Abstract
Cells are organized into a complex network of subcellular compartments that are specialized for various biological functions. Subcellular location is an important attribute of protein function. To facilitate systematic elucidation of protein subcellular location, we analyzed experimentally verified protein localization data of 1,300 Arabidopsis (Arabidopsis thaliana) proteins. The 1,300 experimentally verified proteins are distributed among 40 different compartments, with most of the proteins localized to four compartments: mitochondria (36%), nucleus (28%), plastid (17%), and cytosol (13.3%). About 19% of the proteins are found in multiple compartments, in which a high proportion (36.4%) is localized to both cytosol and nucleus. Characterization of the overrepresented Gene Ontology molecular functions and biological processes suggests that the Golgi apparatus and peroxisome may play more diverse functions but are involved in more specialized processes than other compartments. To support systematic empirical determination of protein subcellular localization using a technology called fluorescent tagging of full-length proteins, we developed a database and Web application to provide preselected green fluorescent protein insertion position and primer sequences for all Arabidopsis proteins to study their subcellular localization and to store experimentally verified protein localization images, videos, and their annotations of proteins generated using the fluorescent tagging of full-length proteins technology. The database can be searched, browsed, and downloaded using a Web browser at http://aztec.stanford.edu/gfp/. The software can also be downloaded from the same Web site for local installation.
Plant cells are organized into a complex network of subcellular compartments that are specialized for a multitude of biological functions. It is not possible to fully understand plant metabolism, physiology, and development without comprehensive knowledge of the expression and localization patterns of proteins within these cellular microenvironments. Likewise, protein localization and function are tightly correlated (Kumar et al., 2002; Huh et al., 2003), and the subcellular location of a protein can provide clues for the molecular functions and biological processes in which a protein may be involved (Liu et al., 2002; Carrari et al., 2003; Hernandez-Munoz et al., 2003; Huq et al., 2003). Unfortunately, our knowledge about subcellular locations of Arabidopsis (Arabidopsis thaliana) proteins is scarce; only about 5% (1,300) of all predicted Arabidopsis proteins have had their subcellular locations determined empirically (The Arabidopsis Information Resource [TAIR], http://arabidopsis.org/tool/bulk/go/index.jsp).
To visualize protein localization and expression within individual cells and tissues in situ or in planta, proteins are usually detected by specific antibodies (Li et al., 2001) or are labeled by genetic fusion to antigentic tags (Dyer and Mullen, 2001), enzyme reporters such as β-glucuronidase (GUS; Kertbundit et al., 1998), or fluorescent proteins (Cutler et al., 2000). Determination of subcellular location by these empirical methods is a time-consuming and costly endeavor. Computational methods could help to make this process more rapid by providing some clues about a protein's potential location (Claros and Vincens, 1996; Cedano et al., 1997; Nakai and Horton, 1999; Emanuelsson et al., 2000; Chou, 2001; Hua and Sun, 2001; Chou and Cai, 2002). To date, there are mainly two computational approaches applied to predict protein subcellular localization. The first class of methods is based on the recognition of known protein sorting signals. TargetP (Emanuelsson et al., 2000), PSORT (Nakai and Horton, 1999), and MitoProt (Claros, 1995) are examples of tools for predicting subcellular localization that use N-terminal sorting sequence information. Methods based on sorting signals are strongly dependent on the quality of the N-terminal sequence assignment. Unfortunately, the number of the empirically determined targeting sequences is frequently not large enough for training and testing these algorithms. In addition, some proteins have no sorting signals, and localization and targeting are dependent on binding to another protein that has the protein-sorting signal (Magae et al., 1996; Lindeman et al., 1997). The second class of methods for predicting subcellular localization uses sequence information such as amino acid composition (Cedano et al., 1997; Hua and Sun, 2001), a combination of conventional amino acid composition and physical-chemical parameters of amino acids such as hydrophobicity value, hydrophilicity value, side chain mass (Chou, 2001), and functional domain composition (Chou and Cai, 2002). This approach does not rely on analysis of sorting signals and thus is complementary to those methods that do. All of these computational methods are limited in that they are developed with the assumption that proteins exist in only one location (Chou and Cai, 2002). In addition, the accuracy of some of these methods is low for predicting proteins localized to the chloroplast (Richly and Leister, 2004). In summary, computer-based prediction programs are not yet sufficiently reliable, and empirical localization methods are labor intensive and time consuming.
To alleviate these problems associated with empirical or computational methods when used alone, an approach that combines computational predictions with a high-throughput experimental validation is needed. Until recently, the subcellular localization of most proteins in Arabidopsis was determined on a case-by-case basis as individual proteins or small groups of proteins were studied. With the advent of efficient methods to tag large numbers of proteins with in vivo markers, a much larger collection of localization data is being accumulated. Cutler et al. (2000) described a general approach to screen for subcellular localization information in Arabidopsis. Libraries of Arabidopsis cDNAs were cloned, at the 3′ end of the coding region, with the green fluorescent protein (GFP) coding sequence and placed under control of the viral 35S promoter. These libraries were transformed stably into plants, and the resulting first generation of transgenic plants was screened for GFP expression and subcellular localization pattern. Escobar et al. (2003) designed a related strategy for high-throughput localization of proteins in Nicotiana benthamiana. A third method, developed by Koroleva et al. (2005), fused selected full-length cDNAs with the GFP coding sequence at the 3′ end of the coding region of the cDNA under the control of the viral 35S promoter using Gateway technology (Invitrogen). While these methods are fairly high throughput, they have a few limitations (Tian et al., 2004). For example, with N-terminal fusions, endoplasmic reticulum (ER) signal peptides, mitochondria, or chloroplast signal peptides may be masked, or they could become stop transfer sequences, thereby generating localization artifacts. In addition, overexpression of GFP fusion protein controlled by 35S viral promoter may disrupt protein complexes or can mask subtle localization patterns due to an overabundance of tagged protein. Finally, ambiguities in the interpretation of localization pattern can result in screens that rely on transient expression of tagged proteins. To address these limitations in the above methods, Tian et al. (2004) developed a high-throughput subcellular localization technology called fluorescent tagging of full-length proteins (FTFLP) to analyze expression pattern and subcellular localization in whole plant seedlings. FTFLP differs from other high-throughput methods applied in Arabidopsis in that proteins are tagged at a selected internal site near the C terminus, and the potential native gene regulatory sequences including 5′ and 3′ genomic and intron sequences are used to drive gene expression. This method generates GFP-tagged genes efficiently using triple-template PCR. Triple-template PCR introduces the GFP tag into the selected site within the target gene without the need for conventional cloning and creates an internally tagged full-length gene ready for Gateway recombination cloning (Invitrogen).
To determine the predictive power of protein localization as an indicator of its function, we analyzed the functional annotations of the collection of Arabidopsis proteins that have been experimentally localized to different subcellular locations. In addition, to support systematic empirical determination of protein subcellular localization using the FTFLP technology, we developed a Web-accessible database, FTFLPdb, which stores and manages GFP insertion positions and primer sequences for all Arabidopsis proteins to facilitate any interested researcher to produce GFP-tagged fusion protein using the FTFLP or related technology. In addition, the database stores subcellular localization images, videos, and their annotations of proteins generated using the FTFLP technology. Here, we describe the results of the characterization of proteins with empirically determined localization data and functionalities of the FTFLPdb.
