Abstract
Sucrose starvation of Arabidopsis (Arabidopsis thaliana) cell culture was used to identify translationally regulated genes by DNA microarray analysis. Cells were starved by subculture without sucrose, and total and polysomal RNA was extracted between 6 and 48 h. Probes were derived from both RNA populations and used to screen oligonucleotide microarrays. Out of 25,607 screened genes, 224 were found to be differentially accumulated in polysomal RNA following starvation and 21 were found to be invariant in polysomal RNA while their total RNA abundance was modified. Most of the mRNA appears to be translationally repressed (183/245 genes), which is consistent with a general decrease in metabolic activities during starvation. The parallel transcriptional analysis identifies 268 regulated genes. Comparison of transcriptional and translational gene lists highlights the importance of translational regulation (mostly repression) affecting genes involved in cell cycle and cell growth, these being overrepresented in translationally regulated genes, providing a molecular framework for the arrest of cell proliferation following starvation. Starvation-induced translational control also affects chromatin regulation genes, such as the HD1 histone deacetylase, and the level of histone H4 acetylation was found to increase during starvation. This suggests that regulation of the global nuclear transcriptional activity might be linked to cytoplasmic translational regulations.
Plants are primary producers of sugars through photosynthesis, and their production and transport regulate many aspects of their growth and metabolism. Suc is the main transported form and is cleaved by the invertase into Glc and Fru in the sink tissues. Suc and other hexoses act not only as nutrients but also as signaling molecules through pathways that are not yet fully understood, among which the activity of hexokinases is involved (Moore et al., 2003). Sugar sensing is connected to the perception of other essential nutrients, such as nitrogen, to coordinate the production of cell components (Stitt, 1999; Coruzzi and Bush, 2001; Coruzzi and Zhou, 2001) with several plant hormone signaling pathways (Roitsch, 1999; Sheen et al., 1999; Smeekens, 2000; Gazzarrini and McCourt, 2001; Finkelstein and Gibson, 2002). High Suc or metabolizable hexoses generally repress photosynthesis and promote carbohydrate storage, while low sugar increases photosynthesis and export. The expression of a large number of genes was found to be modified by changes in sugar levels (Koch, 1996; Yu, 1999; Smeekens, 2000; Rolland et al., 2002; Contento et al., 2004; Price et al., 2004). Transcription profiling in whole plants reveals that Glc regulates nitrogen metabolism genes and induces several stress and ethylene responsive genes (Price et al., 2004). Physiological and cellular changes that occur during the starvation of proliferating tissues were studied in isolated sink organs such as excised maize (Zea mays) root tips (Brouquisse et al., 1991) or in various cell culture systems (Journet et al., 1986; Chen et al., 1994; Aubert et al., 1996; Moriyasu and Ohsumi, 1996; Bassham, 2002). Following sugar starvation, sequential changes are observed: cell growth arrest; consumption of cellular carbohydrate and decrease in respiration rate; degradation of lipids and proteins; increased accumulation of inorganic phosphate, phosphorylcholine, and free amino acids; and a general decline in glycolytic activities. An increase in the size of the vacuole and a decrease in the activity of (vacuolar) enzymes were also observed (Moriyasu and Ohsumi, 1996). A link between growth and the cell cycle was provided by the demonstration that Suc has a positive effect on the transcription of D-type cyclin in Arabidopsis (Arabidopsis thaliana) cells (Riou-Khamlichi et al., 2000), and transcriptional analysis of gene expression in response to sugar starvation also shows repression of several genes coding for ribosomal proteins, translation factors, and cell cycle-related functions (Contento et al., 2004).
Several mRNAs were found to be differentially stabilized in the presence of sugars (Chan and Yu, 1998; Cheng et al., 1999; Ho et al., 2001) and Suc repression of translation of a class of Arabidopsis mRNAs encoding bZIP transcription factors was found to be mediated by a conserved short open reading frame in their 5′ untranslated region (Rook et al., 1998; Wiese et al., 2004). In addition, the transcriptional activation of genes by Glc in whole plants was found to be more affected by cycloheximide treatment than by transcriptional repression, suggesting that the activation of synthesis of specific proteins was required prior to transcriptional activation (Price et al., 2004).
The molecular mechanisms underlying posttranscriptional regulation are generally less well understood than transcriptional ones both at the qualitative and quantitative levels. To improve our understanding of these mechanisms and evaluate their role in plant growth in response to nutrients, it would be useful to identify their targets in a systematic fashion. One approach to study global translational regulation of gene expression is to derive probes from mRNA populations isolated from cell fractions enriched in polysomes and to evaluate their complexities using DNA microarrays.
In this study, we evaluated the effect of Suc starvation on the translational state of a set of 25,607 Arabidopsis putative genes. As we wanted to focus more specifically on the effect of Suc on proliferation, cultured cells were used as a source of mRNA. Furthermore, as translational control is often of lower amplitude than transcriptional regulation, undifferentiated cells form a more homogeneous population than whole plant tissues, allowing an improved detection of translationally regulated mRNAs. This work reveals that a large number of genes involved in the cell cycle are translationally regulated by Suc and suggests a link between cytoplasmic Suc responses and global control of gene transcription through the translational regulation of histone deacetylase.
