Abstract
Growth hormone (GH) signaling is tightly controlled by ubiquitination of GH receptors, phosphorylation levels, and accessibility of binding sites for downstream signaling partners. Members of the suppressors of cytokine signaling (SOCS) family function as key regulators at all levels of this pathway, and mouse knockout studies implicate SOCS2 as the primary suppressor. To elucidate the structural basis for SOCS2 function, we determined the 1.9-Å crystal structure of the ternary complex of SOCS2 with elongin C and elongin B. The structure defines a prototypical SOCS box ubiquitin ligase with a Src homology 2 (SH2) domain as a substrate recognition motif. Overall, the SOCS box and SH2 domain show a conserved spatial domain arrangement with the BC box and substrate recognition domain of the von Hippel–Lindau (VHL) tumor suppressor protein, suggesting a common mechanism of ubiquitination in these cullin-dependent E3 ligases. The SOCS box binds elongin BC in a similar fashion to the VHL BC box and shows extended structural conservation with the F box of the Skp2 ubiquitin ligase. A previously unrecognized feature of the SOCS box is revealed with the burial of the C terminus, which packs together with the N-terminal extended SH2 subdomain to create a stable interface between the SOCS box and SH2 domain. This domain organization is conserved in SOCS1–3 and CIS1, which share a strictly conserved length of their C termini, but not in SOCS4, 5, and 7, which have extended C termini defining two distinct classes of inter- and intramolecular SOCS box interactions.
Keywords: growth hormone receptor, cytokine signaling
The growth hormone (GH) signaling pathway is the main regulator of longitudinal growth in mammals, and it stimulates differentiation and mitogenesis and modulates lipid, nitrogen, and mineral metabolism. Signaling from the GH receptor (GHR) is initiated by GH-induced dimerization followed by Janus kinase (JAK) 2 crossphosphorylation and subsequent phosphorylation of signal transducers and activators of transcription (STATs) (in particular, STAT5b). Phosphorylated STAT5 dimerizes and enters the nucleus, where it initiates transcription of insulin-like growth factor (IGF) I, IGFBP3, suppressors of cytokine signaling (SOCS) 1–3, CIS, and other target genes (recently reviewed in ref. 1). The duration of the GHR signal is a key determinant of the biological response. The signal is attenuated by many regulatory mechanisms, including ubiquitin-dependent receptor internalization and degradation (see, for example, ref. 2), cleavage by TACE (TNF-α-converting enzyme) (3), tyrosine dephosphorylation of receptors and associated JAK kinases (4), and SOCS.
The SOCS family comprises SOCS1–7 and CIS1, of which SOCS1–3 and CIS1 have been shown to regulate GH signaling in vitro (1). The role of SOCS2 in GH signaling in vivo has been convincingly demonstrated, as displayed by the phenotype of SOCS2-deficient mice, which are 30–40% larger than normal littermates (5). This phenotype is similar to mice overexpressing GH (6) and high growth (hg) mice, which have a disrupted socs2 locus (7). The growth characteristics also resemble those of patients with gigantism (8). The effect of SOCS2 is mediated by signaling through the JAK/STAT pathway, as demonstrated by STAT5b/SOCS2 double knockout mice, which have no overgrowth phenotype (9). Furthermore, SOCS2 knockout neural stem cells are 100-fold more sensitive to GH and produce 50% fewer neurons than wild type, pointing to an important role for SOCS2 in cell differentiation (10).
SOCS proteins contain three domains necessary for SOCS2 regulation of GHR signaling: a variable N-terminal domain, a central SH2 domain, and a conserved SOCS box in the C-terminal domain (11). SOCS2 binds directly to tyrosine-phosphorylated GHR but does not interact with or regulate signaling from the nonphosphorylated receptor, consistent with binding of an SH2 domain (11). This interaction requires an additional extended SH2 subdomain (ESS) at the N terminus of unknown function. GHR binding is inhibitory only in the presence of the SOCS box, which mediates interaction with elongin BC. By analogy with the von Hippel–Lindau (VHL) protein, which binds elongin BC to target hypoxia-inducible factor (HIF-1α) for proteasomal degradation (12, 13), it is speculated that many, if not all, SOCS box-containing proteins act as part of an E3 ubiquitin ligase complex (ref. 14; reviewed in ref. 15). Despite sharing <20% sequence identity, mutagenesis points to a common BC box-binding motif, and, indeed, many SOCS box proteins have been shown to assemble with elongin BC and the additional E3 components Cul-5 and Rbx2 (16). Most notably, SOCS1 promotes TEL-JAK2 degradation and inhibits its cellular transformation in a SOCS box-dependent manner (17).
