Abstract
The E6 oncoprotein of human papillomaviruses (HPVs) that are associated with cervical cancer utilizes the cellular ubiquitin–protein ligase E6-AP to target the tumor suppressor p53 for degradation. In normal cells (i.e., in the absence of E6), p53 is also a target of the ubiquitin–proteasome pathway. Under these conditions, however, p53 degradation is mediated by Mdm2 rather than by E6-AP. Here we show in a mutational analysis that, surprisingly, the structural requirements of p53 to serve as a proteolytic substrate differ between E6 proteins derived from different HPV types and, as expected, between Mdm2 and E6 proteins in vitro and in vivo. Stable expression of such mutants in HPV-negative and HPV-positive cell lines demonstrates that in HPV-positive cancer cells, the E6-dependent pathway of p53 degradation is not only active but, moreover, is required for degradation of p53, whereas the Mdm2-dependent pathway is inactive. Because the p53 pathway was reported to be functional in HPV-positive cancer cells, this finding indicates clearly that the ability of the E6 oncoprotein to target p53 for degradation is required for the growth of HPV-positive cancer cells.
On the basis of epidemiological and experimental evidence, it is widely accepted that certain human papillomaviruses (HPVs), including HPV types 16 and 18, play an etiologic role in cervical carcinogenesis. The major oncoproteins of these HPVs are encoded by the E6 and E7 genes, which are the only viral genes that are generally retained and expressed in HPV-positive cancer cells. Furthermore, inhibition of E6/E7 expression in cell-culture systems interferes with the growth of HPV-positive cancer cell lines, indicating that E6 and E7 represent potent targets for therapeutic intervention in the treatment of cervical cancer (reviewed in ref. 1).
The E6 oncoprotein has been shown to recruit the cellular ubiquitin–protein ligase E6-AP to target the tumor-suppressor protein p53 for ubiquitin–proteasome-mediated degradation (2, 3). Although it is commonly assumed that this property of E6 is in part responsible for its oncogenic and antiapoptotic potential, it is also clear that, in addition, E6 has p53-independent transforming and antiapoptotic activities (4–8). E6 has been reported to interact with several cellular proteins, including E6BP (9), hDLG (10), IRF-3 (11), Bak (12), and E6TP1 (13), and it seems likely that at least some of these interactions contribute to HPV-induced cellular transformation.
Under normal growth conditions, p53 is turned over by the ubiquitin–proteasome system also in HPV-negative cells (14–16). Over the past few years, it has become clear that, under these conditions, p53 degradation is mediated mainly by Mdm2, whereas several lines of evidence indicate that E6-AP plays no, or only a minor, role in p53 degradation in the absence of E6 (15–24). This notion may indicate that E6 can target p53 for degradation under conditions when the normal pathway for p53 degradation is inactive (e.g., after DNA damage), by using a ubiquitin–protein ligase (E6-AP), which normally is not involved in p53 degradation. Indeed, it has been reported that after DNA damage, p53 is not stabilized and activated in cells that ectopically express E6 (25). It should be noted, however, that p53 can be activated by DNA-damaging agents in HPV-positive cancer cells, which probably can be explained by the notion that the viral promoter from which E6 is transcribed is shut off after DNA damage (26, 27). Regardless of the exact mechanism, however, this observation indicates that in HPV-positive cancer cells, the ability of E6 to circumvent the normal stability regulation of p53 is limited to certain yet-unknown stress signals.
On the basis of the results obtained with antisense approaches directed against E6-AP expression (20, 23) and by the use of a dominant-negative E6-AP mutant (21), it is clear that in HPV-positive carcinoma cell lines, degradation of p53 involves the E6/E6-AP-dependent pathway. However, whether the Mdm2-dependent pathway of degradation is active also in HPV-positive cells or, alternatively, whether degradation of p53 depends entirely on the E6-dependent pathway in such cells remains unclear. A possibility to address this question without interfering with any other cellular processes (as is probably the case in approaches that generally interfere with E6-AP activity) is the characterization of the turnover rate of p53 mutants that are recognized as proteolytic targets by the normal Mdm2-dependent pathway but not by the E6/E6-AP-dependent pathway and vice versa. Here we report that the structural requirements for p53 to serve as a substrate for ubiquitination/degradation differ between Mdm2 and E6 and, unexpectedly, between E6 proteins derived from different HPVs. Characterization of the turnover rate of p53 mutants that distinguish between these different p53-degradation pathways reveals that in HPV-positive carcinoma cell lines, the degradation of p53 depends entirely on the presence of E6.
Materials and Methods
Plasmids and Protein Expression.
