cell. 2005;20:733–745. doi: 10.1016/j.molcel.2005.09.026. [DOI] [PubMed] [Google Scholar]
- Robert F., Blanchette M., Maes O., Chabot B., Coulombe B. A human RNA polymerase II-containing complex associated with factors necessary for spliceosome assembly. J. Biol. Chem. 2002;277:9302–9306. doi: 10.1074/jbc.M110516200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooke N., Markovtsov V., Cagavi E., Black D.L. Roles for SR proteins and hnRNP A1 in the regulation of c-src exon N1. Mol. Cell. Biol. 2003;23:1874–1884. doi: 10.1128/MCB.23.6.1874-1884.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothrock C.R., House A.E., Lynch K.W. HnRNP L represses exon splicing via a regulated exonic splicing silencer. EMBO J. 2005;24:2792–2802. doi: 10.1038/sj.emboj.7600745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Kan J.L., Ghigna C., Biamonti G., Green M.R. A single polypyrimidine tract binding protein (PTB) binding site mediates splicing inhibition at mouse IgM exons M1 and M2. RNA. 2004;10:787–794. doi: 10.1261/rna.5229704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery E., Dignam J.D., Matsui T., Roeder R.G. Purification and analysis of a factor which suppresses nick-induced transcription by RNA polymerase II and its identity with poly(ADP-ribose) polymerase. J. Biol. Chem. 1983;258:5955–5959. [PubMed] [Google Scholar]
- Stepinski J., Waddell C., Stolarski R., Darzynkiewicz E., Rhoads R.E. Synthesis and properties of mRNAs containing the novel ‘anti-reverse’ cap analogs 7-methyl(3′-O-methyl)GpppG and 7-methyl (3′-deoxy)GpppG. RNA. 2001;7:1486–1495. [PMC free article] [PubMed] [Google Scholar]
- Tian H. RNA ligands generated against complex nuclear targets indicate a role for U1 snRNP in co-ordinating transcription and RNA splicing. FEBS Lett. 2001;509:282–286. doi: 10.1016/s0014-5793(01)03188-x. [DOI] [PubMed] [Google Scholar]
- Wagner E.J., Garcia-Blanco M.A. Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell. Biol. 2001;21:3281–3288. [Google Scholar]
- Wang Z., Rolish M.E., Yeo G., Tung V., Mawson M., Burge C.B. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119:831–845. doi: 10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Wetterberg I., Zhao J., Masich S., Wieslander L., Skoglund U. In situ transcription and splicing in the Balbiani ring 3 gene. EMBO J. 2001;20:2564–2574. doi: 10.1093/emboj/20.10.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonaha M., Proudfoot N.J. Specific transcriptional pausing activates polyadenylation in a coupled in vitro system. Mol. Cell. 1999;3:593–600. doi: 10.1016/s1097-2765(00)80352-4. [DOI] [PubMed] [Google Scholar]
- Yonaha M., Proudfoot N.J. Transcriptional termination and coupled polyadenylation in vitro. EMBO J. 2000;19:3770–3777. doi: 10.1093/emboj/19.14.3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z., Maldonado E., Pillutla R., Cho H., Reinberg D., Shatkin A.J. Mammalian capping enzyme complements mutant Saccharomyces cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA polymerase II. Proc. Natl. Acad. Sci. 1997;94:12898–12903. doi: 10.1073/pnas.94.24.12898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuryev A., Patturajan M., Litingtung Y., Joshi R.V., Gentile C., Gebara M., Corden J.L. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc. Natl. Acad. Sci. 1996;93:6975–6980. doi: 10.1073/pnas.93.14.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Licklider L.J., Gygi S.P., Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- Zhu J., Mayeda A., Krainer A.R. Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol. Cell. 2001;8:1351–1361. doi: 10.1016/s1097-2765(01)00409-9. [DOI] [PubMed] [Google Scholar]