In this study, we confirmed the direct interaction between SOCS2 and elongin BC and crystallized the ternary complex to determine the mechanism by which SOCS2 associates tyrosine-phosphorylated substrates with the ubiquitination machinery. The structure reveals significant differences with current models and provides details of the concerted function of all three SOCS2 subdomains by their respective orientations and interdomain packing. We further characterized the binding affinity of the SOCS2/C/B complex for phosphopeptide substrates derived from GHR and the erythropoietin receptor to assess the most physiologically relevant target.
Results
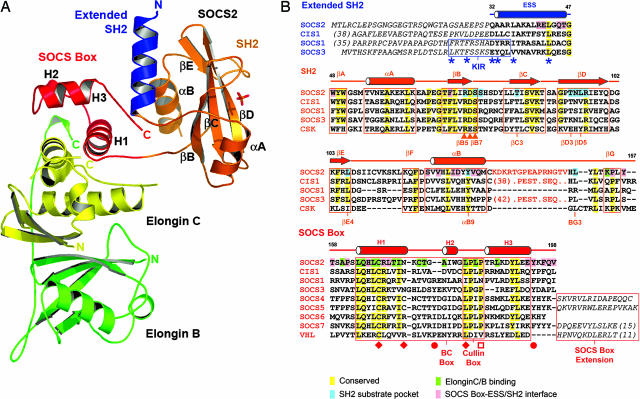
The structure of the human SOCS2–elongin C–elongin B complex was determined by molecular replacement using elongin C and elongin B as search models and refined at 1.9-Å resolution (Fig. 1 and Table 1). Three conserved SOCS family domains are defined, including an N-terminal ESS formed from a single amphipathic helix, a central SH2 domain with a classic phosphotyrosine pocket, and a C-terminal SOCS box, which mediates interaction with elongin BC.
Fig. 1.
Overall structure of the SOCS2–elongin BC ternary complex and structure-based sequence alignment. (A) The different domains and complex components are differentiated by colors showing the SOCS2 SH2 domain in orange, the ESS in blue, the SOCS box in red, and two elongins in yellow and green. Secondary structure elements discussed in the Results are labeled. (B) Conserved and domain interface residues are highlighted by different colors in the sequence alignment. An unstructured 15-residue insertion in the SOCS2 BG loop is represented by orange text and is the site of predicted PEST sequences in CIS1 and SOCS3 (21). Additional N- and C-terminal extensions are defined by italic text. The positions of mutations that disrupt the SOCS1–JAK2 (22), SOCS2–GHR (11), VHL–elongin C (18), and SOCS1–Cul-5 (16) interactions are indicated by asterisks, triangles, diamonds, and open squares, respectively. Filled circles mark SOCS3 phosphorylation sites, which disrupt the SOCS3–elongin C interaction (20). The SOCS2 SH2 structure is most similar to the structures of C-terminal Src kinase (PDB ID code 1K9A) and Csk-homologous kinase (PDB ID code 1JWO).
Table 1.
Crystallographic data and refinement statistics for SOCS2–elongin C–elongin B
| Space group | P3221 |
|---|---|
| Cell dimensions, Å | a = b = 105.29; c = 70.2 |
| Resolution, Å | 1.9 |
| Total observations (unique, redundancy) | 395,240 (33,443, 11.06) |
| Completeness (outer shell) | 98.67 (98.51) |
| Rmerge | 0.0726 |
| I/σ (outer shell) | 18.39 (3.29) |
| Rwork/Rfree, %* | 18.5/22.5 |
| Protein atoms (water) | 2,601 (172) |
| Hetero groups | Sulfate, Ni |
| rms deviation bond length, Å | 0.01 |
| rms deviation bond angle, degrees | 1.209 |
| Average B factor, Å2 | |
| Protein atoms | 41.5 |
| Solvent atoms | 47.3 |
| Other | 49.2 |
| Ramachandran | |
| Allowed, % | |
| SOCS2 | 94.8 |
| Elongin B | 88.6 |
| Elongin C | 95.6 |
| Generously allowed, % | |
| SOCS2 | 5.2 |
| Elongin B | 9.1 |
| Elongin C | 4.4 |
| Dissallowed, % | |
| SOCS2 | 0 |
| Elongin B | 2.3 |
| Elongin C | 0 |
*Using randomly selected 5% of data.
The SOCS Box Is a Conserved Ubiquitin Ligase Motif.