The various chimeric p53 forms consisting of different regions of human p53 (hp53) and the respective regions of murine p53 (mp53; Fig. 2A) were constructed by either restriction-site cloning or PCR-directed mutagenesis (further details will be provided on request). The parental hp53 and mp53 plasmids were kindly provided by T. Unger (Weizmann Institute, Rehovot, Israel). The deletion mutants Δ69 (deletion of amino acids 62–96) and ΔN43 (deletion of the N-terminal 43 amino acids) were described previously (22). For in vitro translation and transient expression, the various p53 forms were cloned into the expression vector pRC/CMV (Invitrogen), respectively. For the generation of cell lines stably expressing the various p53 forms, the respective cDNAs were cloned into the expression vector pEF1/V5-His (Invitrogen).
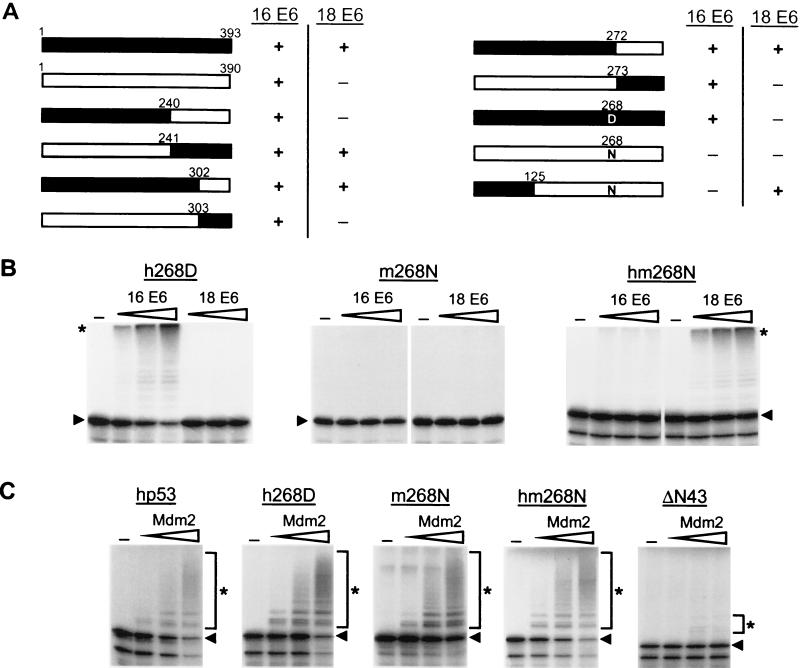
Figure 2.
Amino acid residue 268 of p53 plays a critical role in the interaction with E6. (A) Schematic representation of the p53 mutants generated and the results obtained in E6-dependent ubiquitination assays. Black bars represent hp53 or the respective region of hp53, and white bars represent mp53 or the respective regions of mp53. +, the respective p53 form is ubiquitinated in the presence of the HPV E6 protein indicated; −, the respective p53 form is not recognized as a substrate by the HPV E6 protein indicated. (B and C) The indicated forms of p53 were generated in rabbit reticulocyte lysate in the presence of 35S-labeled methionine. The radiolabeled proteins then were incubated in the presence or absence of increasing amounts of HPV-16 E6 or HP-18 E6 (B) or Mdm2 (C) under standard ubiquitination conditions (Materials and Methods). The running position of ubiquitinated p53 forms is indicated with an asterisk and that of the respective nonmodified p53 forms with an arrowhead.
In transient-expression experiments, HPV-16 E6 and HPV-18 E6 were expressed from the pRC/CMV expression vector. The expression construct for Mdm2 (kindly provided by M. Oren, Weizmann Institute) was described previously (17, 28).
For immunoprecipitations, the HPV E6 proteins and p53 were generated in vitro in the rabbit reticulocyte lysate system in the presence (E6) or in the absence (p53) of [35S]cysteine.
For in vitro ubiquitination experiments, Mdm2, HPV-18 E6, and HPV-16 E6 were expressed as glutathione S-transferase fusion proteins in Escherichia coli DH5α. The ubiquitin-activating enzyme E1 and the ubiquitin-conjugating enzyme UbcH5 were expressed in E. coli BL21 by using the pET expression system as described (29).
Cell Lines, Transfection, and Selection Procedures.
H1299 and RKO cells were grown in RPMI medium 1640 supplemented with 10% (vol/vol) FBS. MCF-7, HeLa, SW756, CaSki, and SiHa cells were grown in DMEM supplemented with 10% (vol/vol) FBS.
To generate cell lines stably expressing the various p53 forms, cells were transfected with the respective expression construct by lipofection {DOTAP, N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium methylsulfate} according to the manufacturer's instructions (Roche Molecular Biochemicals). Cells stably containing the expression construct were selected by resistance to neomycin (Geneticin, Sigma). The neomycin-resistance gene is encoded within the vector used for p53 expression (pEF1/V5-His). For half-life measurements, pooled, rather than single-cell, clones were used as soon as was practical after selection to avoid clonal artifacts.