RESULTS
Establishment of Suc Starvation
An Arabidopsis cell culture was chosen to investigate the translational regulations linked to Suc starvation (Axelos et al., 1992). These cells require Suc for optimal growth, divide rapidly in its presence, and are known to respond to Suc starvation and provide a large amount of homogeneous material for gradient centrifugation and mRNA extraction. In Suc-containing media, their growth curves are identical in the light or in the dark (Fig. 1A). Suc concentration in the culture medium was quantified over time. Starting with a concentration of 30 g/L, its level dropped down to less than 5 g/L after 9 d, and growth arrest occurs at 11 d (Fig. 1B). Although external sugar is low at 9 d, cells are accumulating starch and remain healthy for at least 48 h. When subculture occurs at 9 d in a medium lacking Suc, proliferation is stopped, and signs of cell death are evident after 72 h (Fig. 2, A and B). Cells were therefore routinely subcultured at 9 d when they are still healthy and at the end of the exponential growth phase. For microarray experiments, cells were subcultured in medium without Suc and control cells were subcultured in Suc-containing media.
Figure 1.
Growth characteristics of the Arabidopsis cell line. A, Evolution of the PCV during time expressed in percentage of the volume of growing media (100 mL). B, Evolution of the concentration of Glc and Suc, in grams per 100 mL in the medium during the growth of the culture (n = 4). Cells were routinely subcultured at 9 d into fresh medium or Suc-free medium to establish starvation.
Figure 2.
Establishment of Suc starvation. A, Evolution of PCV during Suc starvation expressed in percentage of the volume of the growing media (100 mL). B, Evolution of the number of living cells during the culture either in the presence or absence of Suc expressed as the fraction of the number of living cells at the initiation of subculture. Measurements have been done at the time of transfer then after 24, 48, and 72 h. C, Molecular validation of the establishment of Suc starvation. The expression levels of the mRNA coding for NR2, MCC, and E1α were quantified by Q-RT-PCR. Results are expressed as mRNA ratios in starved cells over control cells. 18S ribosomal RNA and α-tubulin mRNA were used as control.
The effectiveness of starvation was further monitored by the expression of mRNA previously described (Fujiki et al., 2001) as responding to Suc starvation such as those coding for nitrate reductase 2 (NR2), the biotinylated subunit of β-methylcrotonyl-CoA carboxylase (MCC), and the E1α-subunit of branched-chain α-keto acid dehydrogenase (E1α). This shows that a physiological response to Suc starvation occurs as soon as 6 h after subculture (Fig. 2C). Suc starvation is known to synchronize Arabidopsis cells to some extent (Menges and Murray, 2002). Since the doubling time of the cells is approximately 72 h, we selected 48 h postsubculture for translational control experiments to ensure that a good proportion of control cells had reentered the growth phase.
Changes in Transcriptional Patterns
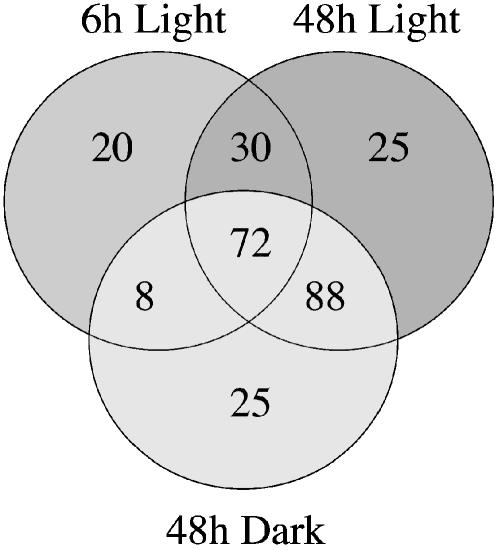
Translational analysis experiments include a built-in comparison with transcriptional analysis. Microarrays containing 25,607 oligonucleotides representing Arabidopsis mRNAs were hybridized with total RNA from control and starved cells. We therefore obtained an image of transcriptional patterns 6 and 48 h after the start of Suc starvation in the light and after 48 h in the dark and compared this to cells subcultured for the same time and in the same light regime in Suc-containing media. For each condition of starvation, two independent biological replicates were analyzed, and genes with an expression ratio of 2-fold and a P value <0.05 were retained for analysis. Out of a total of 8,041 hybridizing genes, we identified 130 mRNAs for which the steady-state level was affected after 6 h of Suc starvation in the light; this number rises to 215 mRNA after 48 h of Suc starvation, while 193 mRNAs are regulated after 48 h of Suc starvation in the dark.
Between the three data sets, an overlap of 72 mRNAs was found, and 160 mRNAs were identical for data obtained after 48 h of starvation in the dark and light. We therefore established a list of 268 genes transcriptionally regulated by sugar starvation in at least one of the experimental points (Fig. 3; Supplemental Table I). As a check of the microarray transcriptional data, the expression level of a set of 33 genes was analyzed by quantitative real-time (Q-RT)-PCR and sugar regulation was confirmed for 31 of them (94%; Table I). Northern analysis was also used to confirm the changes in expression level of five genes (data not shown).
Figure 3.
Venn diagrams showing the number of transcripts regulated in response to Suc starvation after 6 or 48 h under light condition or 48 h under dark condition. Only transcripts for which at least a 2-fold variation in the steady-state level together with P value < 0.05 over control cells are considered.
Table I.