RESULTS
Functional Characterization of Major Compartments of the Cell
Distribution of Protein Localization within the Cell
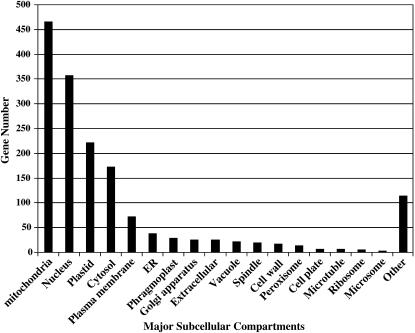
Since the completion of sequencing of the Arabidopsis genome (Arabidopsis Genome Initiative, 2000), postgenomic analyses of this reference plant have progressed rapidly. The current genome annotation (TAIR 6.0) includes 26,751 protein-coding genes, 838 noncoding genes, and 3,818 pseudogenes (ftp://ftp.arabidopsis.org/home/tair/Genes/TAIR6_genome_release). Approximately 48% of the genes have been assigned putative molecular functions, 30% have no predicted molecular functions, and 22% have not yet been annotated. For the majority of these genes that are assigned putative molecular functions, function was predicted from sequence similarity to other genes (Wortman et al., 2003). Only 3.5% (917) of all predicted Arabidopsis proteins have had their molecular functions elucidated empirically. Similarly, only 5% (1,300) of all predicted Arabidopsis proteins have had their subcellular locations determined empirically. These proteins with empirically determined locations are distributed among 40 different subcellular compartments: mitochondria (36%), nucleus (28%), plastid (17%), cytosol (13.3%), plasma membrane (5.5%), ER (3%), phragmoplast (2.2%), Golgi apparatus (2.0%), extracellular (2.0%), vacuole (1.7%), spindle (1.5%), cell wall (1.3%), peroxisome (1.1%), cell plate (0.5%), microtubule (0.5%), ribosome (0.46%), microsome (0.3%), and other (8.8%) that includes 23 different locations (Fig. 1). The complete list of genes with experimentally verified subcellular locations can be found in the Supplemental Table I. Although these results are based on only a portion of the proteome, the general distribution is similar to the results obtained from analyzing localization of the whole yeast (Saccharomyces cerevisiae) proteome by Huh et al. (2003), with the exception of the proportions of proteins observed in the cytosol, mitochondria, and plastid. Forty percent of yeast proteins are found in the cytosol, compared to 13.3% of experimentally localized Arabidopsis proteins. On the other hand, only 10% of the yeast proteins localize to mitochondria, whereas 36% of Arabidopsis proteins were found in this organelle. Also, many Arabidopsis proteins (17%) are found in the plastid, an organelle that yeasts do not have.
Figure 1.
Distribution of empirically derived Arabidopsis protein subcellular localization. Arabidopsis genes whose protein subcellular locations have been experimentally verified were retrieved from TAIR (Berardini et al., 2004). Genes have annotations to GO cellular component and evidence codes IDA, IMP, IEP, IGI, or IPI (Berardini et al., 2004). The Other category includes 15 protein complexes such as PSII antenna complex, PSII associated light-harvesting complex II, and Skp1/Cul1/F-box ubiquitin ligase complex, and eight subcellular locations such as cytoskeleton, lipid storage body (sensu Viridiplantae), and tubulin. Full list of the compartments can be found in Supplemental Table I.
When examining the entire set of proteins with experimentally determined localizations, we found that 19% (250/1,300) are observed in multiple locations. The majority of these proteins (79%) are found in two distinct subcellular locations while the rest (21%) are found associated with three or more locations. The most commonly occurring paired location patterns are nucleus and cytosol (91/197) and plastid and mitochondria (18/197). The most commonly occurring three-location pattern is cytosol, ER, and nucleus (9/37).
Because a high proportion of proteins localize to cytosol and to nucleus, which has nuclear pores through its envelope, we asked whether proteins in this localization class are smaller in size compared with proteins with a single subcellular location. To test this hypothesis, we compared the protein mass between the proteins in those two classes using a two-sample t test for comparing means of two populations (Peck et al., 2001). Proteins localized to both cytosol and nucleus have an average mass (37,405 D) that is 12.4% smaller than proteins with single location (P value = 0.0082).
Functional Characterization of Subcellular Compartments Based on the Localized Proteins
The Gene Ontology (GO) controlled vocabularies are a standard set of terms used for functional annotation of genes (Berriman and Harris, 2004). The terms or descriptors are organized into three categories: molecular function, biological process, and cellular component (http://www.geneontology.org). The molecular function term describes the biochemical activity performed by a gene product such as transcription factor activity. The biological process term describes the ordered assembly of more than one molecular function such as regulation of transcription. The cellular component term describes the macromolecular subcellular constituents of the cell such as the nucleus or the plasma membrane. With each category, terms are organized into a hierarchy where general terms are parents of specific terms (e.g. chloroplast stroma is a child of [a part of] chloroplast). These terms have been used to assign biochemical function, subcellular localization, and involvement in a process for most of the Arabidopsis proteins (Berardini et al., 2004). In addition to the use of controlled, hierarchical terms, these annotations include evidence types that indicate the type of experimental or computational evidence that was used to make the annotations (Berardini et al., 2004). These attributes of structured annotations allow quantitative comparison of protein characteristics.
We asked two related questions utilizing GO annotations and the computational tool GeneMerge (version 1.2; Castillo-Davis and Hartl, 2003; as described in “Materials and Methods”): (1) To what extent can we infer molecular function or biological process of a protein from its subcellular location? and (2) What properties do the major cellular compartments have and share with other compartments? GeneMerge method compares the GO annotations of genes in a subset of genes under investigation (e.g. genes that are induced in expression under a certain condition) with the total set of genes under investigation and returns statistically based rank scores of overrepresented terms in the studied set. We determined overrepresented GO functions and processes of genes localized to one compartment versus all of the genes in our data set. Results are summarized in Table I and detailed results are found in the Supplemental Table II. Most of overrepresented terms on molecular functions and biological processes in each major subcellular compartment were found as expected, but some obvious terms in some subcellular compartments were missed. For example, the terms for antioxidant enzyme activity such as catalase, and antioxidant process such as ascorbate-glutathione cycle were not found in the peroxisome (del Rio et al., 2002). Another example is the term motor activity that was also not found in the microtubule. These missed terms may result from the small data set. For example, there are only 14 and seven Arabidopsis proteins found in peroxisome and microtubule experimentally, respectively (Table I).
Table I.