In transient-expression experiments, cells were transfected with the respective expression constructs in the presence of a reporter construct encoding β-galactosidase by lipofection (DOTAP). Protein extracts were prepared 20 h after transfection as described (22), and transfection efficiency was determined by measuring β-galactosidase activity. Then p53 levels were determined by Western blot analysis (see below) by using transfection-efficiency-adjusted protein amounts.
In Vitro Ubiquitination Assays and Half-Life Measurements.
Binding of the HPV E6 proteins to p53 was measured in an immunoprecipitation analysis by using the p53-specific monoclonal antibody PAb 421 (Dianova, Hamburg, Germany) (2). HPV E6-mediated ubiquitination of p53 was assayed as described (2) by using either in vitro-translated unlabeled E6 proteins or bacterially expressed glutathione S-transferase–E6 fusion proteins. For Mdm2-dependent ubiquitination, 2 μl of rabbit reticulocyte lysate-translated 35S-labeled p53 was incubated in the presence of increasing amounts of bacterially expressed human Mdm2 (100–500 ng), 50 ng E1, 50 ng UbcH5, and 6 μg ubiquitin (Sigma) in 60-μl volumes. In addition, reactions contained 25 mM Tris⋅HCl (pH 7.5), 60 mM NaCl, 1 mM DTT, 2 mM ATP, and 4 mM MgCl2. After incubation at 30°C for 2 h, total-reaction mixtures were electrophoresed in 10% SDS-polyacrylamide gels and 35S-labeled p53 detected by fluorography.
For half-life measurements of the various p53 forms, the respective cells were grown to confluency and then split 1:3 onto 6-cm plates. After 16 h, 60 μg/ml cycloheximide was added, and the cells were lysed for protein extraction after incubation for the indicated time periods. Alternatively, cells were labeled metabolically with an L-[35S]methionine/cysteine mix, and the half-life was determined in a pulse–chase analysis as described (22).
For Western blotting, equal amounts of protein were separated on 10% SDS-polyacrylamide gels and blotted onto poly(vinylidene difluoride) membranes. The mouse monoclonal PAb1801 (Dianova) was used to detect ΔN43. Then, enhanced chemiluminescence was performed according to the manufacturer's instructions (Amersham Pharmacia). For all other p53 forms, the mouse monoclonal antibody DO-1 (Dianova) was used.
Results
mp53 Is Recognized by HPV-16 E6 but Not by HPV-18 E6.
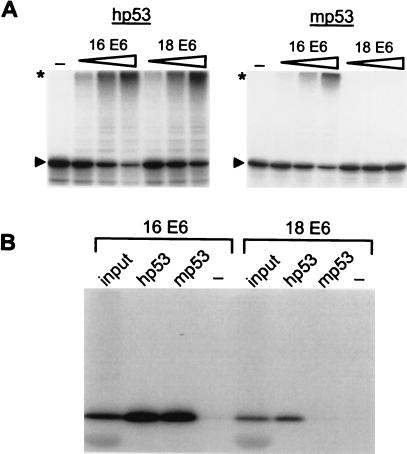
To understand further the mechanism by which E6 can circumvent the normal stability regulation of p53, a series of p53 mutants was recently characterized in vitro and in vivo with respect to their ability to serve as substrates for E6-dependent and E6-independent ubiquitination/degradation (22). During the course of these studies, it was observed that in contrast to hp53, mp53 was recognized and ubiquitinated in vitro by HPV-16 E6 but not by HPV-18 E6 (Fig. 1A). Subsequent coprecipitation experiments revealed that HPV-18 E6 also could not bind to mp53 (Fig. 1B), providing a plausible explanation for the inability of HPV-18 E6 to target mp53 for ubiquitination.
Figure 1.
mp53 is recognized by HPV-16 E6 but not by HPV-18 E6. (A) hp53 and mp53 were generated in rabbit reticulocyte lysate in the presence of 35S-labeled methionine. The radiolabeled proteins then were incubated in the presence or absence of increasing amounts of HPV-16 E6 or HP-18 E6 under standard ubiquitination conditions (Materials and Methods). After 2 h at 25°C, reaction mixtures were subjected to SDS/PAGE and fluorography. The running position of highly ubiquitinated p53 forms is indicated with an asterisk, and that of the respective nonmodified p53 forms with an arrowhead. (B) HPV-16 E6 and HPV-18 E6 were generated in rabbit reticulocyte lysate in the presence of 35S-labeled cysteine and incubated in the absence (−) or presence of in vitro-translated unlabeled hp53 or mp53 as indicated. After 3 h at 4°C, HPV E6 proteins bound to hp53 or mp53 were detected by coimmunoprecipitation by using the anti-p53 monoclonal antibody PAb 421 and then subjected to SDS/PAGE and fluorography. “input” represents 10% of the amount of E6 proteins used in the coimmunoprecipitation analysis.