Q-RT-PCR validation of the transcriptional microarray data for 33 genes
Column 1, AGI numbers of the selected genes; column 2, RNA sample used for Q-RT-PCR (6hL, 6 h light; 48hL, 48 h light; 48hD, 48 h day); column 3, Q-RT-PCR data expressed as the ratio of starved over control; columns 4 to 6, microarray data expressed as ratio of starved over control in the three experiments; column 7, validation yes/no.
| AGI | Sample Used for Q-RT-PCR | Q-RT-PCR | Microarray
|
Validation | ||
|---|---|---|---|---|---|---|
| 6hL | 48hL | 48hD | ||||
| At2g03090 | 6hL | 5.12 | 2.85 | 0.52 | 0.63 | Y |
| At3g10950 | 6hL | 0.52 | 0.50 | 1.91 | 1.75 | Y |
| At3g25250 | 6hL | 3.22 | 3.40 | 1.32 | 1.73 | Y |
| At4g30280 | 6hL | 0.43 | 0.41 | 0.86 | 1.72 | Y |
| At5g13210 | 6hL | 0.26 | 0.50 | 1.84 | 1.56 | Y |
| At5g65110 | 6hL | 0.38 | 0.44 | 0.52 | 0.70 | Y |
| At1g12780 | 48hL | 0.6 | 0.57 | 0.21 | 0.82 | Y |
| At4g02520 | 48hL | 0.22 | 1.05 | 0.30 | 0.86 | Y |
| At3g12970 | 48hD | 7.01 | 1.55 | 1.92 | 2.35 | Y |
| At3g47540 | 48hD | 3.69 | 0.93 | 1.92 | 2.35 | Y |
| At4g11650 | 48hD | 0.09 | 0.89 | 1.41 | 0.30 | Y |
| At4g30490 | 48hD | 5.55 | 0.85 | 1.39 | 2.40 | Y |
| At2g29490 | 48hL | 0.34 | 6.70 | 0.21 | 0.61 | Y |
| At2g38870 | 48hL | 0.32 | 0.39 | 0.39 | 1.52 | Y |
| At3g45970 | 48hL | 0.29 | 0.38 | 0.33 | 0.68 | Y |
| At5g22920 | 48hL | 12.12 | 0.15 | 6.15 | 0.94 | Y |
| At5g39320 | 48hL | 0.25 | 0.46 | 0.43 | 0.77 | Y |
| At1g75380 | 48hD | 0.37 | 0.42 | 0.62 | 0.39 | Y |
| At5g39580 | 48hD | 0.11 | 0.19 | 0.87 | 0.27 | Y |
| At5g57655 | 48hD | 0.18 | 0.39 | 1.16 | 0.35 | Y |
| At1g54100 | 48hD | 0.38 | 1.11 | 0.35 | 0.24 | Y |
| At1g68440 | 48hD | 0.39 | 0.76 | 0.07 | 0.34 | Y |
| At1g76870 | 48hD | 0.99 | 1.20 | 0.18 | 0.09 | N |
| At2g33150 | 48hD | 0.43 | 1.09 | 0.29 | 0.35 | Y |
| At3g04720 | 48hD | 0.22 | 1.11 | 0.11 | 0.16 | Y |
| At3g44300 | 48hD | 0.41 | 1.14 | 0.32 | 0.30 | Y |
| At3g59270 | 48hD | 0.12 | 1.11 | 0.13 | 0.14 | Y |
| At1g51400 | 48hD | 0.48 | 0.48 | 0.42 | 0.31 | Y |
| At2g30860 | 48hD | 2.9 | 2.19 | 2.49 | 3.50 | Y |
| At3g28180 | 48hD | 0.13 | 0.26 | 0.41 | 0.41 | Y |
| At4g37610 | 48hD | 0.07 | 0.12 | 0.18 | 0.25 | Y |
| At5g07440 | 48hD | 5.88 | 0.32 | 0.36 | 0.24 | N |
| At5g49360 | 48hD | 0.25 | 0.15 | 0.33 | 0.31 | Y |
When we compared this gene list with the gene list found in a similar wide transcription study of genes regulated by sugar starvation (Contento et al., 2004), an overlap of 68 genes was observed (Supplemental Table V). In addition, many identified genes can be assigned to the same functions while they do not bear the same Arabidopsis Genome Initiative (AGI) number (e.g. auxin-regulated protein, xyloglucan endotransglycosylase, and glutathione transferase).
Changes in Translational Patterns
The translational analysis was performed with polysomes purified on Suc gradients from cells starved for 48 h in the dark. From each gradient, 11 fractions were collected by monitoring absorbance at 254 nm, and fractions corresponding to polyribosomes (usually fractions 7–11) of the Suc gradients from starved- or control-cell extracts were combined and used for mRNA extraction and labeling. Microarrays were hybridized in parallel with labeled polysomal RNA and with labeled total RNA derived from the same cell cultures. The experiment was repeated with two independent biological samples. Candidate genes were selected in each experiment on the basis of a 2-fold variation in signal intensity and a P value of <0.05. Then, for each mRNA, the ratio of signal intensities of starved over control samples was compared between the total RNA experiment and the polysomal RNA experiment to identify genes potentially translationally regulated.
The mRNAs can be classified into two classes: class 1 consists of 224 mRNAs for which the relative abundance within polysomal RNA changed more than 2-fold with a P value < 0.05, without significant changes in their abundance in total RNA (this category defines genes that are translationally regulated independently of transcriptional control); and class 2 consists of 21 mRNAs for which the relative abundances within polysomal RNA did not change while their relative abundance in total RNA changed by more than 2-fold with a P value < 0.05. This category tentatively defines genes that are cellularly buffered following transcription, preventing or increasing their access to the transcriptional machinery.
