Summary
To establish an infection, the pathogen Candida albicans must assimilate carbon and grow in its mammalian host. This fungus assimilates six-carbon compounds via the glycolytic pathway, and two-carbon compounds via the glyoxylate cycle and gluconeogenesis. We address a paradox regarding the roles of these central metabolic pathways in C. albicans pathogenesis: the glyoxylate cycle is apparently required for virulence although glyoxylate cycle genes are repressed by glucose at concentrations present in the bloodstream. Using GFP fusions, we confirm that glyoxylate cycle and gluconeogenic genes in C. albicans are repressed by physiologically relevant concentrations of glucose, and show that these genes are inactive in the majority of fungal cells infecting the mouse kidney. However, these pathways are induced following phagocytosis by macrophages or neutrophils. In contrast, glycolytic genes are not induced following phagocytosis and are expressed in infected kidney. Mutations in all three pathways attenuate the virulence of this fungus, highlighting the importance of central carbon metabolism for the establishment of C. albicans infections. We conclude that C. albicans displays a metabolic program whereby the glyoxylate cycle and gluconeogenesis are activated early, when the pathogen is phagocytosed by host cells, while the subsequent progression of systemic disease is dependent upon glycolysis.
Introduction
Candida albicans is the major systemic fungal pathogen of humans. This fungus exists as a relatively harmless commensal in the oral cavity and gastrointestinal tracts of most individuals, but when the defences of the host become compromised, it can cause mucocutaneous infections such as oral or vaginal candidiasis (Odds, 1988; Calderone, 2002). In severely immunocompromised individuals, C. albicans can establish deep-seated systemic infections, which in some patient groups are often fatal (Odds, 1988; Calderone, 2002). Bloodstream infections are thought to arise by two major routes: through penetration of mucosal surfaces, or via intravascular catheters (Velasco et al., 2000; Kullberg and Oude Lashof, 2002), and the formation of C. albicans biofilms upon catheters can exacerbate the infection source (Douglas, 2003). In the absence of effective immune defences, C. albicans can then disseminate via the bloodstream and colonize internal organs such as the kidney.
To grow, a microbe must assimilate carbon. Pathogens such as C. albicans, which can thrive within diverse niches such as the skin, mucous membranes, blood and internal organs of its human host and in biofilms (Odds, 1988; Calderone, 2002), must display sufficient metabolic flexibility to assimilate the available nutrients in these niches. Data from several transcript profiling studies are consistent with this notion (reviewed in Brown, 2005). For example, amino acid biosynthetic genes are upregulated in C. albicans cells growing in biofilms (Garcia-Sanchez et al., 2005). Also, following exposure to human neutrophils or cultured macrophages, C. albicans populations upregulate amino acid biosynthetic genes and display a shift from fermentative to non-fermentative metabolism (Rubin-Bejerano et al., 2003; Lorenz et al., 2004; Fradin et al., 2005). This includes the downregulation of glycolytic genes and the activation of glyoxylate cycle genes (ICL1, MLS1), which facilitate the assimilation of two-carbon compounds in concert with gluconeogenic genes (PCK1, FBP1). Further evidence for the activation of glyoxylate cycle genes following exposure to macrophages has been obtained by differential display reverse transcription polymerase chain reaction (PCR) (Prigneau et al., 2003).
Lorenz and Fink (2001) have shown that C. albicans icl1/icl1 mutants display attenuated virulence in the mouse model of systemic candidiasis. This has led to the suggestion that the glyoxylate cycle is required for fungal virulence (Lorenz and Fink, 2001; 2002). However in Saccharomyces cerevisiae, gluconeogenic and glyoxylate cycle genes are exquisitely sensitive to glucose (Yin et al., 2003). These genes are repressed by glucose at concentrations as low as 0.01%, which are well below those present in the bloodstream (3–5 mM, which is equivalent to about 0.06–0.1% glucose). If C. albicans gluconeogenic and glyoxylate cycle genes are regulated in an analogous fashion to those in S. cerevisiae, which is often presumed to be the case, one would expect them to be repressed during systemic infections. How then is the glyoxylate cycle is required for the establishment of systemic C. albicans infections?
To address this apparent paradox, we have revisited the role of the glyoxylate cycle in C. albicans virulence. Furthermore, we have extended this analysis to examine the glycolytic and gluconeogenic pathways. In this study we used GFP fusions, rather than transcript profiling, to monitor gene activity (Barelle et al., 2004). This was done for three main reasons. First, transcript profiling averages the behaviour of potentially heterogeneous cell populations, whereas the use of GFP fusions has allowed us to examine the behaviour of individual C. albicans cells within complex niches. Second, this approach has allowed us to extend our analyses beyond the in vitro and ex vivo models that have been used for transcript profiling, to monitor gene expression levels in the mouse model of systemic candidiasis. Third, while transcript profiling has indicated whether genes are up- or downregulated (by generating expression ratios), our GFP approach has revealed interesting differences in the absolute expression levels for specific gene fusions. All three factors have proved important in describing the behaviour of this pathogenic fungus in vivo. Taken together with our phenotypic analysis of metabolic null mutants, our GFP studies suggest that C. albicans differentially regulates its central pathways of carbon assimilation at different stages and sites of disseminated infections.
Results
Regulation of glycolytic, gluconeogenic and glyoxylate cycle genes in vitro
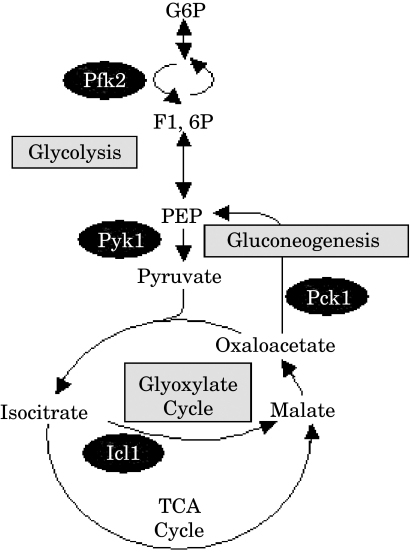
Promoter–GFP fusions were constructed to monitor the regulation of glycolytic, gluconeogenenic and glyoxylate cycle genes in C. albicans (Fig. 1). Most of the enzymes on the glycolytic pathway catalyse reversible reactions that also contribute to gluconeogenesis. However, two steps in glycolysis are essentially irreversible and these are catalysed by the glycolysis-specific enzymes, phosphofructokinase and pyruvate kinase. PFK2 encodes one of two phosphofructokinase subunits, and PYK1 encodes pyruvate kinase. PCK1 encodes the gluconeogenenic-specific enzyme, phosphoenolpyruvate carboxykinase. ICL1 encodes the glyoxylate cycle enzyme, isocitrate lyase (Fig. 1). Therefore, the PFK2, PYK1, PCK1 and ICL1 promoter regions were cloned upstream of the yeast enhanced GFP (yEGFP) gene in the vector pGFP, and these plasmids were stably integrated at the RPS1 locus in the C. albicans genome. The genotype of these strains was confirmed by diagnostic PCR and Southern blotting (not shown).