To identify the region(s) responsible for the observed difference of hp53 and mp53 with respect to HPV-18 E6-mediated ubiquitination, a series of chimeric proteins consisting of different parts of hp53 and the respective regions of mp53 was generated (Fig. 2A). p53-dependent transactivation assays revealed that the transactivation activity of all of the chimeric proteins generated was similar to that of hp53 and mp53, respectively, indicating that the chimeric proteins assume a wild-type (wt)-like conformation (data not shown). The various chimeric proteins were then tested for their ability to serve as substrates for HPV-16- and HPV-18-dependent ubiquitination in vitro. As summarized in Fig. 2A, a chimeric protein containing the N-terminal 240 amino acids of hp53 behaved like mp53 in that it is recognized by HPV-16 E6 but not by HPV-18 E6, whereas the reciprocal chimera behaved like hp53. Conversely, a chimeric protein consisting of the N-terminal 272 amino acids of hp53 was recognized by both HPV-16 E6 and HPV-18 E6, whereas the reciprocal chimera was recognized only by HPV-16 E6 (data not shown). These results indicated that a major difference between hp53 and mp53 with respect to their different interaction with HPV E6 proteins is determined in the region encompassing amino acids 240–272 of hp53.
The amino acid sequence of hp53 and mp53 differs only at one position in the region 240–272 (numbering according to the hp53 sequence), namely at position 268 (N in hp53, D in mp53). Indeed, replacement of N by D resulted in an hp53 form (h268D) that, like mp53, was not ubiquitinated by HPV-18 E6 but was by HPV-16 E6 (Fig. 2B). The reciprocal mp53 mutant (m268N), however, was recognized neither by HPV-18 E6 nor by HPV-16 E6. Finally, additional replacement of the N-terminal 125 amino acids of m268N by the respective human region resulted in a p53 form (hm268N) that was recognized by HPV-18 E6, whereas the ability to interact with HPV-16 E6 was not restored. Together, these results demonstrate that the identity of the residue at position 268 plays a critical role in the E6/p53 interaction. Additional regions, however, are required for this interaction to occur.
p53 Mutants That Are Not Recognized by Either HPV-18 E6 or HPV-16 E6 Are Targeted by Mdm2.
The unexpected difference in the requirements for p53 to be recognized by HPV-16 E6 and HPV-18 E6 prompted us to test the most informative p53 forms (i.e., h268D, m268N, and hm268N) for their ability to serve as substrates in an in vitro Mdm2-dependent ubiquitination assay. As shown in Fig. 2C, h268D, m268N, and hm268N were ubiquitinated in the presence of bacterially expressed Mdm2 with an efficiency similar to that of wt hp53. In contrast, a deletion mutant of p53 devoid of the Mdm2 binding site (ΔN43; refs. 30 and 31) was not, or only poorly, ubiquitinated by Mdm2, demonstrating the specificity of this assay.
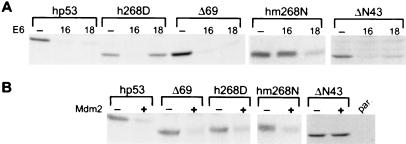
To obtain evidence that the results obtained from in vitro assays also reflect the situation within a cell, transient transfection assays were performed. H1299 cells, which do not express endogenous p53, were transfected with the respective p53 expression plasmids in the absence or presence of expression plasmids encoding HPV-16 E6 or HPV-18 E6 (Fig. 3A) or Mdm2 (Fig. 3B). After 20 h, whole-cell extracts were prepared and p53 levels determined by immunoblot analysis. The results obtained correlated well with the in vitro results in that h268D was degraded in the presence of HPV-16 E6 but not by HPV-18 E6, whereas the converse situation was observed for hm268N. m268N was not degraded by either E6 protein (data not shown), but all three forms of p53 were degraded in the presence of Mdm2. It should be noted that in these experiments, the p53 mutants h268D and hm268N were expressed both in the respective full-length form (data not shown) and in the background of a deletion mutant of hp53 in which amino acids 62–96 were deleted (Δ69; Fig. 3 A and B). The reason for this is that Δ69, or similar deletion mutants of p53, have lost the growth-suppressive properties of full-length p53 and thus can be expressed stably in cells and that the stability regulation of ectopically expressed Δ69 is similar to that of endogenous full-length hp53 (22, 32).
Figure 3.