In addition, for 51 mRNAs the relative abundance within polysomal RNA changed in the same direction as their relative abundance in total RNA (Supplemental Table IV). This category therefore defines genes that are coregulated at the transcriptional and translational level. An overlap of 32 mRNAs was observed between transcriptional and translational experiments: the 21 mRNAs of the class 2 and 11 mRNAs were found to undergo transcriptional variations before 48 h. This is the case for the MCC (At1g03090), which was selected as a control for establishing starvation conditions. The mRNA of this gene undergoes an increased accumulation at 24 h of starvation but returns to basal level after 48 h, although it increases in polysomal RNA only at 48 h as indicated by the microarray experiment. The examination of its variations in polysomal RNA at 6 h post starvation by Q-RT-PCR experiment indeed confirms that it did not vary. Therefore, for this gene, transcriptional activation occurs first and is followed by translational activation with a significant time delay.
Overall, 245 genes can be predicted to undergo some form of translational control during Suc starvation (Supplemental Table II). To further validate the microarray analysis, the relative abundance of 18 mRNAs in total and polysomal RNA samples, with and without Suc starvation, was monitored by Q-RT-PCR in three biological replicates, including the two samples used for microarray analysis (Table II). The variations observed using microarrays were qualitatively confirmed for 13 mRNAs (72.2%) both at the transcriptional and at the translational levels, while for three mRNAs only the modification of polysomal abundance could be confirmed.
Table II.
Q-RT-PCR validation of the translational microarray data for 18 genes
Column 1, AGI identification; columns 2 and 3, Q-RT-PCR data for total RNA (Tot) and polysomal RNA (Poly) samples expressed as the ratio of starved over control; columns 4 and 5, microarray data for total RNA and polysomal RNA expressed as ratio of starved over control; columns 6 and 7, validation Yes/No; column 8, putative nature of gene product.
| The Arabidopsis Information Resource Identification | Q-RT-PCR
|
Microarray
|
Validation
|
Description | |||
|---|---|---|---|---|---|---|---|
| Tot | Poly | Tot | Poly | Tot | Poly | ||
| At1g03090 | 0.92 | 0.25 | 1.17 | 0.28 | Y | Y | 3-Methylcrotonyl-CoA carboxylase-related ESTs gb |
| At1g09415 | 0.99 | 0.25 | 1.58 | 0.46 | Y | Y | Expressed protein |
| At1g35320 | 0.9 | 0.31 | 1.01 | 7.37 | Y | N | Hypothetical protein |
| At1g51400 | 0.48 | 1.08 | 0.31 | 1.05 | Y | Y | PSII 5-kD protein; 100% identical to GI:4836947 (F5D21,10) |
| At1g63800 | 0.79 | 0.37 | 1.34 | 0.43 | Y | Y | Ubiquitin-conjugating enzyme 5 (UBC5) E2; identical to gi:431269, SP:P42749 |
| At1g76410 | 0.01 | 0.04 | 0.74 | 0.32 | N | Y | RING zinc finger protein-related contains Pfam profile: PF00097 zinc finger, C3HC4 type (RING finger) |
| At2g38870 | 1.25 | 0.23 | 1.52 | 0.41 | Y | Y | Protease inhibitor related |
| At3g09840 | 0.95 | 17.23 | 1.01 | 26.84 | Y | Y | Transitional endoplasmic reticulum ATPase-related identical to cell division cycle protein 48 (CDC48) homolog GB:P54609 (EMBO J. 14 (22), 5626–5637 [1995]); similar to transitional endoplasmic reticulum ATPase GB:P46462 (Rattus norvegicus) |
| At3g25250 | 1.1 | 5.71 | 1.73 | 6.23 | Y | Y | Protein kinase family contains protein kinase domain, Pfam:PF00069 |
| At3g46690 | 0.59 | 0.13 | 0.74 | 0.09 | Y | Y | Glucuronosyl transferase-related protein glucuronosyl transferase homolog, ripening-related Lycopersicon esculentum, PIR2:S39507 |
| At3g46720 | 0.24 | 0.57 | 0.83 | 0.23 | N | N | Glycosyltransferase family contains Pfam profile: PF00201 UDP-glucoronosyl and UDP-glucosyl transferase |
| At3g51910 | 0.02 | 0.26 | 1.37 | 0.18 | N | Y | Heat shock transcription factor family contains Pfam profile: PF00447 HSF-type DNA-binding domain |
| At3g55910 | 0.78 | 0.39 | 1.82 | 0.37 | Y | Y | Expressed protein PA26, p53 regulated PA26-T3 nuclear protein, Homo sapiens, EMBL:AF033121 |
| At4g38130 | 0.74 | 10.03 | 1.02 | 14.86 | Y | Y | HD1 histone deacetylase |
| At5g18630 | 1.39 | 0.32 | 0.82 | 0.26 | Y | Y | Lipase (class 3) family low similarity to triacylglycerol acylhydrolase (E,C,3,1,1,3; Rhizomucor miehei) GI:230348; contains Pfam profile PF01764: lipase |
| At5g35790 | 0.8 | 0.34 | 0.99 | 0.38 | Y | Y | Glc-6-P dehydrogenase |
| At5g58720 | 0.99 | 5.17 | 1.04 | 16.61 | Y | Y | PRL1 associated protein related |
| At5g61440 | 0.5 | 0.19 | 1.43 | 0.31 | N | Y | Thioredoxin family low similarity to thioredoxin (Callithrix jacchus) GI:13560979; contains Pfam profile: PF00085 thioredoxin |
Functional Classification: Transcriptional Control
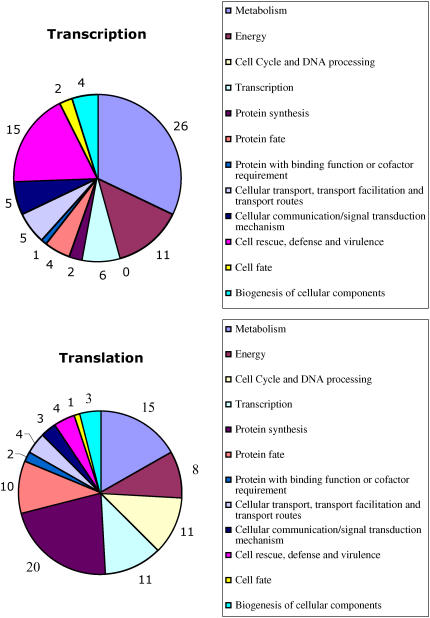
For transcriptional and translational gene lists, tentative functions were assigned using the Munich Information Center for Protein Sequences (MIPS) database and visual inspection (Fig. 4). More than 62% of the genes identified are not yet classified based on known function and/or sequence homology. The distribution of genes regulated transcriptionally is very similar to that obtained by Contento et al. (2004) with more than 50% of genes potentially involved in metabolism and in cell rescue, defense, and virulence. Genes involved in mechanisms of defense and particularly those of the ethylene/wound responsive pathway are largely represented. This includes not only genes coding directly for defense proteins such as protease inhibitors (At5g05110, At2g38870) or defensins (At1g19610, At2g02120), but also signaling to transcription-related proteins such as ethylene responsive element binding factor (At5g61590), WRKY transcription factors (At1g29860, At3g01970), and wound induced genes (At4g28240, At1g75380). These genes appear to be generally activated by starvation. Functions involved in cell wall modification and cytoskeleton reorganization are also well represented. Induction of photosynthesis-related genes is also a dominant scheme in Suc starvation as previously reported (Contento et al., 2004).