Fig. 1.
Cartoon of central carbon metabolism, highlighting the steps analysed in this study.
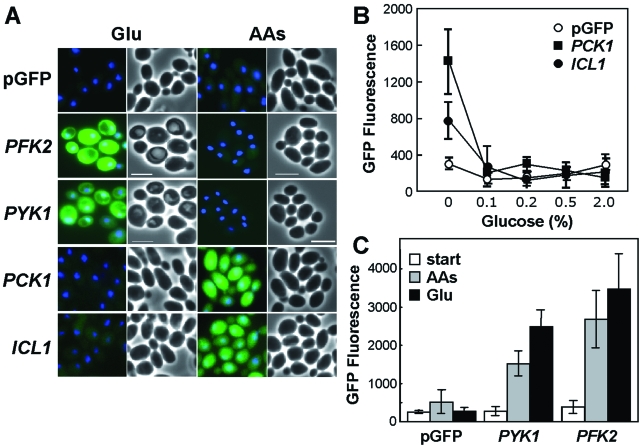
The expression patterns of these promoter fusions were first examined in vitro. To achieve this, we grew the C. albicans strains on different carbon sources and measured the mean GFP fluorescence per cell. As expected (Leuker et al., 1997; Lorenz and Fink, 2001; Murad et al., 2001), the PCK1- and ICL1-GFP fusions were expressed during growth on amino acids and repressed during growth on glucose (Fig. 2A). The PCK1 and ICL1 fusions displayed similar expression patterns in yeast and hyphal cells (not shown). Interestingly, these C. albicans genes were sensitive to low glucose concentrations, like their homologues in S. cerevisiae (Yin et al., 2003). Significantly, the PCK1 and ICL1 fusions were repressed at physiologically relevant glucose concentrations (0.1%), implying that the gluconeogenic pathway and the glyoxylate cycle are inactive under these conditions (Fig. 2B).
Fig. 2.
Differential in vitro regulation of glycolysis, gluconeogenesis and the glyoxylate cycle in C. albicans.A. C. albicans strains containing PFK2-, PYK1-, PCK1- and ICL1-GFP promoter fusions or the empty pGFP control (CJB-1, CJB-2, CJB-3, CLM1-1, CLM3-2: Table 1) were examined after growth overnight on minimal media containing 2% glucose (Glu) or 2% casamino acids (AAs) as sole carbon source. Merged GFP and DAPI images are shown alongside the corresponding light micrographs. Scale bar represents 10 µm.B. Repression of PCK1- and ICL1-GFP by different concentrations of glucose (mean GFP fluorescence intensity per cell).C. Quantification of mean GFP fluorescence levels for C. albicans cells with PFK2- and PYK1 -GFP fusions resuming growth on glucose or amino acids: cells grown overnight on amino acids (start); cells grown for 2 h on glucose (Glu) or amino acids (AAs).
The PFK2- and PYK1-GFP gene fusions, which were designed to monitor glycolytic activity, were expressed at low levels following protracted growth on non-fermentative growth media containing amino acids as sole carbon source (Fig. 2A). However, both fusions were activated when C. albicans cells resumed growth after stationary phase, irrespective of whether amino acids or glucose was available as sole carbon source (Fig. 2C). The PFK2 and PYK1 fusions displayed similar expression patterns in yeast and hyphal cells (not shown). We conclude that PFK2- and PYK1-GFP expression reflects active growth of C. albicans rather than glycolytic activity per se.
To test whether the behaviour of these promoter fusions reflected the expression of the native proteins, we examined the expression levels of the Pfk2, Pyk1, Pck1 and Icl1 proteins during the growth of C. albicans in the presence or absence of glucose. This was done as part of a global proteomics study of C. albicans (in preparation: http://www.abdn.ac.uk/cogeme). The glycolytic enzymes, Pfk2 and Pyk1, were expressed at roughly equivalent levels during exponential growth in the presence or absence of glucose (not shown). In contrast, the expression of the Pck1 and Icl1 proteins was repressed by glucose. These proteins were not detected during growth on glucose. Also, Pck1 expression levels decreased about threefold within 2 h of 0.1% glucose addition to cells growing on amino acids or lactate as sole carbon source (not shown). Therefore, the behaviour of the PFK2-, PYK1-, PCK1- and ICL1-GFP fusions accurately reflected the expression of the native enzymes.
Differential expression of glycolytic, gluconeogenic and glyoxylate cycle genes in ex vivo models
Having established the expression patterns of the PYK1, PFK2, PCK1 and ICL1 fusions in vitro, we examined their behaviour in two medically significant ex vivo models. Neutrophils and macrophages represent first lines of innate host defence when C. albicans cells infect the bloodstream or endothelia respectively. Neutropenic patients are especially susceptible to bloodstream candidaemias (Abi-Said et al., 1997). Transcript profiling has revealed that glyoxylate cycle genes are induced in C. albicans cells following exposure to neutrophils or macrophages (Lorenz et al., 2004; Fradin et al., 2005). Therefore, we profiled single C. albicans cells under similar conditions (Fig. 3). The C. albicans strains were exposed to primary human neutrophils or cultured murine J774A-1 macrophages, and the expression in phagocytosed fungal cells was compared with the expression in non-phagocytosed cells (ratio of mean GFP fluorescence per fungal cell).
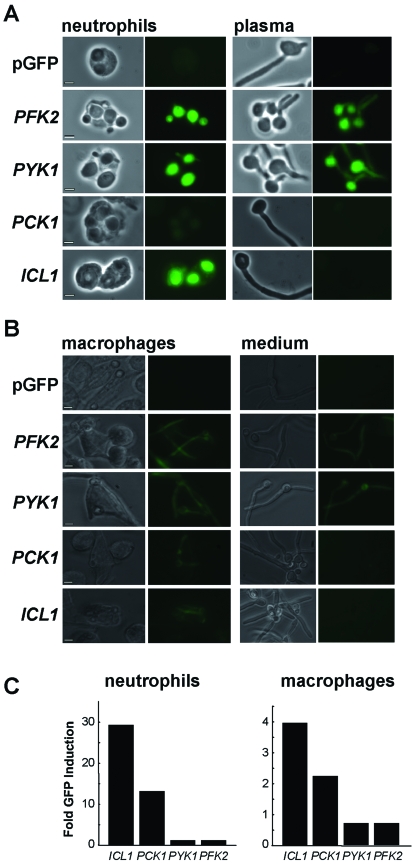
Fig. 3.
Differential regulation of PFK2-, PYK1-, PCK1- and ICL1-GFP fusions in C. albicans following phagocytosis by (A)neutrophils or (B) macrophages.