The structural requirements of p53 to serve as a substrate for degradation within a cell differ between E6 proteins derived from HPV-16 and HPV-18. H1299 cells, which are null for p53 expression, were transfected transiently with a wt p53 (hp53) expression vector or with vectors encoding the indicated mutants in the absence or presence of HPV E6 expression constructs (A) or Mdm2 (B) as indicated. Whole-cell extracts were prepared 20 h after transfection and the levels of p53 determined in a Western blot analysis by using the monoclonal anti-p53 antibodies DO1 (hp53, h268D, Δ69, and hm268N) or 1801 (ΔN43). par., extract prepared from nontransfected H1299 cells. Note that h268D and hm268N were expressed in the Δ69 form.
Degradation of p53 in HPV-Positive Cells Depends Entirely on E6.
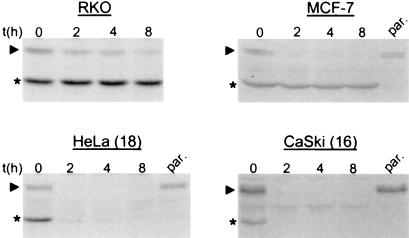
Together with the ΔN43 mutant, which is recognized as a ubiquitination–degradation substrate by HPV E6 proteins (Fig. 3A; ref. 22) but not by Mdm2 (Figs. 2C and 3B), the p53 forms h268D and hm268N enabled us to determine the individual contributions of the E6- and Mdm2-dependent pathways to p53 degradation in HPV-positive cells. If the Mdm2-dependent pathway for p53 degradation were intact in HPV-positive cells, the prediction would be that both h268D and hm268N are degraded in all cell lines independent of their HPV status. If degradation of p53 in HPV-positive cells would depend entirely on E6, however, one would expect that h268D is degraded in HPV-16-positive cells (as well as in HPV-negative cells), and it should have a significantly increased half-life in HPV-18-positive cells. Conversely, hm268N should be degraded in HPV-18-positive cells (as well as in HPV-negative cells), but it would not be recognized efficiently as a proteolytic substrate in HPV-16-positive cells. To test these possibilities, HPV-negative (RKO and MCF-7) and HPV-positive (HeLa, SW756, CaSki, and SiHa) cell lines stably expressing the various p53 mutants (h268D and hm268N as Δ69 forms) were established.
Determination of the half-life of stably expressed ΔN43 revealed that it is a long-lived protein in HPV-negative cells (RKO and MCF-7; Fig. 4) as expected, because this deletion mutant is not recognized by Mdm2 (Figs. 2C and 3B). In HPV-positive cells (HeLa, SW756, CaSki, and SiHa), however, ΔN43 is degraded with a rate similar to the endogenous wt p53 protein (Fig. 4 and data not shown). This result clearly demonstrates that the E6/E6-AP-dependent pathway for p53 degradation is still functional in cell lines derived from HPV-positive cancer cell lines.
Figure 4.
The E6-mediated pathway of p53 degradation is active in HPV-positive cancer cells. The half-life of ΔN43 and of endogenous wt p53 was determined in the indicated cell lines stably expressing ΔN43 after addition of cycloheximide in a Western blot analysis by using the monoclonal anti-p53 antibody 1801. The running position of wt p53 is indicated with an arrowhead, and that of ΔN43 with an asterisk. The HPV status of the various cell lines is: RKO and MCF-7, HPV-negative; HeLa, HPV-18 positive; CaSki, HPV-16 positive. par., extract prepared from the respective parental cell lines. It should be noted that similar results were obtained in pulse–chase analyses by using radioactively labeled cell extracts (see Material and Methods).
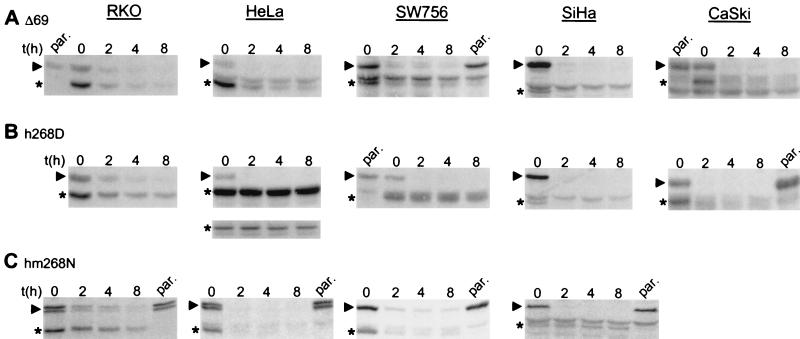
As shown in Fig. 5, determination of the half-life of stably expressed h268D and hm268N revealed that the turnover rate of these mutants is very similar to the turnover rate of the endogenous wt p53 in HPV-negative cell lines (t1/2 of ≈1–2 h in RKO cells and t1/2 of ≈30–60 min in MCF-7 cells) (Fig. 5 B and C and data not shown). In contrast, whereas h268D was turned over rapidly in HPV-16-positive cells (SiHa, CaSki; t1/2 of ≈10–15 min), its turnover rate was extended significantly in HPV-18-positive cells (HeLa, SW756) with a half-life of more than 8 h (Fig. 5B). For hm268N, the inverse was observed. It was rapidly degraded in HPV-18-positive cells, but it was not, or only very slowly, turned over in HPV-16-positive cells (Fig. 5C). Finally, the mutant m268N that was not recognized by either E6 protein in vitro and in transient-transfection assays was long lived in HPV-16 as well as in HPV-18 positive cell lines, whereas it was recognized as a proteolytic substrate in HPV-negative cells (data not shown). Together, these results clearly demonstrate that the E6-dependent pathway for p53 degradation is not only functional in HPV-positive cells but, moreover, p53 degradation is entirely dependent on the presence of E6.