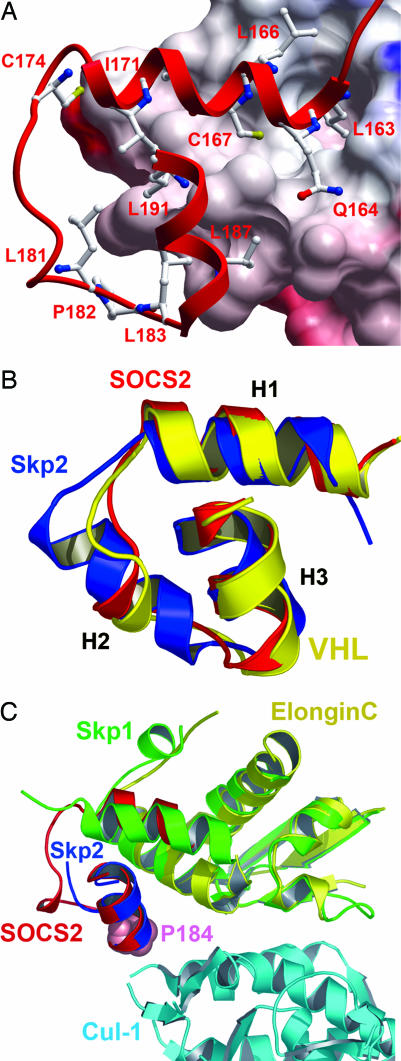
The three core helices (H1–3) of the SOCS box show significant structural conservation with the VHL BC box (rms deviation = 0.75 Å) and pack similarly together with elongin C H4 into a four-helix cluster. Binding is dominated by the burial of SOCS2 H1 into a deep cleft between elongin C loop 5 and H4. In this region, only L163 and C167 are conserved with VHL, in which mutation of these residues destroys the assembly (18). Further hydrophobic interaction is provided by L166, T170, and I171 (Fig. 2A). The extended structural conservation between the SOCS box, VHL BC box, and Skp2 F box defines a common structural motif linking E3 substrate recognition domains with ubiquitin ligase complexes (Fig. 2B).
Fig. 2.
Comparative structural interactions and conservation of the SOCS box, BC box, and F box. (A) SOCS2 binds elongin C (electrostatic surface shown) in a hydrophobic interface of ≈2,200 Å2 (for clarity, elongin B is not shown). Deep pockets accommodate residues from SOCS2 H1 (L163 and C167). (B) The three core helices of the SOCS box (red) show a remarkable structural conservation with the VHL BC box (yellow) and Skp2 F box (blue), forming a common structural motif to link E3 substrate recognition domains with the ubiquitin ligase complex. (C) The crystal structure of the Skp2-Skp1–Cul-1–Rbx1 ternary complex provides a template for the study of SOCS box–elongin C binding to Cul-5 (38). Because of different packing arrangements in their respective complexes, Skp1 shows structural and functional similarity to elongin C and SOCS2 H1, whereas H1 from Skp2 provides equivalence to SOCS2 H3 [consistent with previous Skp2-Skp1 comparison with VHL (39)]. SOCS2 P184 (shown in space-fill) occurs at the likely Cul-5 interface and terminates the “LPXP” cullin box. Mutation of this position in SOCS1 determines Cul-5 interaction (16).
Elongin B makes limited contact with SOCS2 but significantly stabilizes the ternary structure; in its absence, we observe unfolding of the SOCS2–elongin C dimer at physiological temperature (Fig. 5, which is published as supporting information on the PNAS web site). Further assembly with the cullin ubiquitination complex is predicted through the SOCS box LPXP motif, which confers Cul-5 selection (16) and falls at the predicted cullin interface (Fig. 2C).
Structural Basis for Phosphotyrosine Substrate Recognition.