Summary of functional analysis of proteins classified by empirically determined subcellular locations
| Subcellular Locations | No. Genes | Percent Genes with Overrepresented Functions | Overrepresented Function Terms | Percent Genes with Overrepresented Processes | Overrepresented Process Terms |
|---|---|---|---|---|---|
| Ribosome | 6 | 67 | Structural constituent of ribosome | 67 | Protein biosynthesis |
| Extracellular | 26 | 50 | Lipase activity; acyltransferase activity; carboxylic ester hydrolase activity; lipid binding; nutrient reservoir activity | 40 | Sexual reproduction; lipid storage; regulation of meristem organization; cell differentiation; |
| Plasma membrane | 72 | 44 | Protein phosphorylated amino acid binding; l-Pro transporter activity; water channel activity; protein binding; protein Ser/Thr kinase activity; amino acid permease activity; Tyr amino peptidase activity; GTPase activity | 35 | Water transport; pollen tube growth; auxin polar transport; l-Pro transport; root development; abscisic acid mediated signaling; regulation of cell proliferation; transport |
| Microtubule | 7 | 43 | Microtubule binding | 29 | Microtubule cytoskeleton organization and biogenesis |
| Cell wall | 17 | 41 | Hydrolase activity, acting on glycosyl bonds; actin binding | 18 | Unidimensional cell growth |
| Nucleus | 358 | 40 | Transcription factor activity; DNA binding; RNA binding; protein phosphorylated amino acid binding; protein binding; transcriptional activator activity; transcription regulator activity | 31 | Nuclear mRNA splicing, via spliceosome; photomorphogenesis; ubiquitin-dependent protein catabolism; protein deneddylation; red or far-red light signaling pathway; regulation of transcription, DNA dependent; positive regulation of transcription; flower development; response to cytokinin stimulus; red, far-red light phototransduction; response to blue light; negative regulation of photomorphogenesis; negative regulation of transcription; response to auxin stimulus; regulation of meristem organization; response to cold |
| Cell plate | 8 | 38 | SNAP receptor activity; t-SNARE activity | 38 | Cytokinesis by cell plate formation; membrane fusion |
| Vacuole | 22 | 36 | Calcium:hydrogen antiporter activity; calcium:cation antiporter activity; cation:cation antiporter activity; methylammonium transporter activity; water channel activity | 41 | Vacuole organization and biogenesis; calcium ion transport; protein secretion; flavonoid biosynthesis |
| ER | 39 | 31 | ATPase activity; NADPH-hemoprotein reductase activity; oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD or NADH as one donor, and incorporation of one atom of oxygen; farnesyltranstransferase activity | 46 | Ubiquitin-dependent protein catabolism; intracellular transport; N-terminal protein myristoylation; phenylpropanoid metabolism; negative regulation of ethylene mediated signaling pathway; isoprenoid biosynthesis; glucosinolate biosynthesis |
| Plastid | 222 | 28 | Endopeptidase Clp activity; ATP-dependent peptidase activity; metallopeptidase activity; peptidyl-prolyl cis-trans isomerase activity; ATPase activity; FK506 binding; UDP-galactosyltransferase activity; σ factor activity; structural molecule activity; phosphoribosylanthranilate isomerase activity; 1,2-diacylglycerol 3-β-galactosyltransferase activity; protein phosphorylated amino acid binding; dimethylallyltranstransferase activity; Trp synthase activity; Ser-type endopeptidase activity | 26 | ATP-dependent proteolysis; chloroplast organization and biogenesis; galactolipid biosynthesis; protein-chloroplast targeting; Trp biosynthesis; cellular response to phosphate starvation; photosynthetic water oxidation; chloroplast-nucleus signaling pathway; glycolipid biosynthesis; chloroplast thylakoid membrane protein import; transcription initiation; chlorophyll biosynthesis; removal of superoxide radicals; iron-sulfur cluster assembly |
| Spindle | 20 | 25 | Microtubule binding; GTP binding; GTPase activity | 10 | Microtubule cytoskeleton organization and biogenesis |
| Golgi apparatus | 26 | 24 | Hydrogen-translocating pyrophosphatase activity; receptor activity; t-SNARE activity | 69 | Protein-vacuolar targeting; intracellular protein transport; golgi to vacuole transport; retrograde transport, golgi to ER; nucleotide-sugar metabolism; cell wall biosynthesis (sensu Magnoliophyta) |
| Mitochondria | 466 | 24 | Protein translocase activity; succinate dehydrogenase activity; NADH dehydrogenase (ubiquinone) activity; hydrogen-transporting ATP synthase activity, rotational mechanism; isocitrate dehydrogenase (NAD+) activity; translation elongation factor activity; voltage-gated ion-selective channel activity; ATP binding; Gly dehydrogenase (decarboxylating) activity; alternative oxidase activity; ATP-dependent peptidase activity; hexokinase activity; ubiquinol-cytochrome-c reductase activity; aconitate hydratase activity; catalase activity; succinate-CoA ligase (GDP-forming) activity; ATPase activity; malate dehydrogenase activity; cytochrome-c oxidase activity | 19 | Protein-mitochondrial targeting; mitochondrial electron transport, succinate to ubiquinone; electron transport; metabolism; ATP-dependent proteolysis; anion transport; Gly catabolism; Leu catabolism; purine nucleotide transport; mitochondrial electron transport, NADH to ubiquinone; Gly decarboxylation via Gly cleavage system |
| Cytosol | 173 | 21 | GTP binding; kinase activity; protein phosphatase type 2A activity; Ser O-acetyltransferase activity; GTPase activity | 10 | Ubiquitin-dependent protein catabolism |
| Phragmoplast | 29 | 21 | Actin binding; microtubule binding | 7 | Cytoskeleton organization and biogenesis |
| Peroxisome | 14 | 14 | Fatty-acyl-CoA synthase activity; 4-coumarate-CoA ligase activity | 71 | Peroxisome organization and biogenesis; photorespiration; fatty acid β-oxidation; jasmonic acid biosynthesis |
| Microsome | 5 | 0 | 0 |
The results in Table I also show that both overrepresented molecular functions and biological processes in each major subcellular compartment tend to be specific to that compartment, although the proportion of proteins that are annotated to the overrepresented functions and processes is variable among the compartments. The proportion of the ribosome-localized proteins that contributes to overrepresented functions (structural constituent of the ribosome, GO:0003735) and overrepresented processes (protein biosynthesis, GO:0004612) is 67% (4/6). While it is difficult to make a clear conclusion from the small numbers of experimentally determined ribosome-localized proteins, this suggests that the ribosome may be more specialized in function and involved in self-contained processes than other subcellular locations, and is consistent with the ribosome being a subcellular component composed of a single macromolecular complex.
Only about 10% to 28% of proteins localized to plastid, mitochondria, and cytosol contribute to the overrepresented molecular functions and overrepresented biological processes, respectively. For example, about 21% and 10% of cytosol-localized proteins contribute to the overrepresented molecular functions and biological processes, respectively. These results suggest that these subcellular locations may play more diverse functions in more diverse processes than other locations such as the ribosome and nucleus.
Unlike the subcellular locations described above, about 24% of Golgi-localized proteins (6/26) contribute to overrepresented molecular functions such as hydrogen-translocating pyrophosphatase activity and receptor activity. However, a much higher percentage, 69% (18/26), contributes to overrepresented biological processes. The overrepresented biological processes are involved in protein transport except one process that is involved in cell wall biosynthesis. Similarly, about 14% of peroxisome-localized proteins (2/14) contribute to overrepresented molecular functions such as fatty-acyl-CoA synthase activity and 4-coumarate-CoA ligase activity, while a much higher percentage, 71% (10/14), contributes to overrepresented biological processes such as peroxisome organization and biogenesis and oxidations. These results suggest that Golgi apparatus and peroxisome may be subcellular compartments that play more diverse functions but in more specialized or self-contained processes than other compartments. Because the dataset we analyzed is only a small subset of the proteome, we asked whether this pattern would be observed in an organism where the majority of the proteins are localized. In yeast, where 59% of the proteins have empirical localization information, about 22% of Golgi-localized proteins and 33% of peroxisome-localized proteins contribute to overrepresented molecular functions, but 70% of Golgi-localized proteins and 81% of peroxisome-localized proteins contribute to overrepresented biological processes, similar to the findings in Arabidopsis. Together, these results suggest that Golgi apparatus and peroxisome are more specialized in their biological roles than other compartments of the cell and that proteins localized to these compartments may be inferred with a reasonable confidence to play a part in these roles.
To determine whether the overrepresented functions and processes of proteins in a certain subcellular location are specific to that location or are shared by different subcellular locations, we compared the overrepresented molecular functions and processes among the different subcellular locations by determining the Jaccard distance (Han and Kamber, 2001; Supplemental Tables III and IV). Jaccard distance is the proportion of characters that do not match in two data sets over a series of parameters, excluding those characters that are absent in both sets. It has a value between 1 and 0, where 1 indicates the two data sets are 100% dissimilar and 0 indicates the two clusters are identical. The majority of Jaccard distances obtained by comparing overrepresented functions and processes between two subcellular locations in Arabidopsis is 1, indicating that most subcellular compartments have overrepresented functions and processes that are not shared with other subcellular compartments. These results suggest that the predominant functions and processes played by major cellular compartments of the cell are mutually exclusive.