Figure 5.
Degradation of p53 in HPV-positive cancer cells requires the viral E6 oncoprotein. The half-life of the p53 mutants Δ69 (A), h268D (B), hm268N (C), and endogenous wt p53 was determined in the indicated cell lines stably expressing the respective mutants (h268D and hm268N were expressed in the Δ69 form) after addition of cycloheximide in a Western blot analysis by using the monoclonal anti-p53 antibody DO1. The running position of wt p53 is marked with an arrowhead, and the running positions of the respective p53 mutants are marked with an asterisk. The inset in B, below the panel of the half-life analysis in HeLa cells, represents a Western blot analysis with less cell extract of the same experiment to ensure that the proposed increase in half-life of h268D is indeed due to an increased half-life rather than an artifact of the enhanced chemiluminescence system used because of the high expression levels of this particular mutant. Because of the low amounts of the extract loaded, the endogenous wt p53 was not detectable under these conditions. The HPV status of the various cell lines is: RKO, HPV negative; HeLa and SW756, HPV-18 positive; SiHa and CaSki, HPV-16 positive. par., extract prepared from the respective parental cell lines.
Discussion
Certain types of HPVs have been associated etiologically with cervical cancer, and HPV-16 and HPV-18 are the types that are most frequently found in this type of cancer (1). The oncogenic activity of these HPVs is explained partly by the ability of the viral E6 oncoprotein to target p53 for degradation and thus for inactivation. The present study shows that the structural requirements of p53 to serve as a proteolytic substrate differ between the HPV-16 E6 and the HPV-18 E6 oncoprotein. Although hp53 is a good substrate for both HPV-16 E6 and HPV-18 E6, mp53 is recognized only by HPV-16 E6 and not by HPV-18 E6, despite the fact that these E6s are ≈70% similar at the amino acid sequence level. Interestingly, HPV-18 E6 has been reported to have oncogenic properties in mouse cells (4). Thus, the observation that HPV-18 E6 cannot bind to p53 and, consequently, cannot target it for degradation supports the notion that E6 proteins have p53-independent transforming activities (4, 7).
Mutational analysis revealed that the difference between hp53 and mp53 is mainly due to (i) the presence of an asparagine residue at position 268 of hp53 (the corresponding residue in mp53 is an aspartate) and (ii) an undefined region in the N-terminal 125 amino acids of hp53. Interestingly, it was reported previously that alteration of residue 268 of hp53 from N to D results in a p53 protein with a thermodynamically more stable conformation (33). This finding may indicate that the residue at position 268 does not represent a direct contact site for E6, but rather that even subtle changes in the conformation of p53 result in p53 molecules that are not recognized by E6. Furthermore, E6 is bound to p53 in a complex with E6-AP (34). Thus far, however, it is not known whether E6-AP or E6 or both contact p53 directly. We have reported recently that E6-AP can target p53 for ubiquitination in vitro in the absence of E6 (22), indicating that E6-AP can directly interact with p53. Because E6-AP does not distinguish between mp53 and hp53 (unpublished observation), the observed difference between HPV-16 E6 and HPV-18 E6 with respect to their ability to bind to mp53 and hp53 most likely is explained by the notion that the E6 protein also interacts directly with p53.