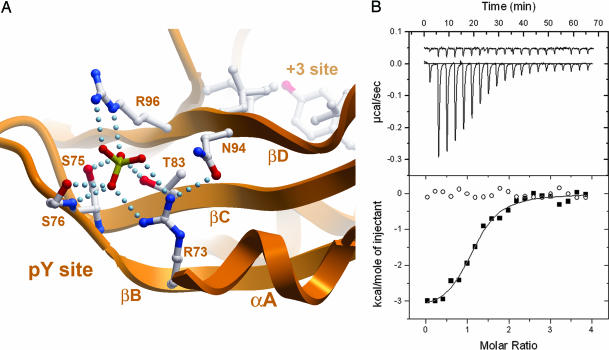
The SOCS family E3 ligases are targeted to their substrates by means of a SH2 interaction domain. Accordingly, GHR regulation by SOCS2 is abolished by mutation of either the SH2 phosphotyrosine pocket (R73K/D74E/S75C) or two GHR phosphotyrosine sites (Y595F/Y487F) (11). The structural basis for specific phosphotyrosine recognition by SOCS2 can be inferred from a bound sulfate ion in the pY pocket, which is coordinated through eight hydrogen bonds that involve those residues identified by mutagenesis (Fig. 3). The common αA2 Arg ligand is replaced by valine, but SOCS2 gains compensatory pY interactions from T83 and R96, which are less conserved in the SH2 family. Peptides derived from these GHR sites bind directly to SOCS2 in a strict phosphorylation-dependent manner. We determined that SOCS2 binds to the primary GHR site (pY595) with 5-fold higher affinity than to the analogous site of the erythropoietin receptor, another suggested SOCS2 target (19) (Fig. 3B and Table 2). Substrate selectivity among different SOCS SH2 domains varies as a result of nonconservative substitutions within the pY recognition site and changes within the hydrophobic +3 pocket. The SOCS2 substrate pocket is framed by large EF and BG loop insertions that hindered previous comparative sequence alignments (Fig. 1B).
Fig. 3.
SOCS2 SH2 domain and binding of GHR phosphopeptide PVPDpYTSIHIV determined by ITC. (A) The SOCS2 substrate pocket has the common hydrophobic cluster at the +3 site, including L95 (βD6), L106 (βE4), Y129 (αB9), and L150 (BG3). The binding site of the phosphotyrosine moiety is indicated by the presence of a bound sulfate ion. Hydrogen bonds are shown as dotted lines. (B) Binding thermodynamic data determined by ITC showed that the GHR-derived phosphopeptide bound with an affinity of 1.6 μM.
Table 2.
ITC-binding data for phosphotyrosine peptides
| Peptide | KD, μM | KB × 105, M−1 | ΔHobs, kcal/mol | TΔS, kcal/mol | ΔG, kcal/mol | n* |
|---|---|---|---|---|---|---|
| pY595 GHR† | 1.6 | 6.08 ± 1.03 | −3.35 ± 0.11 | 4.54 | −7.89 | 1.22 |
| pY401 EpoR‡ | 7.1 | 1.42 ± 0.27 | −3.69 ± 0.46 | 3.34 | −7.03 | 0.73 |
EpoR, erythropoietin receptor.
*Stoichiometry determined from a single-binding-site model.
†PVPDpYTSIHIV amide.
‡ASFEpYTILDPS amide.
A requirement for E3-dependent ubiquitin ligation is a stable protein–protein interaction between the SOCS box and the substrate recognition domain and also a conserved spatial domain arrangement of the substrate and enzyme. The location and orientation of the pY-binding site within the SOCS2/C/B ternary complex is within 3 Å of the VHL-bound hydroxyproline, which serves as a substrate recognition motif for this E3 ligase. This similarity suggests not only that the SOCS2 ternary complex adopts a productive orientation for substrate ubiquitination but also that the mechanism of target recognition and ubiquitination is a common structural feature of these ligases.
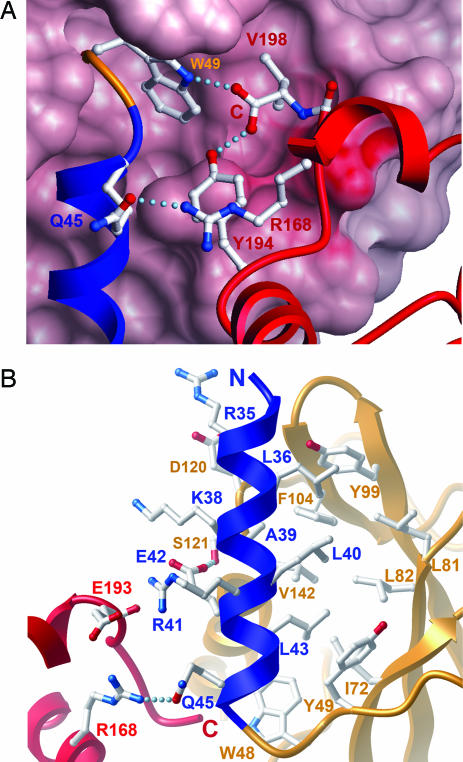
Burial of the C Terminus.