A. C. albicans cells were mixed with primary human neutrophils in a 1:1 ratio, and examined microscopically after 1.5 h. The ratio of GFP expression in phagocytosed to non-phagocytosed C. albicans cells was measured (C). Corresponding light and fluorescence micrographs of phagocytosed C. albicans cells (neutrophils), and light and fluorescence micrographs of control cells in plasma alone (plasma) are shown: bars represent 10 µm. Fold induction was measured by comparing the mean fluorescence intensity for phagocytosed cells with that for non-phagocytosed C. albicans cells (n > 50). C. albicans strains containing PFK2-, PYK1-, PCK1- or ICL1-GFP fusions were compared with the control carrying the empty vector, pGFP (CJB-1, CJB-2, CJB-3, CLM1-1, CLM3-2: Table 1).
B. Cultured murine J774A-1 macrophages were mixed with C. albicans cells in a 1:1 ratio and analysed after 3 h, as described in A: phagocytosed C. albicans cells (macrophages); non-phagocytosed cells (medium). Scale bar represents 20 µm.
C. Fold induction of GFP fluorescence in phagocytosed cells versus non-phagocytosed cells in neutrophils and macrophages respectively.
Three important observations were made. First, the induction of ICL1- and PCK1-GFP was dependent upon the internalization of fungal cells by neutrophils or macrophages, and was not observed in non-phagocytosed cells. In contrast, the glycolytic gene fusions were expressed both inside and outside the neutrophils and macrophages, and no significant induction was observed following phagocytosis. These observations are consistent with transcript profiling experiments which have suggested that glyoxylate cycle and gluconeogenic genes are induced following phagocytosis. However, these studies were unable to distinguish unambiguously between phagocytosed and non-phagocytosed cells within heterogeneous populations (Lorenz et al., 2004; Fradin et al., 2005). Second, GFP expression levels were generally an order of magnitude higher following phagocytosis by neutrophils, compared with macrophages (Fig. 3). This difference is probably attributable to differences in transcription because in vitro experiments have show that GFP stability remains unaffected following exposure of C. albicans cells to an oxidative stress (5 mM H2O2) or low pH (pH 4) (not shown). These data add to the available transcript profiling data, which did not provide information about absolute expression levels in phagocytosed cells (Lorenz et al., 2004; Fradin et al., 2005). Our data are consistent with the idea that genes involved in central carbon metabolism are more active in C. albicans cells engulfed by neutrophils, although neutrophils are thought to be more potent than macrophages at killing fungal cells (Fradin et al., 2005). Third, PCK1-GFP expression levels were reproducibly lower than ICL1-GFP expression levels after phagocytosis by neutrophils (Fig. 3A). This suggests that glyoxylate cycle activity might not always be associated with gluconeogenesis. It is possible that, in some body sites, C. albicans cells might exploit the glyoxylate cycle for the generation of metabolic intermediates rather than hexose anabolism.
Differential expression of glycolytic, gluconeogenic and glyoxylate cycle genes during systemic infections
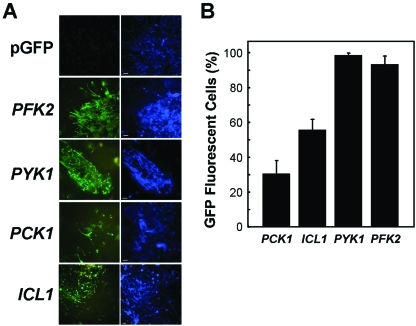
Having characterized the behaviour of the PFK2-, PYK1-, PCK1- and ICL1-GFP fusions in vitro and ex vivo, we analysed their expression in vivo in the mouse model of disseminated Candida infection (Fig. 4). The C. albicans strains were used to establish infections in immunocompetent mice, and the GFP fluorescence examined in the infected kidneys, as described previously (Barelle et al., 2004). The PFK2- and PYK1-GFP fusions were expressed in nearly all of the fungal cells infecting these kidneys, indicating that most C. albicans cells were actively growing in the infected tissue, or at least had been growing shortly before the tissue was harvested and fixed. In contrast, of the thousands of fungal cells we examined in infected kidneys, only about half expressed ICL1-GFP, and about one-third expressed PCK1-GFP. Two significant conclusions can be drawn from these observations. First, it is clear that the population of fungal cells infecting the kidney is highly heterogeneous, suggesting that individual fungal cells occupy subtly different micro-environments and are exposed to different environmental signals within the kidney. Second, only about third of C. albicans cells infecting the kidney appear to assimilate carbon via gluconeogenesis. The analysis of the PFK2- and PYK1-GFP fusions suggested that these cells were actively growing (or had been very recently). Therefore, by inference, most C. albicans cells growing in the kidney were assimilating carbon via glycolytic metabolism. This did not seem consistent with the observation that the glyoxylate cycle is required for C. albicans virulence (Lorenz and Fink, 2001).
Fig. 4.
Differential regulation of PFK2-, PYK1-, PCK1- and ICL1-GFP fusions in C. albicans in the kidney during systemic candidiasis.
A. Corresponding GFP (green) and Calcofluor White-stained (blue) images of C. albicans cells infecting the mouse kidney. C. albicans strains containing PFK2-, PYK1-, PCK1- or ICL1-GFP fusions were compared with the control strain carrying the empty vector, pGFP (CJB-1, CJB-2, CJB-3, CLM1-1, CLM3-2: Table 1).
B. Proportion of C. albicans cells (n > 1000) infecting the kidney that display GFP fluorescence above background levels, in animals displaying clinical signs of infection.
Virulence of C. albicans null mutants with blocks in glycolysis, gluconeogenesis or the glyoxylate cycle
Lorenz and Fink (2001) reported that C. albicans icl1/icl1 mutants display attenuated virulence in the mouse model of systemic candidiasis. However, after their article was published it was shown that URA3 position effects can influence the outcome of such virulence assays (Sundstrom et al., 2002; Brand et al., 2004; Sharkey et al., 2005). As the URA3 marker was used to generate the icl1/icl1 mutant, it was conceivable that the virulence of this mutant was attenuated by the position of the URA3 at the icl1 locus, rather than the disruption of the ICL1 gene itself. Therefore, we re-evaluated the effect of inactivating ICL1 upon C. albicans virulence. Having generated a homozygous icl1/icl1 mutant in C. albicans RM1000 by standard procedures (Wilson et al., 1999), we reintroduced the URA3 and HIS1 markers back into the mutant through integration at the RPS1 locus via the vector, CIp20. CIp20 is derived from CIp10 (Dennison et al., 2005), which has been shown to circumvent URA3 marker position effects (Brand et al., 2004). In addition, we extended this work by generating congenic pck1/pck1 and pyk1/pyk1 null mutants with a view to determining the impact of these mutations upon C. albicans virulence.