Several lines of evidence indicated that the E6/E6-AP-dependent pathway of p53 degradation is functional in cervical carcinoma cell lines. Antisense approaches directed against E6-AP expression resulted in an increase in p53 levels in HPV-positive cells but not in HPV-negative cells (20, 23). Similar results were obtained by overexpression of a catalytically inactive E6-AP mutant (21). Conversely, antisense approaches directed against Mdm2 or expression of Mdm2-inactivating peptides resulted in an increase of p53 levels in HPV-negative cell lines but not in HPV-positive cell lines (23, 24). The latter results clearly indicate that in HPV-positive cells, the E6/E6-AP-dependent pathway is more active in p53 degradation than the Mdm2-dependent pathway, but they do not address the question whether Mdm2 is inactive in HPV-positive cells. By the use of a set of mutant p53s that clearly distinguish between Mdm2-mediated and HPV E6-mediated degradation of p53, the present study demonstrates that in HPV-positive cells, degradation of p53 depends entirely on the presence of E6. The molecular reason why the Mdm2-dependent pathway is not active in HPV-positive cancer cells is presently unknown. There are several possibilities to explain this observation. For instance, Mdm2 may not, or only poorly, be expressed. Indeed, Western blot analysis revealed that Mdm2 levels are ≈2- to 4-fold lower in HPV-positive cells than in other wt p53-containing cells (data not shown). Another but not mutually exclusive possibility is that p14ARF, a known negative regulator of Mdm2-mediated p53 degradation, may be overexpressed, possibly because of the presence of viral oncoproteins (35–39). In support of these notions, preliminary results indicate that, at least in some HPV-positive cell lines, p14ARF levels are increased significantly. In addition, in transient-transfection experiments, ectopically expressed Mdm2 facilitates the efficient degradation of cotransfected hp53, h268D, and hm268N in the HPV-positive cell lines used (data not shown).
With respect to cancer therapy, the viral etiology of cervical cancer provides the advantage that therapeutic approaches can be developed to eliminate selectively HPV-positive cells and leave the normal tissue unaffected. The finding that in HPV-positive cancer cell lines degradation, and thus probably inactivation, of p53 entirely depends on the action of E6 supports the notion that the E6/E6-AP-dependent pathway for p53 degradation constitutes an attractive target for the development of novel therapeutic strategies for the treatment of cervical cancer. Indeed, it was reported recently that the expression of peptides that specifically bind to E6 results in p53 accumulation and in apoptosis in HPV-positive cancer cells, but the growth of HPV-negative cells is not affected (40). This observation indicates that the wt p53 present in HPV-positive cancer cells is still functional with respect to its growth-suppressive properties.
Acknowledgments
We thank S. Jentsch (MPI for Biochemistry, Munich, Germany) for comments on the manuscript. This work was supported by the Zentrum für Molekulare Medizin Köln, the Cooperation Program in Cancer Research of the Deutsches Krebsforschungszentrum and the Israeli Ministry of Science, and the German–Israeli Foundation for Scientific Research and Development.
Abbreviations
- HPV
human papillomavirus
- wt
wild-type
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.031470698.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.031470698
References
- 1.zur Hausen H. J Natl Cancer Inst. 2000;92:690–698. doi: 10.1093/jnci/92.9.690. [DOI] [PubMed] [Google Scholar]
- 2.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 3.Scheffner M, Huibregtse J M, Vierstra R D, Howley P M. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 4.Pim D, Storey A, Thomas M, Massimi P, Banks L. Oncogene. 1994;9:1869–1876. [PubMed] [Google Scholar]
- 5.Pan H, Griep A E. Genes Dev. 1994;8:1285–1299. doi: 10.1101/gad.8.11.1285. [DOI] [PubMed] [Google Scholar]
- 6.Pan H, Griep A E. Genes Dev. 1995;9:2157–2169. doi: 10.1101/gad.9.17.2157. [DOI] [PubMed] [Google Scholar]
- 7.Liu Y, Chen J J, Gao Q, Dalal S, Hong Y, Mansur C P, Band V, Androphy E J. J Virol. 1999;73:7297–7307. doi: 10.1128/jvi.73.9.7297-7307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas M, Pim D, Banks L. Oncogene. 1999;18:7690–7700. doi: 10.1038/sj.onc.1202953. [DOI] [PubMed] [Google Scholar]
- 9.Chen J J, Reid C E, Band V, Androphy E J. Science. 1995;269:529–531. doi: 10.1126/science.7624774. [DOI] [PubMed] [Google Scholar]