The SOCS2 structure reveals the function and packing arrangements that distinguish the SOCS protein family from the VHL system. The SOCS box contains a shorter C terminus that lacks the H4 helix that is present in VHL. To maintain analogous stabilizing interactions with its substrate recognition domain, the C terminus is buried in the interface between the SH2 domain and the SOCS box. The C-terminal carboxyl group forms a hydrogen bond network with W48 (βA) and Y194 (H3) to link both elements, and the valine (V198) side chain binds to a deep hydrophobic pocket in the back of the SH2 domain (Fig. 4A). This configuration is further stabilized by R168, which forms a hydrophobic stacking interaction between Y190 and Y194 as well as further polar contacts that tether the H1, H3, and ESS helices. The addition of a phosphate moiety to Y194 would break these interactions and forms part of a regulatory mechanism in SOCS3 (Y221) that results in the loss of elongin C binding (20). The additional phosphorylation of SOCS3 Y204 (absent in SOCS2) is required and may enable kinase access to this inaccessible location. Similarly, the packing does not allow for C-terminal sequence extensions, explaining the strictly conserved length of the C termini in SOCS1–3 and CIS1. The more distantly related SOCS4–7 have larger structured N-terminal domains and variable C-terminal domains and therefore form a distinct SOCS box subfamily with alternative interdomain interactions (Fig. 1B).
Fig. 4.
Unpredicted packing arrangements and burial of the SOCS2 N and C termini. (A) The side chain of V198 at the C terminus packs into a deep pocket in the back of the SH2 domain, and the terminal carboxyl forms hydrogen bond interactions with W48 (SH2) and Y194 (SOCS box). This conformation is stabilized by R168, which is strictly conserved within the SOCS family but not within the VHL BC box or Skp2 F box. (B) The ESS forms a single amphipathic helix (blue) that packs between the SH2 (orange) and SOCS box (red) domains with hydrophobic and electrostatic interactions, respectively, that bury a surface area of ≈1,200 Å2.
Function of the ESS.
The SOCS2 N terminus comprises an ESS, and further N-terminal sequence is predicted to lack secondary structure (21). Deletion of this region inhibits SOCS2 regulation of GHR, but the mechanism has yet to be assigned (11). Mutagenesis of the same ESS region in SOCS1 has identified conserved hydrophobic positions that are critical for SH2 function and a preceding DY motif (QA in SOCS2) that forms a pseudosubstrate kinase inhibitory region (KIR) for JAK2 (22). Previous structural predictions placed this subdomain adjacent to the SH2 pY pocket (23). This arrangement was anticipated to facilitate cooperation between the SH2 and KIR domains for binding to the JAK2 activation loop (pY1007) and the catalytic pocket, respectively.
The SOCS2 structure provides a structural basis for examining this hypothesis. In contrast to the preferred model (23), the SOCS2 N-terminal subdomain packs alongside the C terminus as a single amphipathic helix, forming hydrophobic interactions (L36/L40/L43) with the SH2 site most distal to the pY pocket and electrostatic interactions with the SOCS box (Q45–R168, Fig. 4B). The sequence similarity throughout this surface and the conservation of the C termini suggest that this structural feature is conserved in the SOCS1–3/CIS1 subfamily. Thus, the ESS can now be defined as a SOCS-specific element used as a structural subdomain to bridge the interface between the SOCS box and the SH2 domain and to link E3 ligase activity to substrate capture. The kinase inhibitory region DY motif in SOCS1 maps to the exposed N terminus of this helix. This position would allow it to function as an independent protein interaction site, consistent with its continued inhibition of JAK2 in the presence of a mutated SH2 domain (22).
Discussion
Members of the SOCS protein family have been shown to regulate GH signaling in vitro through multiple mechanisms involving all three SOCS functional domains (24). For example, SOCS1 can inhibit JAK2 through either its N-terminal kinase inhibitory region or SH2 domains and also mediate its degradation by means of the SOCS box (17). Downstream inhibition of STAT3 and STAT5 may also be effected by SOCS7 to suppress multiple cytokine pathways (25). Direct target specificity for GHR appears to be restricted to the SH2 domains of SOCS3, which binds pY333 and pY338, and SOCS2 and CIS1, which target the membrane-distal pY487 and pY595 sites (1). SOCS2 is uniquely identified as a primary GHR inhibitor in vivo by its overgrowth knockout phenotype in mice (5); other SOCS family knockout mice show no overgrowth phenotype or are growth-retarded (25–27).