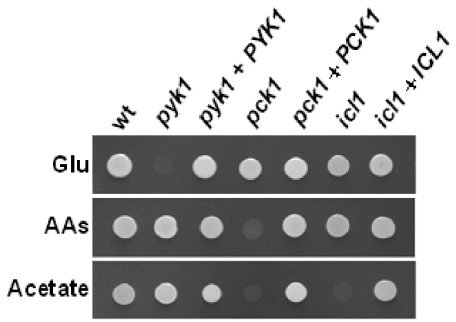
The growth phenotype of each mutant was examined in vitro (Fig. 5). As expected the pyk1/pyk1 mutant did not grow on glucose, but grew on non-fermentable carbon sources. The pck1/pck1 mutant displayed growth defects on non-fermentable carbon sources, but grew on glucose. As expected (Lorenz and Fink, 2001), the icl1/icl1 mutant did not grow on acetate, but grew on the other carbon sources. These phenotypes were suppressed by the reintroduction of the corresponding wild-type gene (Fig. 5).
Fig. 5.
Growth of C. albicans pyk1/pyk1, pck1/pck1 and icl1/icl1 null mutants on different carbon sources. C. albicans wild type, pyk1/pyk1, pck1/pck1 and icl1/icl1 strains (wt, CLM19-3; pyk1, CLM44-5; pck1, CLM56-4; icl1, CLM25-5: Table 1) and the corresponding control strains containing a reintegrated copy of the wild-type gene (pyk1 + PYK1, CLM45-9; pck1 + PCK1, CLM57-2; icl1 + ICL1, CLM26-1: Table 1) were grown on minimal medium containing 2% glucose (Glu), 2% amino acids (AAs) or 2% acetate as sole carbon source.
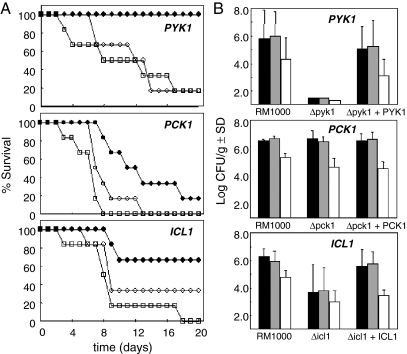
The virulence of these C. albicans mutants and the control reintegrant strains was then tested in the mouse model of systemic candidiasis (MacCallum and Odds, 2005). The icl1/icl1 and pck1/pck1 cultures were prepared in NGY medium, which contains glucose. However, the pyk1/pyk1 mutant did not grow on glucose (Fig. 5), and therefore this strain was grown on a derivative of NGY medium containing amino acids instead of glucose. Control experiments, in which the parental and reintegrant (pyk1/pyk1/PYK1) strains were prepared under identical conditions, confirmed that this change had a minimal effect upon the virulence of C. albicans (Fig. 6).
Fig. 6.
Virulence of C. albicans pyk1/pyk1, pck1/pck1 and icl1/icl1 null mutants in the mouse model of systemic infection.
A. Survival of immunocompetent female BALB/c mice following tail vein injection of C. albicans cells: open squares, first control representing the parental strain, RM1000 containing CIp20 (CLM19-3: Table 1); closed circles, experiment representing the homozygous null mutant containing CIp20 (icl1/icl1, CLM25-5; pck1/pck1, CLM56-4; pyk1/pyk1, CLM44-5: Table 1); open circles, second control representing the null mutant containing CIp20 and the wild-type gene (ICL1, CLM26-1; PCK1, CLM57-2; PYK1, CLM45-9: Table 1).
B. Tissue burdens in mouse organs infected with the same C. albicans strains. Fungal colony forming units per gram of tissue were measured in the left kidney (black), right kidney (grey) and brain (white).
Several observations were made in the virulence assays. First, the inactivation of ICL1 did attenuate the virulence of C. albicans, albeit partially, and this defect was suppressed by the reintroduction of the ICL1 gene (Fig. 6A). This confirms the original study of Lorenz and Fink (2001), which showed that the glyoxylate cycle (or at least the ICL1 gene itself) is required for normal levels of virulence in C. albicans. Second, the C. albicans pck1/pck1 and pyk1/pyk1 mutants also displayed attenuated virulence, and these defects were suppressed by the reintroduction of the wild-type PCK1 or PYK1 genes respectively (Fig. 6A). Hence these glycolytic and gluconeogenic genes are also required for the virulence of C. albicans. Third, there was a strong correlation between the virulence data and the corresponding fungal burdens observed in the tissues of infected animals (Fig. 6B). This lends weight to conclusions relating to the relative effects of the gene knockouts upon virulence. Four, the degree of virulence attenuation was significantly greater for pyk1/pyk1 cells than for icl1/icl1 or pck1/pck1 cells. The inactivation of PCK1 or ICL1 did attenuate virulence (Fig. 6A), but did not prevent fungal growth in the host (Fig. 6B). We conclude that neither gluconeogenesis nor the glyoxylate cycle is essential for C. albicans to thrive in the host. However, glycolysis is important for the development of a systemic infection.
Discussion
Our data confirm and reinforce previous observations relating to the role of the glyoxylate cycle, and the ICL1 gene in particular, in C. albicans pathogenesis. However, we have significantly extended these observations by describing the relative contributions of glycolysis and gluconeogenesis to the development of systemic candidiasis, and by providing significant insights into the heterogeneous behaviour of C. albicans cells within complex micro-environments in its mammalian host.
Several transcript profiling studies have indicated that C. albicans glyoxylate cycle genes (ICL1, MLS1) are induced following exposure to host defences (Fradin et al., 2003; 2005; Lorenz et al., 2004). This was the case following exposure to macrophages, which play an important defensive role during infections of parenchyma, and to neutrophils, which kill fungal cells present in the bloodstream. As a large proportion of C. albicans cells in these samples had been phagocytosed by the mammalian cells, it was presumed that the fungal glyoxylate cycle genes were induced after phagocytosis rather than in response to cell-cell contact. Our data support the view that these genes are induced after phagocytosis, when the fungal cells enter an environment relatively free of glucose (Fig. 3).
The inactivation of ICL1 has been reported to be required for fungal virulence (Lorenz and Fink, 2001). However, C. albicans ICL1 was strongly repressed in response to low levels of glucose (0.1%: Fig. 2) that are equivalent to those found in the bloodstream in humans and mice. Furthermore, although the gene is induced following phagocytosis (Fig. 3), it is not expressed in most C. albicans cells infecting the kidney (Fig. 4). How then can ICL1 be required for the development of systemic infections? Several possible explanations might account for this apparent paradox. First, the interpretation of the C. albicans virulence assays performed by Lorenz and Fink (2001) might have been compromised by URA3 position effects that were described after their findings were published (Sundstrom et al., 2002; Brand et al., 2004; Sharkey et al., 2005). We have shown that this is not the case by testing the virulence a new homozygous icl1/icl1 mutant alongside appropriate controls that excluded potential URA3 position effects (Fig. 4). Second, ICL1 might contribute to the virulence of C. albicans without being essential for the establishment of systemic infections. We show that this is the case. ICL1 is required for normal levels of C. albicans virulence. However, the virulence of C. albicans icl1/icl1 cells is only partially attenuated, and these cells are capable of establishing lethal systemic infections, albeit more slowly than wild-type cells (Fig. 6).