Protein Localization Database for Facilitating Fluorescent Tagging of Full-Length Arabidopsis Proteins
Content of the Database
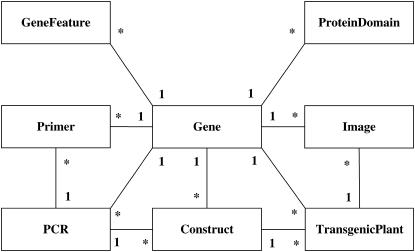
Empirically determined subcellular localization data allows not only the development and improvement of prediction algorithms, but also quantitative analysis of protein function and properties of organelles and other cellular compartments. To facilitate empirical determination of protein subcellular localization in Arabidopsis by the entire plant research community, we developed a database (FTFLPdb) and software to efficiently select genes of interest and download predesigned primers to clone the target genes. In addition, researchers can upload and update construct, transgenic plant, and localization image and video data to share the results with the rest of the community. The nine top-level objects in the database and relationships among them are shown in Figure 2. Table Gene stores attributes of a gene such as genomic sequence, protein sequence, exon/intron positions, gene length, orientation of transcription, full-length cDNA and expressed sequence tag (EST) information, protein Mr, predicted subcellular localization, and taxon grouping of genes such as plant-specific gene or Arabidopsis-specific gene. Table GeneFeature stores the coordinates of gene features such as intron, exon, and coding sequence on a chromosome. Table ProteinDomainFeature stores the mapping positions of functional domains of a protein from TAIR. The primer sequences and relevant primer information such as length, melting temperature (Tm), and GFP insertion position are stored in the table Primer. Tables PCR, Construct, Plant, and Image store the information about PCR products, clones, transgenic plants, and still and video images of the GFP localization in planta, respectively. GO annotations are stored in the Table GOAnnotation. Gene family data are stored in Table TribelGeneFamily.
Figure 2.
Diagram of FTFLPdb relational database schema. The rectangles represent core data object tables and lines represent relationships between the tables. The cardinality between tables is one (denoted as 1) to many (denoted as *) relationship. Noncore data object tables are not shown in the diagram for simplification. Table Gene stores genomic sequence of genes, protein sequence, and annotation of the sequences. Table GeneFeature stores the coordinates of gene features. Table ProteinDomain stores the mapping positions of functional domains to a protein. Table Primer stores the primer sequences and relevant information about the primers. Tables PCR, Construct, Plant, and Image store the information about PCR products, clones, transgenic plants, and still images as well as videos, respectively. Table GOAnnotation stores the GO annotations of Arabidopsis genes. Table TribelGeneFamily stores Gene family relationships.
The current FTFLPdb includes all 26,530 protein-coding genes of Arabidopsis from the latest genome release (TAIR 6; only one splicing form is included). There are 7,142 genes with unknown molecular functions. A total of 19,466 genes are supported by full-length cDNA data and 2,433 genes are supported only with EST data. A total of 23,013 proteins have domains predicted by InterproScan and 7,026 proteins have transmembrane (TM) domains predicted by THMM 2.0. A total of 1,300 proteins have subcellular localization determined by experiments. Summary of the gene data statistics is shown in Supplemental Table V.
Primer Design
In most cases, attaching GFP to the N or C terminus of the protein may abrogate targeting signals. For example, with N-terminal fusions, ER signal peptides may be masked, and they could become stop transfer sequences, generating localization artifacts. N-terminal tagging also obscures chloroplast transit peptides and signal peptides of type I integral membrane proteins. C-terminal fusions can also mask targeting motifs (e.g. farnesylation sites for membrane targeting). Therefore, N- or C-terminal fusions may not only block correct localization, but more importantly, might also misdirect the fusion protein to an artificial location. To mitigate these potential problems, we wrote a Perl script called Primer4FTTLP.pl to identify a suitable GFP insertion position internal to the protein sequence and design primers (four primers named P1, P2, P3, and P4) as described in “Materials and Methods.” The script considers protein domains including targeting sequences, position of each primer within the genomic sequence, annealing temperature, primer length, and primer secondary structure in an iterative fashion. This method has at least one drawback. The identification of GFP insertion site is based on the predicted functional domains, structural domains, and targeting sequences. If signal sequences or functional domains have not been identified in the sequence, the insertion position identified by this method may be inappropriate. For example, no common motif was found in the vacuolar sorting signals identified in the C terminus of vacuolar proteins (Matsuoka and Neuhaus, 1999). To minimize masking a potential targeting sequence, our program avoided 10 amino acid residues from the N and C termini unless there were no other regions in the protein that met the rest of the criteria for primer selection.
We predesigned four primers for the FTFLP protocol: P1, P2, P3, and P4, for each gene. The PCR amplification success rate of these primers was found to be approximately 80% (G. Tian, personal communication). P1 is located at the 5′ end of the gene, which includes up to 3 kb of 5′ flanking genomic sequence from the start codon. About 80% of the genes allow GFP insertion sites (P2 and P3 positions) to be near the C terminus between amino acids 1 and 30 from the C terminus; 15% of the genes have GFP insertion sites near the N terminus between amino acids 1 and 30 from the N terminus. The remaining 5% of the genes did not have suitable insertion sites near the termini. P4 is located at the 3′ end of the gene, which includes up to 1 kb of 3′ flanking genomic sequence from the stop codon. Of the 25,099 genes with predicted PCR primers, 24,910 genes are less than 8 kb in length (including the GFP sequence), which is a length that can be amplified by PCR reliably (Tian et al., 2004).
Software Functionalities
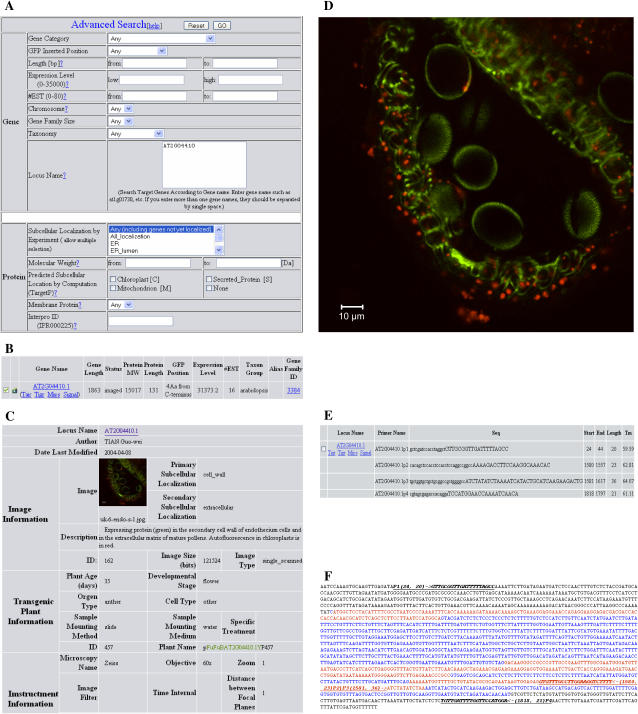
The database is available via the World Wide Web at http://aztec.stanford.edu/gfp. The Web user interface (Fig. 3) was implemented using Perl and Common Gateway Interface (Christiansen and Torkington, 1998; Birznieks et al., 2000). Software functionalities include Search, Submit, Edit, Get Primers, Download Clone Submission Data, and Download Seed Submission Data. The Search function allows users to retrieve genes of interest by using parameters associated to the gene and the protein by entering a list of Arabidopsis Genome Initiative locus names. The gene-specific parameters include locus name, GFP insertion position, expression level, EST number, chromosome number, and gene family size. Protein-specific parameters include Mr, experimentally verified subcellular localization (if any), TargetP-predicted subcellular locations, predicted TM domains, and taxon group information (Fig. 3A). Taxon group is a classification of Arabidopsis proteins based on homologs in other species. Upon submitting a query, the general annotation of queried genes is returned on the search result page (Fig. 3B), which includes gene length, protein Mr, protein length, GFP insertion position, whether it is a membrane protein, gene expression level, EST number, taxon group, gene alias, and membership in a gene family. Camera or book icon indicates that the gene product has been localized in the cell empirically. Clicking on the camera icon brings up an image detail page. An example of image detail page is shown on Figure 3C. This image was taken from the anther at flowering stage of 35 d after germination under Zeiss fluorescent microscopy with objective 60×. A larger version of the image is shown in Figure 3D. This image shows the protein product of gene AT2G04410 (unknown function), which localizes to the cell wall and extracellular matrix. This GFP fusion was expressed under its native promoter. The green fluorescence shows that the protein is localized to the secondary cell wall of endothelium cells and in the extracellular matrix of mature pollen grains. Autofluorescence of chlorophyll is shown in red. The Arabidopsis Genome Initiative number on the gene search result page links to the page that shows the primer information (Fig. 3E) and the map of primers on the target gene (Fig. 3F). The primer information includes sequence, length, and Tm. The primer map shows the positions of primers on the target gene's nucleotide sequence. Alternatively, the database can be browsed using subcellular location information (see sidebar on the Web site called Browse Images). Get Primers function is found on the gene search result page and is used to retrieve detailed information on predesigned primers whose sequences can be downloaded in an Excel format. Submit and Edit functions are used to submit or edit data, respectively. Construct information, transgenic plant data, and subcellular localization images and videos can be submitted and edited online. These two functions are protected by password. Download Clone Submission Data and Download Seed Submission Data functions are used to download detailed information on clones and transgenic seeds with protein localization images in formats suitable for submitting to the Arabidopsis Biological Resource Center at Ohio State University (http://www.biosci.ohio-state.edu/∼plantbio/Facilities/abrc/abrchome.htm).