Figure 4.
Distribution of genes into functional categories. For each gene, a functional category has been assigned based on the known or putative function of the corresponding protein, according to the MIPS Functional Category database. Pie charts show the number of probes identified in each category.
Functional Classification: Translational Control
When analyzed for their abundance in polysomes during sugar starvation, the majority of mRNA appears to be translationally repressed (183/245 genes), which is consistent with a general decrease in metabolic activities during starvation. This is also supported by the fact that about 18 ribosomal protein-encoding genes are translationally repressed. The functional classification of genes for which translational regulation was found to be prevalent is also in contrast to that of genes regulated by transcription (Supplemental Tables I and II). The main observation is that functions potentially involved in metabolism and in cell rescue, defense, and virulence, as defined by MIPS, drop down from 52% to 25%, while those linked to protein synthesis, cell cycle, and growth increased from 2.2% up to 34%. In particular, the cell cycle and DNA processing class, which is not represented in the transcription experiment, contains 11 genes in the translation experiment.
Beside the genes coding for ribosomal proteins, several mRNAs coding for RNA metabolism and translational functions were identified, such as diverse classes of RNA binding proteins and subunits of translation initiation factor 3, which have been described recently as being involved in the translational control of mRNA with specific 5′ leader sequences (Kim et al., 2004). Translationally regulated genes were also found to be involved in the function of different cell structures such as the cytosol, chloroplasts, mitochondria, and the nucleus (Supplemental Table III).
Interestingly, a few translationally repressed genes were found to code for chromatin components, such as histones, and for chromatin regulating proteins such as the AtHD1 histone deacetylase (At4g38130). This prompted us to further study the consequence of this regulation.
Sucrose Starvation Affects Histone Deacetylase Activity
The AtHD1 histone deacetylase is one of the genes that is the most translationally repressed by Suc starvation. Histone acetylation and deacetylation activate or repress transcription, yet the physiological relevance of reversible changes in chromatin structure and gene expressions is poorly understood. The disruption of AtHD1 induces large developmental abnormalities and induces acetylation changes of histone H3 and H4 (Tian et al., 2005). We therefore analyzed the state of acetylation of histone H4 by chromatin immunoprecipitation following Suc starvation. Figure 5 shows that the amount of DNA immunoprecipitated with an antiacetylated histone H4 increases significantly in starved cells compared to control cells, establishing therefore a direct link between translational control and chromatin activity.
Figure 5.
Quantification of DNA immunoprecipitated with an anti-histone H4 antibody from control or Suc-starved cells extracts expressed as the fraction of the DNA fluorescence in control extracts. Results are the average from three independent experiments from which three immunoprecipitations were performed.
DISCUSSION
The physiological consequences of sugar starvation have been studied in whole plants, isolated organs, and cultured cells and include a general decrease in enzymes and proteins involved in anabolic processes such as sugar metabolism, nitrogen assimilation, protein synthesis, and cell division and an increase in catabolic processes such as proteolysis, and amino acids and lipid degradation (Yu, 1999). Gene regulation mechanisms underlying these physiological changes have also been partially uncovered recently using large-scale analysis of transcriptional gene expression in Arabidopsis cells and plants (Contento et al., 2004; Price et al., 2004). In this work, we focused on translational mechanisms and more specifically on those concerning the ribosome loading of individual mRNA during starvation. To perform this study we analyzed the abundance of individual transcripts in polysomes between control and starved cells and compared it to the abundance of the same transcripts in the total RNA population. Therefore, this study included a complete analysis of the steady-state level of each mRNA (transcriptome analysis) that can be compared to that of Contento et al. (2004). The results of this analysis were validated by Q-RT-PCR of 33 genes, and for 31 of them (94%) the microarray data were confirmed. Out of 268 genes transcriptionally affected by sugar starvation, 68 were identical to the study of Contento et al. (2004; Supplemental Table V). Their regulation was also found to follow the same pattern during starvation. Overall, these studies confirm that Suc starvation induces the recycling of cellular components and the activation of stress and defense pathways to maintain cell viability.