How might ICL1 contribute to C. albicans virulence without being essential for the establishment of systemic infections? The transcript profiling and GFP data indicate that ICL1 is induced following phagocytosis (Fig. 3: Lorenz et al., 2004; Fradin et al., 2005). Therefore, the glyoxylate cycle might help to protect C. albicans against host antimicrobial defences by facilitating anabolic metabolism in the absence of fermentable carbon sources (Lorenz and Fink, 2002). The absence of a functional glyoxylate cycle might lead to a reduction in the number of viable fungal cells that are capable of colonizing host tissues. Also, ICL1 might contribute to the growth of some fungal cells at infection sites (Fig. 4).
The same rationale might apply to the PCK1 gene and gluconeogenesis. Like ICL1, PCK1 was induced following phagocytosis (Fig. 3: Lorenz et al., 2004; Fradin et al., 2005). Like the C. albicans icl1/icl1 strain, the virulence of the pck1/pck1 mutant was partially attenuated in the mouse model of systemic candidiasis. C. albicans pck1/pck1 cells are unable to generate the five and six carbon sugars required for the biosynthesis of cell wall and nucleic acids, for example, from two or three carbon substrates (Fig. 1), and hence were unable to grow on such carbon sources (Fig. 5). Therefore, the fact that the icl1/icl1 and pck1/pck1 mutants are able to colonize kidney tissue (Fig. 6) indicates that, while growing in vivo, C. albicans depends mainly upon the assimilation of fermentable carbon sources in vivo, such as blood glucose for example. This idea is reinforced by the observation that pyk1/pyk1 cells are avirulent (Fig. 6), indicating that C. albicans depends upon glycolysis for growth in the host. Hence in principle, some drugs that inhibit fungal glycolytic enzymes without affecting their mammalian counterparts could prove to be effective antifungal agents.
Our analyses of GFP fusions have provided direct evidence for a high degree of metabolic heterogeneity within fungal cell populations in vivo (Fig. 4). Almost all C. albicans cells infecting the mouse kidney expressed the PFK2- and PYK1-GFP fusions, indicating that most of these fungal cells were actively growing, or had recently been actively growing. [The half-life of GFP in growing C. albicans is about 1.5 h (Barelle et al., 2004)] However, fewer than half of these cells expressed the PCK1- and ICL1-GFP fusions (Fig. 4). This implies that, while most cells were assimilating carbon via the glycolytic pathway, some had switched to gluconeogenic metabolism. Therefore, glucose appears to become limiting in some micro-environments within the infected kidney. Currently methods are being developed for the transcript profiling of C. albicans cells isolated from infected tissues. The heterogeneity of fungal populations within foci of infection has major implications for the interpretation of these transcript profiling data sets.
In conclusion, our data reveal that the pathogen C. albicans regulates central carbon metabolism in a niche-specific manner during disease establishment and progression. During the early stages of a systemic infection, C. albicans activates the glyoxylate cycle and gluconeogenesis in response to phagocytosis. In the latter stages of an infection, when the fungus is colonizing tissue, glycolytic metabolism predominates. Presumably, this increases the biological fitness of this pathogen within its host.
Experimental procedures
Strains and growth conditions
Candida albicans strains (Table 1) were grown in YPD or synthetic complete medium (Sherman, 1991; Kaiser et al., 1994) or in minimal medium (0.67% Yeast Nitrogen Base) containing 2% glucose, 2% casamino acids or 2% acetate as sole carbon source.
Table 1.
C. albicans strains
| Strain | Genotype | Parent | Source |
|---|---|---|---|
| SC5314 | Wild type | Gillum et al. 1984 | |
| CAF2-1 | URA3/ura3::λ imm434 | SC5314 | Fonzi and Irwin (1993) |
| CAI4 | ura3::λ imm434/ura3::λ imm434 | CAF2-1 | Fonzi and Irwin (1993) |
| CJB-1 | ura3::λ imm434/ura3::λ imm434, pGFP | CAI4 | Barelle et al. (2004) |
| CJB-2 | ura3::λ imm434/ura3::λ imm434, pPCK1-GFP | CAI4 | Barelle et al. (2004) |
| CJB-3 | ura3::λ imm434/ura3::λ imm434, pICL1-GFP | CAI4 | This study |
| CLM1-1 | ura3::λ imm434/ura3::λ imm434, pPYK1-GFP | CAI4 | This study |
| CLM3-2 | ura3::λ imm434/ura3::λ imm434, pPFK2-GFP | CAI4 | This study |
| RM1000 | ura3::λ imm434/ura3::λ imm434, his1::hisG/his1::hisG | CAI4 | Wilson et al. (1999) |
| CLM19-3 | ura3::λ imm434/ura3::λ imm434, his1::hisG/his1::hisG, CIp20 (URA3, HIS1) | RM1000 | This study |
| CLM4-10 | ura3::λ imm434/ura3::λ imm434, his1::hisG/his1::hisG, ICL1/icl1::URA3 | RM1000 | This study |
| CLM18-1 | ura3::λ imm434/ura3::λ imm434, his1::hisG/his1::hisG, icl1::HIS1/icl1::URA3 | RM1000 | This study |
| CLM21-1 | ura3::λ imm434/ura3::λ imm434, his1::hisG/his1::hisG, icl1::HIS1/icl1::ura3 | RM1000 | This study |
| CLM25-5 | ura3::λ imm434/ura3::λ imm434, his1::hisG/his1::hisG, icl1::HIS1/icl1::ura3, CIp20 (URA3, HIS1) | RM1000 | This study |
| CLM26-1 | ura3::λ imm434/ura3::λ imm434, his1::hisG/his1::hisG, icl1::HIS1/icl1::ura3, CIp20-ICL1 | RM1000 | This study |
| CLM17-1 | ura3::λ imm434/ura3::λ imm434, his1::hisG/his1::hisG, PCK1/pck1::URA3 | RM1000 | This study |
| CLM53-1 | ura3::λ imm434/ura3::λ imm434, his1::hisG/his1::hisG, pck1::HIS1/pck1::URA3 | RM1000 | This study |
| CLM54-1 | ura3::λ imm434/ura3::λ imm434, his1::hisG/his1::hisG, pck1::HIS1/pck1::ura3 | RM1000 | This study |
| CLM56-4 | ura3::λ imm434/ura3::λ imm434, his1::hisG/his1::hisG, pck1::HIS1/pck1::ura3, CIp20 (URA3, HIS1) | RM1000 | This study |
| CLM57-2 | ura3::λ imm434/ura3::λ imm434, his1::hisG/his1::hisG, pck1::HIS1/pck1::ura3, CIp20-PCK1 | RM1000 | This study |
| PD24 | ura3::λ imm434/ura3::λ imm434, his1::hisG/his1::hisG, PYK1/pyk1::URA3 | RM1000 | This study |
| CLM5-1 | ura3::λ imm434/ura3::λ imm434, his1::hisG/his1::hisG, pyk1::HIS1/pyk1::URA3 | RM1000 | This study |
| CLM29-2 | ura3::λ imm434/ura3::λ imm434, his1::hisG/his1::hisG, pyk1::HIS1/pyk1::ura3 | RM1000 | This study |
| CLM44-5 | ura3::λ imm434/ura3::λ imm434, his1::hisG/his1::hisG, pyk1::HIS1/pyk1::ura3, CIp20 (URA3, HIS1) | RM1000 | This study |
| CLM45-9 | ura3::λ imm434/ura3::λ imm434, his1::hisG/his1::hisG, pyk1::HIS1/pyk1::ura3, CIp20-PYK1 | RM1000 | This study |
Strain construction
The plasmids pGFP and pPCK1-GFP have been described previously (Barelle et al., 2004). To generate pICL1-GFP, the CaICL1 promoter region (−983 to −1, relative to the start codon) was PCR-amplified using the primers ICL1-5′ and ICL1-3′, and cloned between the XhoI and HindIII sites of pGFP. To make pPYK1-GFP, the CaPYK1 promoter region (−1576 to −3) was PCR-amplified using the primers PYK5′-2 and PYK3′-3, and cloned between the BstEII and MluI sites of pGFP. To produce pPFK2-GFP, the CaPFK2 promoter region (−992 to −1) was PCR-amplified using primers PFK2-5′ and PFK2-3′ and cloned between the XhoI and HindIII sites of pGFP. These plasmids were linearized with StuI and integrated at the RPS1 locus in C. albicans CAI4 (Murad et al., 2000). The genotypes of transformants were confirmed by PCR and Southern blotting, as described previously (Barelle et al., 2004).