Figure 3.
Web user interfaces. FTFLPdb and associated Web applications have several functionalities including Search, Submit, Edit, Assign, Submit/Edit, Get Primers, Download Clone Submission Data, and Download Seed Submission Data. A, A search page provides two categories of parameters, gene related and protein related, to search for genes of interest. B, Search result page shows general annotation of queried genes, which includes gene length, protein Mr, protein length, GFP insertion position, whether it is a membrane protein, gene expression level, EST number, taxon group, gene alias, and membership in a gene family. Camera or book icon indicates that the gene product has been localized in the cell empirically. C, Detailed information of one of the subcellular localization images of protein AT2G04410.1 is shown. D, The subcellular localization image of protein AT2G04410.1, which localizes to the cell wall and extracellular matrix, is shown. AT2G00410-GFP was expressed under a native promoter. The green shows expression of the protein in the secondary cell wall of endothelium cells and in the extracellular matrix of mature pollens. Autofluorescence in chlorophyll is shown in red. E, Primer information page provides primer sequence, length, and Tm. P1 and P4 primers contain sequences 5′-gctcgatccacctaggct-3′ and 5′-cgtagcgagaccacagga-3′, respectively, that partially overlap the Gateway primers, whereas P2 and P3 primers contained sequences 5′-cacagctccacctccacctccaggccggcc-3′ and 5′-tgctggtgctgctgcggccgctggggcc-3′, respectively, that partially overlap the GFP tag linkers. The capital letters are primer sequences matching the target gene sequence. F, Primer map shows the positions of primers on the target gene (red, coding region; blue, noncoding region). The two numbers within the parentheses show the start position (first no.) and the primer length (second no.). The italic letters show primer sequence. The underline indicates the primer position.
In summary, we developed a database and Web application for researchers to search, browse, submit, edit, and download data to facilitate empirical determination of protein localization. All of the database and software are available from http://aztec.stanford.edu/gfp/.
DISCUSSION
Subcellular localization is a fundamental attribute of proteins. To function together in a common physiological process such as a metabolic pathway or a signal transduction cascade, proteins must be localized to a proper subcellular location. To investigate the distribution of protein subcellular localization, we retrieved 1,300 proteins from TAIR, which have subcellular location verified by experiments. The general distribution of protein subcellular localization in Arabidopsis is similar to the distribution of the entire yeast proteome that has been experimentally verified, except for the distribution in cytosol and mitochondria (Kumar et al., 2002; Huh et al., 2003). In yeast, many proteins are found in the cytosol (40%), but only about 13.3% of the proteins are localized to the cytosol in Arabidopsis. Koroleva et al. (2005) found that 16% of their studied proteins were localized to the cytosol, which is greater than that in our data set, but it is still lower than the proportion of cytosol-localized proteins in yeast. Many Arabidopsis proteins (17%) are also found in the plastid, which is unique to plant cells. The difference in the proportion of cytosol-localized proteins between Arabidopsis and yeast may reflect a bias toward reporting subcellular localization to specific organelles in the Arabidopsis literature and having a complete inventory of subcellular localization of the proteome may alter the distribution. It is also possible that the relative reduction of Arabidopsis proteins localizing to the cytosol, together with the large number of proteins found in the plastid, may reflect a redistribution of cytosolic proteins into this plant-specific organelle during plant evolution. To test this hypothesis, we retrieved the protein ortholog data between Arabidopsis and yeast from the InParanoid Web site (http://inparanoid.cgb.ki.se/download/current/sqltables/longsqltable.ncbAT-modSC). It was found that 15 out of 1,818 cytosolic proteins of yeast have their orthologs in plastid proteins of Arabidopsis, but only 10 out of 2,468 noncytosolic proteins of yeast have their orthologs in plastid proteins of Arabidopsis. The proportion of Arabidopsis plastid protein orthologs in yeast cytosolic proteins is statistically significantly higher than that in yeast noncytosolic proteins (P value = 0.028; Peck et al., 2001), which supports our hypothesis.
On the other hand, many proteins are found in the mitochondria (36%) in Arabidopsis, but only about 10% of the yeast proteins are localized to the mitochondria. One possibility is that Arabidopsis mitochondrion is more complex than yeast mitochondrion. Alternatively, it may be due to the fact that a large proportion of mitochondria-localized proteins in TAIR come from large-scale proteomic data, which may have a higher rate of false positives than individual published studies.
We found that a significant number of Arabidopsis proteins (19%) localize to more than one compartment, similar to the results from the whole proteome analysis in yeast. These proteins are involved in dynamic processes such as transport, regulation of transcription, and signal transduction. The majority of the proteins (79%) that localize to multiple locations are found in two subcellular locations. The most commonly occurring paired location is cytosol and nucleus (46%). The high proportion of proteins that are localized to both the cytosol and nucleus suggests that these two compartments might be tightly coordinated in the processes that they carry out. These proteins are involved in processes such as regulation of transcription, DNA and protein metabolism, protein posttranslational modification, cell organization and biogenesis, signal transduction, response to oxidative stress, transport, and developmental processes. It is possible that translocation to a site of action upon a cue or a signal may be a general regulatory mechanism. It is also possible that the large proportion of dual localization in cytosol and nucleus may be due to the diffusion of proteins into/out of the nuclear pore and cytosol. The average Mr of proteins localized to both the cytosol and nucleus is statistically significantly smaller than that with unique localization (P value = 0.0082; Table II), which suggests that high proportion of proteins that are localized to both the cytosol and nucleus may result from some contribution of protein size. Protein import from the cytosol into the nucleus is a necessary and important step in the signal transduction processes. For small proteins (less than approximately 40–60 kD), this import could take place through nuclear pores by diffusion, but most proteins require nuclear localization signals (Dingwall and Laskey, 1991; Raikhel, 1992; Laskey and Dingwall, 1993) or protein-protein interaction (Moriuchi et al., 2004; Uhrig et al., 2004). It remains to be determined whether the smaller size of the nucleus-cytosol localized proteins contributes to a mechanism by which they move into/out of the nucleus or whether it is an artifact of overexpression of the proteins in the localization experiments.
Table II.