The binding affinity of SOCS2 for the primary pY595 site is typical for physiological SH2–ligand interactions determined by isothermal titration calorimetry (ITC) and 5-fold higher than for the erythropoietin receptor (Table 2). Binding to this site involves competition with the phosphatase SHP2 and the effector STAT5b, providing one potential mechanism for SOCS2-dependent suppression. However, SOCS2 regulation of GHR also requires a functional SOCS box (11). The SOCS2 structure presented here supports its function as an E3 ubiquitin ligase, analogous to the VHL and Skp2 E3 ligases (Fig. 6, which is published as supporting information on the PNAS web site). The closely related CIS1 is also associated with the ubiquitination and internalization of GHR, and this activity is lost upon mutation of its SH2 domain (R107K) or in the presence of proteasome inhibitors (28). The involvement of SOCS3 in GHR regulation remains unclear; its mouse knockout is embryonic lethal (29).
The burial of the C terminus and packing of all three SOCS2 domains in a shared core interface are highly unexpected structural features. Their apparent structural and functional interdependence provides a cautionary note for domain deletion studies as well as a rationale for the loss of SOCS protein activities upon mutation of previously unrecognized core positions in the ESS (22). Such disruption is, in fact, a control mechanism that is used to regulate the activity and interplay between SOCS family members. Double phosphorylation of the SOCS3 C terminus (Y204/Y221) will prevent its core interaction and indeed inhibits elongin C binding, resulting in SOCS3 proteasomal degradation (20).
Such control mechanisms define potential target sites for protein interaction inhibitors. Because of the key role of SOCS2 as a major negative regulator of GH signaling, the development of SOCS2 antagonists has been proposed as an alternative treatment regime to GH injections. The direct interaction between the SOCS box and elongin C constitutes such an intervention point, because the signal for GHR (as well as several other cytokine receptors) is prolonged in the presence of proteasome inhibitors (30). Indeed, we identify one potential site in proximity to the buried SOCS2 C terminus (Fig. 7, which is published as supporting information on the PNAS web site) that is sufficiently large (340 Å3) to be targeted by low-molecular-weight inhibitors that would not only find application as potential drug candidates but would also be valuable reagents to delineate complex control mechanisms in cytokine signaling.
Materials and Methods
Protein Expression and Crystallization.
Human SOCS2 (amino acids 32–198), elongin C (amino acids 17–112), and elongin B were coexpressed in BL21(DE3) from the plasmids pLIC-SOCS2 and pACYCDUET-ElCB. Ternary complex was purified by nickel-affinity, size-exclusion, and anion-exchange chromatography and concentrated to 22 mg/ml in 50 mM Hepes, pH 7.5/250 mM NaCl/2.5% glycerol/10 mM DTT. The N-terminal hexahistidine tag was cleaved by using tobacco etch virus protease before size-exclusion chromatography. The protein complex was judged to be at least 95% pure by SDS/PAGE, and the correct molecular weight of all three proteins was confirmed by using liquid chromatography electrospray ionization MS. Crystals were grown at 4°C in 1-μl sitting drops by using 0.08 M Na-cacodylate (pH 6.5), 0.16 M NaCl, and 1.6 M (NH4)2SO4.
Structure Determination.
SOCS2 diffraction data were collected on a frozen crystal (100 K) at the Swiss Light Source Beamline 10 (Villigen, Switzerland). Images were indexed and integrated by using mosflm and scaled by using scala within the ccp4 program package (31). The structure was solved by using molecular replacement carried out with the program phaser (32) and Src and elongin BC as search models (Protein Data Bank ID codes 1O4H and 1LM8). Iterative rounds of rigid-body refinement and restrained refinement with TLS (translation–libration–screw) against maximum likelihood targets were interspersed by manual rebuilding of the model using coot (33) and xfit/xtalview (34, 35). Figures were prepared by using pymol (36) and icm-pro (37). Atomic coordinates and structure factors have been deposited in the Protein Data Bank (ID code 2C9W).
ITC.
Experiments were carried out in 50 mM Hepes, pH 7.5/150 mM NaCl/1 mM DTT at 25°C, injecting 0.4 mM peptide solution into 15 μM protein solution. Blank titrations were subtracted from binding data, and data were processed by using origin software provided with the instrument.
Supplementary Material
Acknowledgments
We thank members of the Structural Genomics Consortium for assistance with plasmid preparation and diffraction data collection. The Structural Genomics Consortium is a registered charity (no. 1097737) funded by the Wellcome Trust, GlaxoSmithKline, Genome Canada, the Canadian Institutes of Health Research, the Ontario Innovation Trust, the Ontario Research and Development Challenge Fund, the Canadian Foundation for Innovation, the Swedish Governmental Agency for Innovation Systems, the Knut and Alice Wallenberg Foundation, the Swedish Foundation for Strategic Research, and the Karolinska Institutet.