The differences between these results and those of Contento et al. (2004) might be due to differences in the conditions used to establish starvation. In this study, cells were cultured in the presence of Suc for 9 d, where the Suc concentration has already dropped down to one-tenth of its original concentration, and then subculture was initiated in Suc-free medium. In the study of Contento et al. (2004), cells were cultured in the presence of Suc for 3 d then transferred to Suc-free medium. Therefore, starvation-induced cell stress and damage was probably more severe in this study. Although a similar percentage of genes related to cell defense and rescue was found activated in the two studies (15%–18.5%), their identities are often different. In this work, stress- and damage-related genes were frequently found activated, for example, wound-induced protein (At4g28240, At1g75380), harpin-induced protein (At5g53730), and nuclease (At1g68290). In addition, several genes related to osmotic stress are found activated, such as dehydrins (At1g20440, At1g54410), osmotin-like (At4g11650), and osmotic stress-induced Pro dehydrogenase (At3g30775). This suggests that the increased starvation might be accompanied to some extent by osmotic stress linked to increased lack of sugar from the culture media.
Translational Analysis Reveals the Dynamic Status of mRNA between Untranslated and Translated Fractions
The level of 21 mRNAs (class 2) was modified in the total RNA fraction, while their abundance within polysomes was found unchanged. This class therefore contains transcripts for which some form of translational buffering may occur. For transcripts that were found to increase in total RNA without increase in polysomal RNA, for example, UDP-Glc 4 epimerase (At1g63180) or Glc transporter (At1g11260), they might accumulate within ribonucleoprotein complexes, preventing them for being translated. Fifty-one genes were found to be coregulated at the total RNA and polysome levels, while the largest gene class (class 1, 224 genes) contains those for which a variation in polysomal RNA levels was observed without variation in total mRNA levels. In this class, most genes (183) were found to decrease in the polysomal RNA. This suggests that translational control represents the largest part of the gene regulation response to sugar starvation. Similarly, in a study performed with Arabidopsis seedlings submitted to hypoxia, about 70% of the detected transcripts show a decrease in polysomal RNA levels without a decrease in their total mRNA levels (Branco-Price et al., 2005). Conversely, in yeast (Saccharomyces cerevisiae) treated with the antiproliferative drug rapamycin the majority of gene variations in polysomal and total RNA were found homodirectional (Preiss et al., 2003). Interestingly, in yeast cells starved for amino acids, homodirectional variations in polysomal and total RNA were also mainly observed, while in the response to butanol stress, little correlation was found between the variations in total RNA and in polysomal RNA (Smirnova et al., 2005). Altogether, this suggests that the extent of the translational response might differ markedly depending on the physiological conditions and stresses, with some stresses inducing rapid and massive change in the populations of polysome-loaded mRNA without, or well before, transcriptional genome reorganization, while the cellular adaptation to other environmental changes are mainly orchestrated transcriptionally. In any case, however, it should be noted that the amount of translationally regulated mRNA is significant. Despite the fact that transcription is the source of all mRNAs, this shows that the fraction of a single mRNA species that undergoes translation can be dynamically tuned independently of the transcriptional activity.
Translational Control Affects Genes Involved in Cell Proliferation and Chromatin Structure
Transcripts affected by Suc starvation were assigned to functional categories. The relative representation of each functional category can then be compared between genes undergoing a variation of their total RNA abundance and those undergoing variation of their polysomal representation. This reveals that metabolic functions represent a large part of genes regulated either transcriptionally and translationally, which is in line with a large reorganization of primary and secondary metabolisms associated with modification of exogenous Suc status (Lloyd and Zakhleniuk, 2004; Price et al., 2004). Cell rescue and defense and energy are the second and third most abundant categories in the transcriptional analysis, respectively, a similar result to that observed by Contento et al. (2004). These categories are less represented in the translational analysis. Stress- and defense-related genes were found to be affected by sugar status in many studies and are known to be largely regulated at the transcriptional level (Singh et al., 2002). The most striking observation when comparing the transcriptional and translational gene lists is the increased relative abundance of translationally regulated transcripts related to the protein synthesis machinery and to the cell cycle. In this latter functional category, we did not detect any transcriptionally regulated mRNA, while 11 (12%) are found translationally controlled. Similarly, the number of genes linked to protein synthesis increased 10 times (2.5%–21.7%). This includes many genes coding for ribosomal proteins (16) as well as translation initiation (eIF3) and elongation factors (eEF2). Therefore, a large part of the regulation leading to the arrest of cell proliferation during starvation appears to act through translational repression of specific cell cycle-related mRNA and of mRNA coding for components of the translation machinery itself.