Comparison of the Mr of proteins between proteins with dual subcellular locations of cytosol and nucleus and proteins with unique subcellular localization
Proteins were classified into two groups. One group includes proteins localized to both cytosol and nucleus. The other group contains proteins with unique subcellular localization. Two-sample t test for comparing two-population means (Peck et al., 2001) was used for statistical analysis. Protein size was assessed by the calculated Mr of a protein (D). The Mr of proteins localized to both the cytosol and nucleus is statistically significantly smaller than that of proteins with a unique localization (P value = 0.0081).
| Protein Category (Protein No.) | Average Molecular Weight | sd | Min Molecular Weight | Max Molecular Weight | Median Molecular Weight | *P Value |
|---|---|---|---|---|---|---|
| D | D | D | D | |||
| Proteins with unique location (1,050) | 51,144 | 36,376 | 5,958 | 428,084 | 42,703 | 0.0082 |
| Proteins localized to both cytosol and nucleus (119) | 42,141 | 21,596 | 6,533 | 130,891 | 37,450 |
The second most commonly occurring paired compartments are mitochondria and plastid (9% of the proteins), which suggests that there is a significant amount of interaction between these organelles. These proteins are involved in processes such as electron transport or energy pathways, response to stress, hypersensitive response, biosynthesis of galactolipid, Met, steroid, thiamin, and Thr, tricarboxylic acid cycle intermediate metabolism, DNA and protein metabolism, transport, and cell organization and biogenesis. There are beneficial interactions of metabolism between mitochondria and chloroplast (Raghavendra and Padmasree, 2003). For example, mitochondria metabolism such as the bioenergetic processes of oxidative electron transport and phosphorylation are active in the light and indispensable for supporting photosynthetic carbon assimilation. Some studies showed that proteins are targeted to both organelles because they have an ambiguous signal that can be recognized by both import system (Chew et al., 2003; Silva-Filho, 2003). Comparison of such ambiguous signal sequences showed an overall similarity to mitochondria- or chloroplast-specific signals but contained additional Leu and phenylalaine residues, which result in an overall increase in hydrophobicity compared with either mitochondria-specific or chloroplast-specific signals. Some proteins (8.1%) in two locations have a partner location with plasma membrane and are involved in transport, signal transduction, cell organization and biogenesis, and developmental processes. Fifteen percent of the proteins that localize to multiple subcellular locations are found in three locations. The most commonly occurring triple location is cytosol, ER, and nucleus. These proteins (nine) among the triple-location cytosol, ER, and nucleus are found in the proteasome regulatory particle and have functions such as ATPase activity, calmodulin binding, ion channel activity, and peptide receptor activity. They are involved in ubiquitin-dependent protein catabolism, regulation of apoptosis, and N-terminal protein myristoylation. The proteasome regulatory particle is a component of the proteasome that is a large multisubunit complex and has been found to diffuse freely between the cytosol and the nucleus and is involved in degrading both cytoplasmic and nuclear proteins (Reits et al., 1997). Biederer et al. (1996) demonstrated that an ER degradation pathway of abnormal or unassembled membrane proteins was initiated at the cytoplasmic side of the ER.
Multiple-compartment localization may occur via posttranslational modifications. For example, plant G-box binding factors can translocate from the cytosol to the nucleus after they are phosphorylated by casein kinase II (Klimczak et al., 1992; Harter et al., 1994). Similarly, the N-myristoylation of proteins was found to result in protein translocation from cytosol to plasma membrane (Yalovsky et al., 1999). Another possible mechanism for multiple-compartment localization is through interaction between proteins. For example, it was found that nuclear translocation of the MADS-box transcription factors APETALA3 (AP3) and PISTILLATA (PI) from the cytosol was mutually interdependent (McGonigle et al., 1996). Transient expression of AP3-GUS or PI-GUS fusion proteins alone in onion (Allium cepa) epidermal cells resulted in cytosolic localization of the fusion proteins, but when one fusion was expressed together with the native form of the other, nuclear localization of the GUS fusion protein was observed. Another example is the rooting-locus gene B (rolB) in Agrobacterium rhizogenes, which is encoded by the gene on the T-DNA of the root-inducing plasmid. Moriuchi et al. (2004) showed that the RolB protein of pRi1724 (1724RolB) is localized to the nucleus even though it does not have a nuclear localization signal domain. 1724RolB directly interacts with tobacco (Nicotiana tabacum) 14-3-3-like protein ωII (Nt14-3-3 ωII) in tobacco Bright-Yellow 2 cells. 14-3-3 proteins are involved in the translocation of nuclear-encoded chloroplast precursor proteins into the chloroplast (May and Soll, 2000) and of the transcriptional activator RSG (repression of shoot growth) in plants (Igarashi et al., 2001). Paul et al. (2005) showed the subcellular distributions of 14-3-3 isoforms in Arabidopsis can be driven by client interactions and that those interactions are isoform specific in nature. Another example is the P19 protein of Tomato bushy stunt virus, which is a multifunctional pathogenicity determinant involved in suppression of posttranscriptional gene silencing, virus movement, and symptom induction. After infection of plants by Tomato bushy stunt virus or expression of P19 from Agrobacterium, the nuclear always early (ALY) protein from Arabidopsis was relocalized to the cytoplasm by its interaction with P19 protein (Uhrig et al., 2004). ALY is an uncharacterized family of plant proteins that, in animals, are involved in transcriptional coactivation and export of RNAs from the nucleus. These studies suggest the molecular functions and biological processes of proteins that exist in several subcellular locations might be involved in dynamic or signal-transducing processes mediated by protein translocation via interacting proteins or posttranslational modification.
Fusing GFP to proteins may have possible effects on protein subcellular localization. As discussed earlier, fusing GFP at the 3′ end of the coding region may block the targeting signal of some proteins. For example, in the study of yeast protein localization (Huh et al., 2003), the small GTP-binding protein Ras2 was localized to the nucleus and the cytoplasm by fusing with GFP at its C terminus, but it is known to be localized to the plasma membrane due to modification of its C terminus with palmitoyl and farnesyl groups. Proteins localized to the cell wall and subsets of proteins localized to the peroxisome also contain C-terminal targeting signals, and these are often mislocalized when fused to GFP at the C terminus. With N-terminal fusions, ER signal peptides, mitochondria, or chloroplast signal peptides may be masked and result in localization artifacts. Finally, overexpression of GFP fusion protein controlled by 35S viral promoter may produce abnormal multimer of protein or can also mask subtle localization patterns. To overcome these problems of GFP fusion construct, our FTFLPdb provides a predicted GFP insertion position internal from the termini where there is not any potential functional domain predicted based on the protein sequence. Such a GFP fusion construct resulted in correct subcellular localization of a number of proteins whose location had been determined previously (Tian et al., 2004). It is possible that even internal GFP fusions may prevent correct localization by interfering with protein interactions and/or function. It will be important to assess additional lines of evidence for the proper function of the fused proteins before definitively concluding the location of the fused proteins.
MATERIALS AND METHODS
Retrieval of Arabidopsis Genomic Sequence, Predicted Protein Sequence, and Functional Annotation Data
The Arabidopsis (Arabidopsis thaliana) genomic sequences and the predicted protein amino acid sequences were downloaded from TAIR (ftp://ftp.arabidopsis.org/home/tair/home/tair/Genes/TAIR6_genome_release). TargetP-predicted subcellular localization data, predicted functional domains, and protein superfamily annotation data (SCOP) were also downloaded from TAIR (ftp://ftp.arabidopsis.org/home/tair/home/tair/Proteins/). The data for gene structures, full-length cDNA, and EST were downloaded from TAIR (ftp://ftp.arabidopsis.org/home/tair/home/tair/Maps/seqviewer_data/). The Arabidopsis functional annotation data were also downloaded from TAIR (ftp://ftp.arabidopsis.org/home/tair/home/tair/Ontologies/Gene_Ontology/, 10/08/2005/). GO terms were obtained from GO Consortium (http://www.geneontology.org, 04/05/2005). Taxonomic data were downloaded from the National Center for Biotechnology Information (ftp://ftp.ncbi.nih.gov/pub/taxonomy/; 2/28/2004). Subcellular localization data of yeast (Saccharomyces cerevisiae) proteins were downloaded from http://yeastgfp.ucsf.edu. The yeast functional annotation data were also downloaded from GO Consortium (http://www.geneontology.org, version 1.418). Arabidopsis genes whose protein subcellular location have been experimentally verified were retrieved from TAIR (http://arabidopsis.org/tool/bulk/go/index.jsp, 9/10/2004) using the following criteria: genes were selected if they have functional annotation to the cellular component aspect of GO with any of the following evidence codes: inferred from direct assay (IDA), inferred from mutant phenotype (IMP), inferred from expression pattern (IEP), inferred from genetic interaction (IGI), or inferred from physical interaction (IPI; Berardini et al., 2004). The genomic sequence of genes includes 5′ UTR, up to 3 kb of 5′ flanking genomic sequence from the start codon to the adjacent gene, the coding sequence with introns, 3′ UTR, and up to 1 kb of 3′ flanking genomic sequence from the stop codon to the adjacent gene. Most of the intergenic distances in the Arabidopsis genome are around 2 kb, which suggests that the promoter sequences are contained in a relatively short region (Arabidopsis Genome Initiative, 2000). Therefore, the genomic sequence of a gene in the database includes up to 3 kb upstream of the translation initiation codon and up to 1 kb downstream of the stop codon before the start or end of the adjacent gene to ensure that all of the transcriptional regulatory sequences are included. TM domains of Arabidopsis proteins were predicted by THMM 2.0, which is a program that uses Hidden Markov model to predict TM domains of proteins (Krogh et al., 2001).