Abbreviations
- SOCS
suppressors of cytokine signaling
- GH
growth hormone
- GHR
GH receptor
- SH2
Src homology 2
- VHL
von Hippel–Lindau
- STAT
signal transducers and activators of transcription
- JAK
Janus kinase
- ESS
extended SH2 subdomain
- ITC
isothermal titration calorimetry.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2C9W).
References
- 1.Flores-Morales A., Greenhalgh C. J., Norstedt G., Rico-Bautista E. Mol. Endocrinol. 2005;20:241–253. doi: 10.1210/me.2005-0170. [DOI] [PubMed] [Google Scholar]
- 2.van Kerkhof P., Smeets M., Strous G. J. Endocrinology. 2002;143:1243–1252. doi: 10.1210/endo.143.4.8755. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y., Jiang J., Black R. A., Baumann G., Frank S. J. Endocrinology. 2000;141:4342–4348. doi: 10.1210/endo.141.12.7858. [DOI] [PubMed] [Google Scholar]
- 4.Ram P. A., Waxman D. J. J. Biol. Chem. 1997;272:17694–17702. doi: 10.1074/jbc.272.28.17694. [DOI] [PubMed] [Google Scholar]
- 5.Metcalf D., Greenhalgh C. J., Viney E., Willson T. A., Starr R., Nicola N. A., Hilton D. J., Alexander W. S. Nature. 2000;405:1069–1073. doi: 10.1038/35016611. [DOI] [PubMed] [Google Scholar]
- 6.Kopchick J. J., Bellush L. L., Coschigano K. T. Annu. Rev. Nutr. 1999;19:437–461. doi: 10.1146/annurev.nutr.19.1.437. [DOI] [PubMed] [Google Scholar]
- 7.Horvat S., Medrano J. F. Genomics. 2001;72:209–212. doi: 10.1006/geno.2000.6441. [DOI] [PubMed] [Google Scholar]
- 8.Colao A., Merola B., Ferone D., Lombardi G. J. Clin. Endocrinol. Metab. 1997;82:2777–2781. doi: 10.1210/jcem.82.9.4257. [DOI] [PubMed] [Google Scholar]
- 9.Greenhalgh C. J., Bertolino P., Asa S. L., Metcalf D., Corbin J. E., Adams T. E., Davey H. W., Nicola N. A., Hilton D. J., Alexander W. S. Mol. Endocrinol. 2002;16:1394–1406. doi: 10.1210/mend.16.6.0845. [DOI] [PubMed] [Google Scholar]
- 10.Turnley A. M., Faux C. H., Rietze R. L., Coonan J. R., Bartlett P. F. Nat. Neurosci. 2002;5:1155–1162. doi: 10.1038/nn954. [DOI] [PubMed] [Google Scholar]
- 11.Greenhalgh C. J., Rico-Bautista E., Lorentzon M., Thaus A. L., Morgan P. O., Willson T. A., Zervoudakis P., Metcalf D., Street I., Nicola N. A., et al. J. Clin. Invest. 2005;115:397–406. doi: 10.1172/JCI22710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iwai K., Yamanaka K., Kamura T., Minato N., Conaway R. C., Conaway J. W., Klausner R. D., Pause A. Proc. Natl. Acad. Sci. USA. 1999;96:12436–12441. doi: 10.1073/pnas.96.22.12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lisztwan J., Imbert G., Wirbelauer C., Gstaiger M., Krek W. Genes Dev. 1999;13:1822–1833. doi: 10.1101/gad.13.14.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kibel A., Iliopoulos O., DeCaprio J. A., Kaelin W. G., Jr Science. 1995;269:1444–1446. doi: 10.1126/science.7660130. [DOI] [PubMed] [Google Scholar]
- 15.Kile B. T., Schulman B. A., Alexander W. S., Nicola N. A., Martin H. M., Hilton D. J. Trends Biochem. Sci. 2002;27:235–241. doi: 10.1016/s0968-0004(02)02085-6. [DOI] [PubMed] [Google Scholar]
- 16.Kamura T., Maenaka K., Kotoshiba S., Matsumoto M., Kohda D., Conaway R. C., Conaway J. W., Nakayama K. I. Genes Dev. 2004;18:3055–3065. doi: 10.1101/gad.1252404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frantsve J., Schwaller J., Sternberg D. W., Kutok J., Gilliland D. G. Mol. Cell. Biol. 2001;21:3547–3557. doi: 10.1128/MCB.21.10.3547-3557.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stebbins C. E., Kaelin W. G., Jr, Pavletich N. P. Science. 1999;284:455–461. doi: 10.1126/science.284.5413.455. [DOI] [PubMed] [Google Scholar]