Protein synthesis is one of the most energy-consuming processes in proliferating cells and is an essential component of cell cycle progression. It is regulated by nutrient availability through evolutionary conserved signal transduction pathways, such as the TOR (Target of Rapamycin) pathway, acting directly at the level of translation of specific mRNAs involved in cell cycle progression and on the biogenesis of the ribosome (Martin and Hall, 2005). Components of these pathways are known to exist in plants, but their mode of action have not been yet thoroughly studied (Menand et al., 2002; Menand and Robaglia, 2004).
Among the mRNAs that were strongly translationally repressed during Suc starvation, we found the mRNA coding for the HD1 histone deacetylase (At4g38130). We further show that, in response to sugar starvation, an increased amount of acetylated histone H4 is associated with DNA (Fig. 5). HD1 is known to possess a reversible histone H4 deacetylase activity, and its blockage through antisense RNA or gene disruption induces a pleiotropic change in gene regulations leading to early senescence and a large array of developmental defects (Tian and Chen, 2001; Tian et al., 2003). Histone acetylation has been linked to modification of the transcriptional activity of several genes affected by changes in the structure of chromatin. Increased acetylation of histones leads to activation of gene expression (de Ruijter et al., 2003; Thiel et al., 2004). The regulation of the activity of chromatin components, such as histone deacetylases, in response to the environment or to developmental stimuli is yet poorly understood. Our results therefore suggest that at least some of the changes in gene expression during starvation are due to modifications of chromatin structure through translational control of HD1 histone deacetylase mRNA.
MATERIALS AND METHODS
Culture of Cell Suspension
The Arabidopsis (Arabidopsis thaliana) cell suspension culture described previously by Axelos et al. (1992) was grown in sterile culture medium containing Murashige and Skoog salts (Sigma), 0.5 mm kinetin, 0.34 mm 2,4-dichlorophenoxyacetic acid, vitamin mix (4 mm nicotinic acid, 1.26 μm calcium d-pantothenate, 2.66 mm Gly, 150 μm thiamine-HCl, 110 μm folic acid, 0.25 mm pyridoxine-HCl, 20 μm biotine, and 28 mm myoinositol), and 3% (w/v) Suc with pH adjusted to 5.6 using 1 n KOH. Cells in 100 mL of medium were incubated in a 250-mL conical flask and shaken at 125 rpm at 25°C in an orbital shaker (Infors) in continuous dark or light (50 μE). Every 9 d, the cells were sedimented by centrifugation at 700 rpm for 10 min, and the packed cell volume (PCV) was measured. After centrifugation (800g) for 5 min, the cell volume was determined in comparison to the culture volume. Subculturing was subsequently carried out by pipetting approximately 10 mL of the suspension (5% PCV) into 90 mL of fresh medium. The amount of Suc in the medium during the culture was determined using a Suc assay kit (Sigma). Control samples were transferred in Suc-containing media, while for starvation, cells were transferred in media without Suc. After 0, 6, 12, 24, 48, and 72 h of starvation, samples were removed for determination of PCV, viability assay, and/or total and polysomal RNA extraction.
Culture Viability Assay
After Suc starvation of suspension cultures for 0, 24, 48, and 72 h as described above, samples were taken and diluted. The viability of cells was determined using the CellTiter 96 Aqueous One Solution Cell Proliferation assay (Promega) that contains tetrazolium compound and an electron coupling reagent.
Polysome Fractionation
The cells were incubated with 0.35 mm of cycloheximide per milliliter for 10 min to arrest ribosome movement on polysomes before the cells were collected by filtration. Frozen cell pellet (300 mg) was ground into a fine powder in liquid nitrogen and then resuspended in 1 mL of lysis buffer (100 mm Tris-HCl, pH 8.4, 50 mm KCl, 25 mm MgCl2, 5 mm EGTA, 15.4 units/mL heparin, 18 μm cycloheximide, 15.5 μm chloramphenicol, and 10% detergent mix [20% (v/v) Triton X-100, 20% (v/v) Brij 35, 20% (v/v) Tween 40, 20% (v/v) NP-40, 20% (v/v) Polyethylene 10 Tridecyl Ether, and 10% (w/v) sodium deoxycholate]). Detergents help to disrupt cytoskeleton-associated polysomes. Nuclei and cell debris were eliminated by centrifugation at 7,000 rpm for 15 min in Eppendorf centrifuge at 4°C, the cytoplasmic extracts thus obtained were loaded on 11 mL 0.8 to 1.5 m Suc gradient (40 mm Tris-HCl, pH 8.4, 20 mm KCl, and 10 mm MgCl2). After centrifugation at 32,000g in a Beckman SW41 rotor for 135 min, gradients were fractionated into 1-mL fractions, with continuous monitoring of A260. Fractions 1 to 5 contain 80S monosomes and mRNP complexes and fractions 6 to 11 contain disomes and complexes of greater density. For microarray analysis, gradient fractions 7 to 11 were combined and used as the polysome sample.
Nucleic Acids Extraction
Suspension cell samples were collected by filtration and the cells were stored at −80°C until RNA extractions were performed. For total RNA extraction, a frozen cell pellet was ground into a fine powder in liquid nitrogen and RNA was isolated using a TRIzol extraction method. For polysomal samples, RNA was purified from Suc gradient fractions using an equal volume of TRIzol and precipitated with ethanol. RNA was fractionated by denaturing agarose gel electrophoresis.
Oligonucleotide Primer Design
Oligonucleotide primers were designed using Primer 3 and Amplify software. The amplicons were 150 to 180 nt. All primers pairs produced a single band of the expected size. Target genes are summarized in Table III.