The C. albicans pyk1/pyk1 null mutant was constructed using the parental strain RM1000 (Table 1). Each PYK1 allele was inactivated sequentially with loxP-URA3-loxP (LUL) and loxP-HIS1-loxP (LHL) disruption cassettes (Dennison et al., 2005) generated by PCR amplification with the oligonucleotide primers PYK-L1 and PYK-L2 (Table 2). The pyk1::LUL and pyk1::LHL disruption cassettes contained about 65 bp of homology to the 5′ and 3′ ends of the PYK1 locus, and deleted codons 19–490 of the PYK1 open reading frame. The pyk1::LUL cassette was transformed into RM1000, transformants selected on the basis of their uridine protrophy, and integration at the correct locus confirmed by PCR diagnosis with the primers PYK-5′-DIAG and URA3-REV-DIAG (Table 2), as described previously (Wilson et al., 1999). The resultant heterozygote (pyk1::loxP-URA3-loxP/PYK1: PD24) was then transformed with the pyk1::LHL cassette, and the introduction of this allele and the removal of the last wild-type PYK1 allele from the C. albicans genome was confirmed by PCR using the primer PYK-5′-DIAG with HIS1-REV-DIAG or PYK-IP, respectively (Table 2), thereby generating the homozygous null mutant, CLM5-1 (Table 1).
Table 2.
Oligonucleotides
| Primer | Sequence (5′ to 3′) | Use |
|---|---|---|
| ICL1-5′ | CCCTCGAGGTCATGGAATCG | Cloning of ICL1 promoter (XhoI site underlined) |
| ICL1-3′ | GGAAGCTTTTATTAATGTTTATTC | Cloning of ICL1 promoter (HindIII site underlined) |
| PYK5′-2 | CCCTCGAGTATAGAAATTGGGTTGC | Cloning of PYK1 promoter (XhoI site underlined) |
| PYK3′-3 | AAACTGCAGTGATGTAGTTTGTAGG | Cloning of PYK1 promoter (PstI site underlined) |
| PFK2-5′ | CCGCTCGAGATTCTATGACGTGGGT | Cloning of PFK2 promoter (XhoI site underlined) |
| PFK2-3′ | CCCAAGCTTGGTGGTTGTTTTTCGT | Cloning of PFK2 promoter (HindIII site underlined) |
| ICL1-L1 | TCCGGATGGCAATGTTCTTCTACTGCTTCCACTTCTAACGAACCATCTCCAGATTTGGCTCCAGGGTTTTCCCAGTCACG | PCR amplification of icl1 disruption cassettes (homology to pLHL and pLUL plasmids underlined) |
| ICL1-L2B | CTTGATGTAAGTTTCTTGTTCGTCAGCTGGCATGGCCTTGTTCCAGTTGAAAGATGGAGACTCACTAAAGGGAACAAAAGC | PCR amplification of icl1 disruption cassettes (homology to pLHL and pLUL plasmids underlined) |
| ICL1-IP | TTCTGGGTTAGTGGCACCAA | Diagnosis of icl1 alleles, internal 3’ primer |
| ICL1-5′-DIAG | CAGGTGAAGAGTCACTTCTA | Diagnosis of icl1 alleles, upstream primer |
| PCK-L1 | AAATAATTATCAATCATGGCTCCTCCTACTGCTGTTGAATCTTCAATCAATTTCGGAGGTCACCCAGGGTTTTCCCAGTCACG | PCR amplification of pck1 disruption cassettes (homology to pLHL and pLUL plasmids underlined) |
| PCK1-L2 | GACAATTCACCAGAGTGGATAGCATCCAAGATAGCTCTGGTGTATTTCAATGGACATCTCACTAAAGGGAACAAAAGC | PCR amplification of pck1 disruption cassettes (homology to pLHL and pLUL plasmids underlined) |
| PCK-5′-DIAG | GACATCACCTTTTTCAACC | Diagnosis of pck1 alleles, internal 3’ primer |
| PCK-IP | CAACAATACCGTCAACTC | Diagnosis of pck1 alleles, upstream primer |
| PYK-L1 | CTACATCAACAATGTCTCACTCATCTTTATCTTGGTTATCCAACTTCAATGTTGAAACTGTTCCAGGGTTTTCCCAGTCACG | PCR amplification of pyk1 disruption cassettes (homology to pLHL and pLUL plasmids underlined) |
| PYK-L2 | ATCAAGGCTTCTTTTCCACTTAAGCTTGGACGATTCTAACAGTGTTAGAGTGACCAGAACCTCACTAAAGGGAACAAAAGC | PCR amplification of pyk1 disruption cassettes (homology to pLHL and pLUL plasmids underlined) |
| PYK-IP | TCCTTACCTTCTTCACC | Diagnosis of pyk1 alleles, internal 3’ primer |
| PYK-5′-DIAG | TTAGAAAGGAAACAAAGGG | Diagnosis of pyk1 alleles, upstream primer |
| HIS1-REV-DIAG | CACAAGAAGCCTCAACGG | Diagnosis of LHL alleles, downstream primer |
| URA3-REV-DIAG | TAGTGTTACGAATCAATGGC | Diagnosis of LUL alleles, downstream primer |
| ICL1-CIp20-5′ | CCATCGATCAAAAACTTAATTTAACCAGCG | Cloning of ICL1 locus (ClaI site underlined) |
| ICL1-CIp20-3′ | CGACGCGTTAAAGGCTCAAATTGTTCCCG | Cloning of ICL1 locus (MluI site underlined) |
| PYK-CIp20-5′ | CCATCGATTATAGAAATTGGGTTGC | Cloning of PYK1 locus (ClaI site underlined) |
| PYK-CIp20-3′ | CGACGCGTGCAGAATTAACGTATG | Cloning of PYK1 locus (MluI site underlined) |
| PCK-CIp20-5′ | CCATCGATTTGGTCCATTGCCGT | Cloning of PCK1 locus (ClaI site underlined) |
| PCK-CIp20-3′ | CGACGCGTCCATAGGATTATGAGAC | Cloning of PCK1 locus (MluI site underlined) |
To generate the prototrophic reintegrant strain CLM45-9 (pyk1/pyk1/PYK1), ura3– segregants were selected on YPD containing 1 mg ml−1 5- fluoroorotic acid (Fonzi and Irwin, 1993), generating the strain CLM29-2. The plasmid CIp20-PYK1 was then transformed into CLM29-2, selecting for the URA3 marker, to create the strain CLM45-9. CIp20-PYK1 contained the PYK1 locus from coordinates −1576 upstream from the start codon to +354 downstream from the stop codon, and was generated using primers PYK-CIp20-5′ and PYK-CIp20-3′ (Table 2). As a control, the empty CIp20 vector was transformed into CLM29-2, to create CLM44-5 (Table 1). CIp20-based plasmids, which contain both URA3 and HIS1 markers, were integrated at the RPS1 locus, and correct integration confirmed by PCR diagnosis (Dennison et al., 2005).