Analysis of Overrepresented Functions and Processes
Significantly overrepresented molecular functions and biological processes of genes localized to different subcellular locations were evaluated using GeneMerge1.2 software (Castillo-Davis and Hartl, 2003), an ontology file from the GO Consortium (http://www.geneontology.org, version 1.418), and annotations from TAIR (ftp://ftp.arabidopsis.org/home/tair/Genes/Gene_Ontology/ version 1.418). A Bonferroni-corrected P value (e-score) of 0.01 was used as a threshold for measuring significance.
Selection of Genes with Unknown Function
Genes were classified into two functional categories, those with known functions and those of unknown functions. The gene product that did not have a significant match to any known sequences and was supported by either full-length cDNA or by stringently matched EST evidence is designated as expressed protein, while the gene product that has no database matches of any kind is called hypothetical protein (Wortman et al., 2003). We defined genes of unknown function as those that were annotated as expressed protein or hypothetical protein.
Genome-Wide GFP Insertion Site Selection and Primer Design
We wrote a Perl script called Primer4FTTLP.pl (http://aztec.stanford.edu/gfp/) using part of the Primer3 software (http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi) to identify the GFP insertion position and design primers (four primers named P1, P2, P3, and P4). The script considers annealing temperature, position of each primer within the genomic sequence, protein domains, primer length, and primer secondary structure, in an iterative fashion. Our default position for inserting GFP is 10 amino acids upstream of the stop codon. However, if a functional domain, including targeting sequences and TM domains, is predicted to exist at this position, the GFP insertion position is iteratively shifted downstream or upstream until no functional domain is detected. After this temporary GFP insertion position is determined, P2 and P3 sequences were generated. P1 sequence at the 5′ end of the gene was determined using P2 sequence as the right primer sequence. P4 sequence at the 3′ end of the gene was determined using P3 sequence as the left primer sequence. The quality of the four primers was checked by the script according to the default parameters in Primer3. The GFP insert position was iteratively shifted until a suitable GFP insertion site was determined. We first chose the GFP insertion site to be near the C terminus of a protein (amino acids 1–30). If no suitable position was found in this region, we used the same strategy to select the GFP insertion position and design primers near the N terminus of a protein between amino acids 1 and 30.
Protein Family Classification
TribeMCL software (version 4-189) was developed by Enright et al. (2002) using Markov cluster algorithm to classify proteins into families based on overall protein sequence similarity. A similarity matrix was generated from an all-against-all comparison of the Arabidopsis protein sequences using the National Center for Biotechnology Information BLASTP version 2.2.6 (Altschul et al., 1997) with a threshold E-value of 10−5 for matches. This similarity matrix was imported into TribeMCL software to generate protein families by setting parameters -I to 1.1 and -scheme to 4 according to the software manual.
Database and Web Interface Implementation
The database was designed and implemented using MySQL3.0 (DuBois, 2000), a freely available relational database management system. The Web interfaces were programmed using Perl-CGI (Christiansen and Torkington, 1998; Birznieks et al., 2000). We developed a database and software to allow querying for genes of interest, downloading predesigned primers, and uploading and updating construct, transgenic plant, and image (including video) data. We implemented a number of functions to minimize human error and to streamline information management, including automatic generation of nomenclature for constructs and plants and tracking of the work in progress. Both the MYSQL database and relevant software package can be downloaded from http://aztec.stanford.edu/gfp/DOWNLOAD/.
Supplementary Material
Acknowledgments
We thank people in both TAIR and FTFLP projects who curated and generated data used in this study and provided many useful suggestions for the database and software development. We are also grateful to Dr. Tanya Berardini and Dr. Leonore Reiser for their helpful comments on the manuscript.
This work was supported by the Arabidopsis 2010 Project of the National Science Foundation (grant no. 0210992).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Seung Y. Rhee (rhee@acoma.stanford.edu).
The online version of this article contains Web-only data.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhan Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Berardini TZ, Mundodi S, Reiser L, Huala E, Garcia-Hernandez M, Zhang P, Mueller LA, Yoon J, Doyle A, Lander G, et al (2004) Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiol 135: 745–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berriman M, Harris M (2004) Annotation of parasite genomes. Methods Mol Biol 270: 17–44 [DOI] [PubMed] [Google Scholar]
- Biederer T, Volkwein C, Sommer T (1996) Degradation of subunits of the Sec61p complex, an integral component of the ER membrane, by the ubiquitin-proteasome pathway. EMBO J 15: 2069–2076 [PMC free article] [PubMed] [Google Scholar]
- Birznieks G, Guelich S, Gundavaram S (2000) CGI Programming with Perl. O'Reilly & Associates, Sebastopol, CA
- Carrari F, Nunes-Nesi A, Gibon Y, Lytovchenko A, Loureiro ME, Fernie AR (2003) Reduced expression of aconitase results in an enhanced rate of photosynthesis and marked shifts in carbon partitioning in illuminated leaves of wild species tomato. Plant Physiol 133: 1322–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Davis CI, Hartl DL (2003) GeneMerge: post-genomic analysis, data mining, and hypothesis testing. Bioinformatics 19: 891–892 [DOI] [PubMed] [Google Scholar]
- Cedano J, Aloy P, Perez-Pons JA, Querol E (1997) Relation between amino acid composition and cellular location of proteins. J Mol Biol 266: 594–600 [DOI] [PubMed] [Google Scholar]
- Chew O, Rudhe C, Glaser E, Whelan J (2003) Characterization of the targeting signal of dual-targeted pea glutathione reductase. Plant Mol Biol 53: 341–356 [DOI] [PubMed] [Google Scholar]
- Chou KC (2001) Prediction of protein cellular attributes using pseudo-amino acid composition. Proteins 43: 246–255 [DOI] [PubMed] [Google Scholar]
- Chou KC, Cai YD (2002) Using functional domain composition and support vector machines for prediction of protein subcellular location. J Biol Chem 277: 45765–45769 [DOI] [PubMed] [Google Scholar]
- Christiansen T, Torkington N (1998) Perl Cookbook. O'Reilly & Associates, Sebastopol, CA
- Claros MG (1995) MitoProt, a Macintosh application for studying mitochondrial proteins. Comput Appl Biosci 11: 441–447 [DOI] [PubMed] [Google Scholar]
- Claros MG, Vincens P (1996) Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem 24: 779–786 [DOI] [PubMed] [Google Scholar]
- Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR (2000) Random GFP∷cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci USA 97: 3718–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Rio LA, Corpas FJ, Sandalio LM, Palma JM, Gomez M, Barroso JB (2002) Reactive oxygen species, antioxidant systems and nitric oxide in peroxisomes. J Exp Bot 53: 1255–1272 [PubMed] [Google Scholar]