- 19.Eyckerman S., Verhee A., der Heyden J. V., Lemmens I., Ostade X. V., Vandekerckhove J., Tavernier J. Nat. Cell Biol. 2001;3:1114–1119. doi: 10.1038/ncb1201-1114. [DOI] [PubMed] [Google Scholar]
- 20.Haan S., Ferguson P., Sommer U., Hiremath M., McVicar D. W., Heinrich P. C., Johnston J. A., Cacalano N. A. J. Biol. Chem. 2003;278:31972–31979. doi: 10.1074/jbc.M303170200. [DOI] [PubMed] [Google Scholar]
- 21.Babon J. J., Yao S., DeSouza D. P., Harrison C. F., Fabri L. J., Liepinsh E., Scrofani S. D., Baca M., Norton R. S. FEBS J. 2005;272:6120–6130. doi: 10.1111/j.1742-4658.2005.05010.x. [DOI] [PubMed] [Google Scholar]
- 22.Yasukawa H., Misawa H., Sakamoto H., Masuhara M., Sasaki A., Wakioka T., Ohtsuka S., Imaizumi T., Matsuda T., Ihle J. N., Yoshimura A. EMBO J. 1999;18:1309–1320. doi: 10.1093/emboj/18.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giordanetto F., Kroemer R. T. Protein Eng. 2003;16:115–124. doi: 10.1093/proeng/gzg015. [DOI] [PubMed] [Google Scholar]
- 24.Ram P. A., Waxman D. J. J. Biol. Chem. 1999;274:35553–35561. doi: 10.1074/jbc.274.50.35553. [DOI] [PubMed] [Google Scholar]
- 25.Martens N., Uzan G., Wery M., Hooghe R., Hooghe-Peters E. L., Gertler A. J. Biol. Chem. 2005;280:13817–13823. doi: 10.1074/jbc.M411596200. [DOI] [PubMed] [Google Scholar]
- 26.Starr R., Metcalf D., Elefanty A. G., Brysha M., Willson T. A., Nicola N. A., Hilton D. J., Alexander W. S. Proc. Natl. Acad. Sci. USA. 1998;95:14395–14399. doi: 10.1073/pnas.95.24.14395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krebs D. L., Uren R. T., Metcalf D., Rakar S., Zhang J. G., Starr R., De Souza D. P., Hanzinikolas K., Eyles J., Connolly L. M., et al. Mol. Cell. Biol. 2002;22:4567–4578. doi: 10.1128/MCB.22.13.4567-4578.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Landsman T., Waxman D. J. J. Biol. Chem. 2005;280:37471–37480. doi: 10.1074/jbc.M504125200. [DOI] [PubMed] [Google Scholar]
- 29.Marine J. C., McKay C., Wang D., Topham D. J., Parganas E., Nakajima H., Pendeville H., Yasukawa H., Sasaki A., Yoshimura A., Ihle J. N. Cell. 1999;98:617–627. doi: 10.1016/s0092-8674(00)80049-5. [DOI] [PubMed] [Google Scholar]
- 30.Alves dos Santos C. M., van Kerkhof P., Strous G. J. J. Biol. Chem. 2001;276:10839–10846. doi: 10.1074/jbc.M003635200. [DOI] [PubMed] [Google Scholar]
- 31.Bailey S. Acta Crystallogr. D. 1994;50:760–763. [Google Scholar]
- 32.Storoni L. C., McCoy A. J., Read R. J. Acta Crystallogr. D. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 33.Emsley P., Cowtan K. Acta Crystallogr. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 34.McRee D. E. J. Struct. Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 35.Murshudov G. N., Vagin A. A., Dodson E. J. Acta Crystallogr. D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 36.DeLano W. L. pymol. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- 37.Abagyan R. A., Totrov M., Kuznetsov D. J. Comp. Chem. 1994;15:488–506. [Google Scholar]
- 38.Zheng N., Schulman B. A., Song L., Miller J. J., Jeffrey P. D., Wang P., Chu C., Koepp D. M., Elledge S. J., Pagano M., et al. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 39.Schulman B. A., Carrano A. C., Jeffrey P. D., Bowen Z., Kinnucan E. R., Finnin M. S., Elledge S. J., Harper J. W., Pagano M., Pavletich N. P. Nature. 2000;408:381–386. doi: 10.1038/35042620. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.