Congenic C. albicans icl1/icl1 and pck1/pck1 null mutants (CLM18-1 and CLM53-1, respectively: Table 1) were constructed using the same approach. In the icl1/icl1 mutants, codons 121–430 of the ICL1 open reading frame were deleted. In the pck1/pck1 mutants, codons 18–459 of the PCK1 open reading frame were deleted. Reintegrant and control strains were generated, as described above (Table 1). CIp20-ICL1 contained the ICL1 locus from −999 upstream from the start codon to +367 downstream from the stop codon, and CIp20-PCK1 contained the PCK1 locus from −977 to +500. The genotypes of all strains were confirmed by Southern blotting, as well as by PCR (Dennison et al., 2005).
DNA manipulations
Cloning, PCR amplification and Southern analysis were performed as described previously (Sambrook et al., 1989; Dennison et al., 2005).
Proteomics
Protein extracts were prepared from mid-exponential C. albicans CLM19-3 cells (Table 1) growing on 2% glucose, casamino acids, lactate or oleic acid as sole carbon source. Extracts were also prepared 2 h after the addition of 0.1% glucose to cultures growing on 2% casamino acids, lactate or oleic acid. Protein extracts were subjected to 2D-gel electrophoresis, and the spots corresponding to the Pfk2, Pyk1, Pck1 and Icl1 proteins identified by peptide mass fingerprinting (Yin et al., 2004). The relative level of each protein was measured by determining their mean spot volumes under each of the growth conditions in three independent replicate experiments.
Microscopy
Fungi were stained with Calcofluor White as described previously (Gow and Gooday, 1982). Nuclei were stained by overlaying mounting media containing DAPI (Vector Laboratories, Peterborough, UK). For phase contrast and fluorescence microscopy an Axioplan 2 microscope (Carl Zeiss, UK) was used with filter sets; XF 66, XF 67 and XF 77 (Omega Optical, Brattleboro, VT). Images were generated with a Hamamatsu CCD camera, and analysed with Openlab 3.0.9 software (Improvision, Coventry, UK). Relative GFP fluorescence intensities were quantified for individual yeast cells or hyphal compartments by measurement of regions of interest. Mean fluorescence intensities (± standard deviation) were then calculated for at least 50 individual cells, as described before (Barelle et al., 2004). GFP fluorescence levels never exceeded the quantitative range of the assay.
Ex vivo analysis ofC. albicans
Neutrophils were isolated from human blood (Fradin et al., 2005), C. albicans cells mixed with them in a 1:1 ratio, and GFP fluorescence measured after 1.5 h (Barelle et al., 2004). Murine J774A-1 macrophages (Ralph et al., 1975) were mixed with C. albicans cells in a 1:1 ratio and analysed after 3 h. The ratio of GFP expression in phagocytosed to non-phagocytosed C. albicans cells was measured. Fold induction was measured by comparison of mean fluorescence intensity for phagocytosed cells with that for non-phagocytosed C. albicans cells (n > 50).
Murine model of systemic candidiasis
Immunocompetent female BALB/c mice (Harlan Sera-lab, Loughborough, UK) were challenged intravenously with C. albicans. The C. albicans icl1/icl1 and pck1/pck1 strains and their controls (CLM19-3, CLM25-5, CLM26-1, CLM56-4 and CLM57-2: Table 1) were grown with shaking for 18–24 h at 30°C in NGY medium (0.1% Neopeptone, 0.4% glucose and 0.1% Yeast Extract). The pyk1/pyk1 null mutant and its controls (CLM44-5, CLM45-9 and CLM19-3: Table 1) were grown on NGY containing 0.4% Casamino acids instead of glucose. Groups of five or six mice were inoculated via the lateral tail vein with 1.2–1.9 × 104 cfu per gram of body weight (Brand et al., 2004; MacCallum and Odds, 2005). Mice were monitored over 28 days and animals showing signs of distress or illness were humanely terminated and deaths recorded as occurring the following day. The kidneys and brain were removed post mortem, homogenized in 0.5 ml of water, and C. albicans tissue burdens determined by viable counting. Viable counts were performed on glucose-containing medium for the icl1/icl1 and pck1/pck1 mutants, and on amino acid-containing medium for the pyk1/pyk1 mutant. For the analysis of GFP expression in vivo, frozen sections of kidney tissue from infected mice were examined as described previously (Barelle et al., 2004). All experimentation was carried out under the terms of the UK Home Office licenses for research on animals.
Acknowledgments
We are grateful to Zhikang Yin, David Stead, Laura Selway and Janet Walker (COGEME Proteomics Service Facility in Aberdeen) for providing protein expression data prior to publication. We thank Paul Dennison for help with the construction of the PYK1/pyk1 mutant, and Susan Budge for excellent technical assistance. This work was funded by the Wellcome Trust (055015; 063204, 72263), the BBSRC (1/P17124, BBS/B/06679), the European Commission (MRTN-CT-2003-504148) and the University of Aberdeen.