- Dingwall C, Laskey RA (1991) Nuclear targeting sequence: a consensus? Trends Biochem Sci 16: 478–481 [DOI] [PubMed] [Google Scholar]
- DuBois P (2000) MySQL. New Riders, Indianapolis
- Dyer JM, Mullen RT (2001) Immunocytological localization of two plant fatty acid desaturases in the endoplasmic reticulum. FEBS Lett 494: 44–47 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA (2002) An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res 30: 1575–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar NM, Haupt S, Thow G, Boevink P, Chapman S, Oparka K (2003) High-throughput viral expression of cDNA-green fluorescent protein fusions reveals novel subcellular addresses and identifies unique proteins that interact with plasmodesmata. Plant Cell 15: 1507–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Kamber M (2001) Cluster analysis. In Data Mining: Concepts and Techniques. Morgan Kaufmann Publishers, San Francisco, pp 341–343
- Harter K, Kircher S, Frohnmeyer H, Krenz M, Nagy F, Schafer E (1994) Light-regulated modification and nuclear translocation of cytosolic G-box binding factors in parsley. Plant Cell 6: 545–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Munoz I, Benet M, Calero M, Jimenez M, Diaz R, Pellicer A (2003) rgr oncogene: activation by elimination of translational controls and mislocalization. Cancer Res 63: 4188–4195 [PubMed] [Google Scholar]
- Hua S, Sun Z (2001) Support vector machine approach for protein subcellular localization prediction. Bioinformatics 17: 721–728 [DOI] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK (2003) Global analysis of protein localization in budding yeast. Nature 425: 686–691 [DOI] [PubMed] [Google Scholar]
- Huq E, Al-Sady B, Quail PH (2003) Nuclear translocation of the photoreceptor phytochrome B is necessary for its biological function in seedling photomorphogenesis. Plant J 35: 660–664 [DOI] [PubMed] [Google Scholar]
- Igarashi D, Ishida S, Fukasawa J, Takahashi Y (2001) 14-3-3 proteins regulate intracellular localization of the bZIP transcriptional activator RSG. Plant Cell 13: 2483–2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertbundit S, Linacero R, Rouze P, Galis I, Macas J, Deboeck F, Renckens S, Hernalsteens JP, De Greve H (1998) Analysis of T-DNA-mediated translational beta-glucuronidase gene fusions. Plant Mol Biol 36: 205–217 [DOI] [PubMed] [Google Scholar]
- Klimczak LJ, Schindler U, Cashmore AR (1992) DNA binding activity of the Arabidopsis G-box binding factor GBF1 is stimulated by phosphorylation by casein kinase II from broccoli. Plant Cell 4: 87–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koroleva OA, Tomlinson ML, Leader D, Shaw P, Doonan JH (2005) High-throughput protein localization in Arabidopsis using Agrobacterium-mediated transient expression of GFP-ORF fusions. Plant J 41: 162–174 [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer ELL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305: 567–580 [DOI] [PubMed] [Google Scholar]
- Kumar A, Agarwal S, Heyman JA, Matson S, Heidtman M, Piccirillo S, Umansky L, Drawid A, Jansen R, Liu Y, et al (2002) Subcellular localization of the yeast proteome. Genes Dev 16: 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskey RA, Dingwall C (1993) Nuclear shuttling: the default pathway for nuclear proteins? Cell 74: 585–586 [DOI] [PubMed] [Google Scholar]
- Li Y, Kandasamy MK, Meagher RB (2001) Rapid isolation of monoclonal antibodies: monitoring enzymes in the phytochelatin synthesis pathway. Plant Physiol 127: 711–719 [PMC free article] [PubMed] [Google Scholar]
- Lindeman GJ, Gaubatz S, Livingston DM, Ginsberg D (1997) The subcellular localization of E2F-4 is cell-cycle dependent. Proc Natl Acad Sci USA 94: 5095–5100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Amy V, Liu G, McKeehan WL (2002) Novel complex integrating mitochondria and the microtubular cytoskeleton with chromosome remodeling and tumor suppressor RASSF1 deduced by in silico homology analysis, interaction cloning in yeast, and colocalization in cultured cells. In Vitro Cell Dev Biol Anim 38: 582–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magae J, Wu CL, Illenye S, Harlow E, Heintz NH (1996) Nuclear localization of DP and E2F transcription factors by heterodimeric partners and retinoblastoma protein family members. J Cell Sci 109: 1717–1726 [DOI] [PubMed] [Google Scholar]
- Matsuoka K, Neuhaus JM (1999) Cis-elements of protein transport to the plant vacuoles. J Exp Bot 50: 165–174 [Google Scholar]
- May T, Soll J (2000) 14-3-3 proteins form a guidance complex with chloroplast precursor proteins in plants. Plant Cell 12: 53–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGonigle B, Bouhidel K, Irish VF (1996) Nuclear localization of the Arabidopsis APETALA3 and PISTILLATA homeotic gene products depends on their simultaneous expression. Genes Dev 10: 1812–1821 [DOI] [PubMed] [Google Scholar]
- Moriuchi H, Okamoto C, Nishihama R, Yamashita I, Machida Y, Tanaka N (2004) Nuclear localization and interaction of RolB with plant 14-3-3 proteins correlates with induction of adventitious roots by the oncogene rolB. Plant J 38: 260–275 [DOI] [PubMed] [Google Scholar]
- Nakai K, Horton P (1999) PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci 24: 34–36 [DOI] [PubMed] [Google Scholar]
- Paul AL, Sehnke PC, Ferl RJ (2005) Isoform-specific subcellular localization among 14-3-3 proteins in Arabidopsis seems to be driven by client interactions. Mol Biol Cell 16: 1735–1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck R, Olsen C, Devore J (2001) Comparing two populations or treatments. In Introduction to Statistics and Data Analysis. DUXBURY Thomson Learning, Pacific Grove, CA, pp 535–598
- Raghavendra AS, Padmasree K (2003) Beneficial interactions of mitochondrial metabolism with photosynthetic carbon assimilation. Trends Plant Sci 8: 546–553 [DOI] [PubMed] [Google Scholar]
- Raikhel N (1992) Nuclear targeting in plants. Plant Physiol 100: 1627–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reits EA, Benham AM, Plougastel B, Neefjes J, Trowsdale J (1997) Dynamics of proteasome distribution in living cells. EMBO J 16: 6087–6094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richly E, Leister D (2004) An improved prediction of chloroplast proteins reveals diversities and commonalities in the chloroplast proteomes of Arabidopsis and rice. Gene 329: 11–16 [DOI] [PubMed] [Google Scholar]
- Silva-Filho MC (2003) One ticket for multiple destinations: dual targeting of proteins to distinct subcellular locations. Curr Opin Plant Biol 6: 589–595 [DOI] [PubMed] [Google Scholar]
- Tian GW, Mohanty A, Chary SN, Li S, Paap B, Drakakaki G, Kopec CD, Li J, Ehrhardt D, Jackson D, et al (2004) High-throughput fluorescent tagging of full-length Arabidopsis gene products in planta. Plant Physiol 135: 25–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhrig JF, Canto T, Marshall D, MacFarlane SA (2004) Relocalization of nuclear ALY proteins to the cytoplasm by the tomato bushy stunt virus P19 pathogenicity protein. Plant Physiol 135: 2411–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wortman JR, Haas BJ, Hannick LI, Smith RK Jr, Maiti R, Ronning CM, Chan AP, Yu C, Ayele M, Whitelaw CA, et al (2003) Annotation of the Arabidopsis genome. Plant Physiol 132: 461–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalovsky S, Rodr Guez-Concepcion M, Gruissem W (1999) Lipid modifications of proteins: slipping in and out of membranes. Trends Plant Sci 4: 439–445 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.