- 10.Lee S S, Weiss R S, Javier R T. Proc Natl Acad Sci USA. 1997;94:6670–6675. doi: 10.1073/pnas.94.13.6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ronco L V, Karpova A Y, Vidal M, Howley P M. Genes Dev. 1998;12:2061–2072. doi: 10.1101/gad.12.13.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomas M, Banks L. Oncogene. 1998;17:2943–2954. doi: 10.1038/sj.onc.1202223. [DOI] [PubMed] [Google Scholar]
- 13.Gao Q, Srinivasan S, Boyer S N, Wazer D E, Band V. Mol Cell Biol. 1999;19:733–744. doi: 10.1128/mcb.19.1.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maki C G, Huibregtse J M, Howley P M. Cancer Res. 1996;56:2649–2654. [PubMed] [Google Scholar]
- 15.Oren M. J Biol Chem. 1999;274:36031–36034. doi: 10.1074/jbc.274.51.36031. [DOI] [PubMed] [Google Scholar]
- 16.Ashcroft M, Vousden K H. Oncogene. 1999;18:7637–7643. doi: 10.1038/sj.onc.1203012. [DOI] [PubMed] [Google Scholar]
- 17.Haupt Y, Maya R, Kazaz A, Oren M. Nature (London) 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 18.Kubbutat M H, Jones S N, Vousden K H. Nature (London) 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 19.Bottger A, Bottger V, Sparks A, Liu W L, Howard S F, Lane D P. Curr Biol. 1997;7:860–869. doi: 10.1016/s0960-9822(06)00374-5. [DOI] [PubMed] [Google Scholar]
- 20.Beer-Romero P, Glass S, Rolfe M. Oncogene. 1997;14:595–602. doi: 10.1038/sj.onc.1200872. [DOI] [PubMed] [Google Scholar]
- 21.Talis A L, Huibregtse J M, Howley P M. J Biol Chem. 1998;273:6439–6445. doi: 10.1074/jbc.273.11.6439. [DOI] [PubMed] [Google Scholar]
- 22.Hengstermann A, Whitaker N J, Zimmer D, Zentgraf H, Scheffner M. Oncogene. 1998;17:2933–2941. doi: 10.1038/sj.onc.1202282. [DOI] [PubMed] [Google Scholar]
- 23.Traidej M, Chen L, Yu D, Agrawal S, Chen J. Antisense Nucleic Acid Drug Dev. 2000;10:17–27. doi: 10.1089/oli.1.2000.10.17. [DOI] [PubMed] [Google Scholar]
- 24.Hietanen S, Lain S, Krausz E, Blattner C, Lane D P. Proc Natl Acad Sci USA. 2000;97:8501–8506. doi: 10.1073/pnas.97.15.8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kessis T D, Slebos R J, Nelson W G, Kastan M B, Plunkett B S, Han S M, Lorincz A T, Hedrick L, Cho K R. Proc Natl Acad Sci USA. 1993;90:3988–3992. doi: 10.1073/pnas.90.9.3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Butz K, Shahabeddin L, Geisen C, Spitkovsky D, Ullmann A, Hoppe-Seyler F. Oncogene. 1995;10:927–936. [PubMed] [Google Scholar]
- 27.Butz K, Geisen C, Ullmann A, Spitkovsky D, Hoppe-Seyler F. Int J Cancer. 1996;68:506–513. doi: 10.1002/(SICI)1097-0215(19961115)68:4<506::AID-IJC17>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 28.Haupt Y, Barak Y, Oren M. EMBO J. 1996;15:1596–1606. [PMC free article] [PubMed] [Google Scholar]
- 29.Scheffner M, Huibregtse J M, Howley P M. Proc Natl Acad Sci USA. 1994;91:8797–8801. doi: 10.1073/pnas.91.19.8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin J, Chen J, Elenbaas B, Levine A J. Genes Dev. 1994;8:1235–1246. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- 31.Bottger A, Bottger V, Garcia-Echeverria C, Chene P, Hochkeppel H K, Sampson W, Ang K, Howard S F, Picksley S M, Lane D P. J Mol Biol. 1997;269:744–756. doi: 10.1006/jmbi.1997.1078. [DOI] [PubMed] [Google Scholar]
- 32.Walker K K, Levine A J. Proc Natl Acad Sci USA. 1996;93:15335–15340. doi: 10.1073/pnas.93.26.15335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nikolova P V, Henckel J, Lane D P, Fersht A R. Proc Natl Acad Sci USA. 199;95:14675–14680. doi: 10.1073/pnas.95.25.14675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huibregtse J M, Scheffner M, Howley P M. EMBO J. 1991;10:4129–4135. doi: 10.1002/j.1460-2075.1991.tb04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pomerantz J, Schreiber-Agus N, Liegeois N J, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee H W, et al. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y, Xiong Y, Yarbrough W G. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 37.Honda R, Yasuda H. EMBO J. 1999;18:22–27. doi: 10.1093/emboj/18.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Stanchina E, McCurrach M E, Zindy F, Shieh S Y, Ferbeyre G, Samuelson A V, Prives C, Roussel M F, Sherr C J, Lowe S W. Genes Dev. 1998;12:2434–2442. doi: 10.1101/gad.12.15.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bates S, Phillips A C, Clark P A, Stott F, Peters G, Ludwig R L, Vousden K H. Nature (London) 1998;395:124–125. doi: 10.1038/25867. [DOI] [PubMed] [Google Scholar]
- 40.Butz K, Denk C, Ullmann A, Scheffner M, Hoppe-Seyler F. Proc Natl Acad Sci USA. 2000;97:6693–6697. doi: 10.1073/pnas.110538897. . (First Published May 30, 2000; 10.1073/pnas.110538897) [DOI] [PMC free article] [PubMed] [Google Scholar]