References
- Abi-Said D, Anaissie E, Uzun O, Raad I, Pinzcowski H, Vartivarian S. The epidemiology of hematogenous candidiasis caused by different Candida species. Clin Infect Dis. 1997;24:1122–1128. doi: 10.1086/513663. [DOI] [PubMed] [Google Scholar]
- Barelle CJ, Manson C, MacCallum D, Odds FC, Gow NAR, Brown AJP. GFP as a quantitative reporter of gene regulation in Candida albicans. Yeast. 2004;21:333–340. doi: 10.1002/yea.1099. [DOI] [PubMed] [Google Scholar]
- Brand A, MacCallum DM, Brown AJP, Gow NAR, Odds FC. Ectopic expression of URA3 can influence the virulence phenotypes and proteome of Candida albicans but can be overcome by targeted reintegration of URA3 at the RPS10 locus. Eukaryot Cell. 2004;3:900–909. doi: 10.1128/EC.3.4.900-909.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AJP. Integration of metabolism with virulence in Candida albicans. In: Brown AJP, editor. Fungal Genomics. Heidelberg: Mycota XIII, Springer-Verlag; 2005. pp. 185–203. [Google Scholar]
- Calderone RA. Candida and Candidiasis. Washington DC: American Society for Microbiology Press; 2002. [Google Scholar]
- Dennison PMJ, Ramsdale M, Manson CL, Brown AJP. Gene disruption in Candida albicans using a synthetic, codon-optimised Cre-loxP system. Fungal Genet Biol. 2005;42:737–748. doi: 10.1016/j.fgb.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Douglas LJ. Candida biofilms and their role in infection. Trends Microbiol. 2003;11:30–36. doi: 10.1016/s0966-842x(02)00002-1. [DOI] [PubMed] [Google Scholar]
- Fonzi WA, Irwin MY. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradin C, Kretschmar M, Nichterlein T, Gaillardin C, d'Enfert C, Hube B. Stage-specific gene expression of Candida albicans in human blood. Mol Microbiol. 2003;47:1523–1543. doi: 10.1046/j.1365-2958.2003.03396.x. [DOI] [PubMed] [Google Scholar]
- Fradin C, De Groot P, MacCallum D, Schaller M, Klis F, Odds FC, Hube B. Granulocytes govern the transcriptional response, morphology and proliferation of Candida albicans in human blood. Mol Microbiol. 2005;56:397–415. doi: 10.1111/j.1365-2958.2005.04557.x. [DOI] [PubMed] [Google Scholar]
- Garcia-Sanchez S, Mavor A, Russell CL, Argimon S, Dennison P, Enjalbert B, Brown AJP. Global roles of Ssn6 in Tupl- and Nrgl-dependent gene regulation in the fungal pathogen, Candida albicans. Mol Biol Cell. 2005;16:2913–2925. doi: 10.1091/mbc.E05-01-0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillum AM, Tsay EY, Kirsch DR. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet. 1984;198:179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- Gow NAR, Gooday GW. Growth kinetics and morphology of colonies of the filamentous form of Candida albicans. J Gen Microbiol. 1982;128:2187–2194. doi: 10.1099/00221287-128-9-2187. [DOI] [PubMed] [Google Scholar]
- Kaiser C, Michaelis S, Mitchell A. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Kullberg BJ, Oude Lashof AM. Epidemiology of opportunistic invasive mycoses. Eur J Med Res. 2002;7:183–101. [PubMed] [Google Scholar]
- Leuker CE, Sonneborn A, Delbruck S, Ernst JF. Sequence and promoter regulation of the PCK1 gene encoding phosphoenolpyruvate carboxykinase of the fungal pathogen Candida albicans. Gene. 1997;192:235–240. doi: 10.1016/s0378-1119(97)00069-3. [DOI] [PubMed] [Google Scholar]
- Lorenz MC, Fink GR. The glyoxylate cycle is required for fungal virulence. Nature. 2001;412:83–86. doi: 10.1038/35083594. [DOI] [PubMed] [Google Scholar]
- Lorenz MC, Fink GR. Life and death in a macrophage: role of the glycoxylate cycle in virulence. Eukaryot Cell. 2002;1:657–662. doi: 10.1128/EC.1.5.657-662.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz MC, Bender JA, Fink GR. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot Cell. 2004;3:1076–1087. doi: 10.1128/EC.3.5.1076-1087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCallum DM, Odds FC. Temporal events in the intravenous challenge model for experimental Candida albicans infections in female mice. Mycoses. 2005;48:151–161. doi: 10.1111/j.1439-0507.2005.01121.x. [DOI] [PubMed] [Google Scholar]
- Murad AMA, Lee PR, Broadbent ID, Barelle CJ, Brown AJP. CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast. 2000;16:325–327. doi: 10.1002/1097-0061(20000315)16:4<325::AID-YEA538>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Murad AMA, Leng P, Straffon M, Wishart J, Macaskill S, MacCallum D, et al. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 2001;20:4742–4752. doi: 10.1093/emboj/20.17.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odds FC. Candida and Candidosis. 2. London: Bailliere Tindall; 1988. [Google Scholar]
- Prigneau O, Porta A, Poudrier JA, Colonna-Romano S, Noel T, Maresca B. Genes involved in β-oxidation, energy metabolism and glyoxylate cycle are induced by Candida albicans during macrophage infection. Yeast. 2003;20:723–730. doi: 10.1002/yea.998. [DOI] [PubMed] [Google Scholar]
- Ralph P, Prichard J, Cohn M. Reticulum cell sarcoma: an effector cell in antibody-dependent cell-mediated immunity. J Immunol. 1975;114:898–905. [PubMed] [Google Scholar]
- Rubin-Bejerano I, Fraser I, Grisafil P, Fink GR. Phagocytosis by neutrophils induces an amino acid deprivation response in Saccharomyces cerevisiae and Candida albicans. Proc Natl Acad Sci USA. 2003;100:11007–11012. doi: 10.1073/pnas.1834481100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sharkey LL, Liao W -I, Ghosh AK, Fonzi WA. Flanking direct repeats of hisG alter URA3 marker expression at the HWP1 locus of Candida albicans. Microbiology. 2005;151:1061–1071. doi: 10.1099/mic.0.27487-0. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Sundstrom P, Cutler JE, Staab JF. Reevaluation of the role of HWP1 in systemic candidiasis by use of Candida albicans strains with selectable marker URA3 targeted to the ENO1 locus. Infect Immun. 2002;70:3281–3283. doi: 10.1128/IAI.70.6.3281-3283.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco E, Thuler LC, Martins CA, Nucci M, Dias LM, Goncalves VM. Epidemiology of bloodstream infections at a cancer centre. Sao Paulo Med J. 2000;118:131–138. doi: 10.1590/S1516-31802000000500004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RB, Davis D, Mitchell AP. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol. 1999;181:1868–1874. doi: 10.1128/jb.181.6.1868-1874.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z, Wilson S, Hauser NC, Tournu H, Hoheisel JD, Brown AJP. Glucose triggers different global responses in yeast depending on the strength of the signal, and transiently stabilises ribosomal protein mRNAs. Mol Microbiol. 2003;48:713–724. doi: 10.1046/j.1365-2958.2003.03478.x. [DOI] [PubMed] [Google Scholar]
- Yin Z, Stead D, Selway L, Walker J, Riba-Garcia I, Mclnerney T, et al. Proteomic response to amino acid starvation in Candida albicans and Saccharomyces cerevisiae. Proteomics. 2004;4:2425–2436. doi: 10.1002/pmic.200300760. [DOI] [PubMed] [Google Scholar]