Table III.
Accession numbers of genes studied by Q-RT-PCR
| Constitutive Control Genes | Genes Known to Be Regulated by Suc | Genes Transcriptionally Regulated
|
Genes Translationally Regulated
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Regulated in Three Conditions | Regulated at 6 h Only | Regulated at 48 h Only | Regulated in Light Conditions Only | Regulated at 48 h and Light Condition | Regulated in Dark Condition Only | Variation in Total RNA and Not in Polysomal RNA | Variation in Polysomal RNA and Not in Total RNA | ||
| 18S, Actin 8, αTubulin, Glyceraldehyde-3-P Dehydrogenase A, and Glyceraldehyde-3-P Dehydrogenase B | NR2, At1g37130; MCC, At1g03090; Branched-chain α keto-acid dehydrogenase E1α) | At1g51400, At2g03090, At2g30860, At4g37610, At5g07440, At5g22920 | At1g65180, At4g30280, At5g06140 | At1g54100, At3g04720, At3g16150, At3g12970, At3g44300, At3g59270, At4g02520, At4g11650, At5g13210, At5g65110 | At3g16240, At3g28180 | At1g68440, At2g33150, At5g49360 | At1g09720, At1g23360, At1g67120, At1g76870, At2g03090, At2g34360, At2g38640, At3g07690, At3g10950 At3g47540, At4g25100, At5g27410 | At1g51400 | At1g03090, At1g09415, At1g35320, At1g63800, At1g76410, At2g38870, At3g09840, At3g25250, At3g46690, At3g46720, At3g51910, At3g55910, At4g38130, At5g18630, At5g35790, At5g58720, At5g61440 |
Q-RT-PCR
The Q-RT-PCR was analyzed in three independent biological replicates, two of three replicates being the two samples used for microarray analysis. Two technical replicates were performed for each gene analyzed. cDNA was synthesized at 42°C for 1 h in 20 μL reaction mixture using AMV (Roche) according to the manufacturer's protocol. The reaction included oligo(dT) and 4 μg of DNase-treated total or polysomal RNA as template. Q-RT-PCR was performed in a 10-μL reaction using a Quantitect SYBR Green PCR kit (Qiagen) in ABI PRISM 7700 sequence detection system (Applied Biosystems) under the following conditions: 95°C, 10 min; 95°C, 15 s; and 60°C, 1 min. The reaction was performed for up to 40 cycles. Five genes (Table III) with a constitutive expression were used for data normalization.
Microarray Analysis
Arabidopsis Genome Oligo Set V1 (Operon, http://omad.operon.com/arabidopsis/index.php) were spotted onto type 7 star slides (Amersham no. RPK2331) using a Lucidea spotting robot (Amersham).
Total RNA Profiling
Additional RNA cleanup and DNase treatment were performed on Qiagen RNAeasy mini column (catalog no. 74106) according to the manufacturer's instructions. One microgram of total RNA was amplified using the aminoallyl MessageAmp aRNA kit (Ambion no. 1752). Antisense RNA synthesis was performed over 13 h, quality was controlled using the Agilent 2100 Bioanalyzer, and RNA were quantified with a Nanodrop ND-100. Five micrograms of lyophilized Aminoallyl RNA were labeled by coupling of NHS ester dyes (Cy5 Mon-Reactive Dye pack, Amersham no. PA25001 and Cy3 Mon-Reactive Dye pack, Amersham no. PA23001) and purified according to the Ambion Aminoallyl MessageAmp aRNA kit protocol. A total of 50 pmol of each of the labeled aRNA (Cy5 and Cy3, respectively) were vacuum dried to a final volume of 9 μL and mixed with 1 μL of Ambion fragmentation buffer (catalog no. 8740), the solution was incubated 15 min at 70°C and supplemented with 1 μL of the Stop solution (included in the fragmentation buffer packaging) then stored on ice. The two fragmented solutions were mixed to 2.5 μL of 0.1 mg mL−1 sonicated herring sperm DNA (Sigma), denatured 2 min at 95°C, and stored on ice. Then, 7.5 μL of hybridization buffer 2 (Amersham RPK0325) and 9 μL of 100% deionized formamide were added to the denatured probe and directly used for hybridization. Hybridizations were performed overnight at 42°C in Corning hybridization chambers (no. 2551). For each biological replicate a dye swap was performed. Slides were washed once in 1 ± SSC/0.2% SDS (10 min at 42°C), twice in 0.1 ± SSC/0.2% SDS (each 10 min at 42°C), twice in 0.1 ± SSC (each 1 min at room temperature), and 10 s in water at room temperature.
Polysomal RNA Profiling
Ten micrograms of polysomal RNA and 1 μg of random nanomers (RPKO158, Amersham) were incubated at 70°C for 10 min followed by 10 min incubation at room temperature. This solution was added to the labeling mix, which included 4 μL 5× first-strand buffer SSII (Life Technology), 2 μL 0.1 dithiothreitol (Amersham), 1 μL 10 mm dNTP mix (Amersham RPK0147), 1 μL Cy3 or Cy5 dCTP (Amersham PA55021), and 1 μL of SSH2 (Life Technology). The reaction was incubated for 4 h at 42°C. RNA was hydrolyzed by adding 2 μL of 2.5 m NaOH and incubating for 10 min at 37°C. The solution was neutralized by adding 10 μL of 2 m HEPES, pH 8.0. The probe was purified on a Qiagen Qiaquick PCR column according to the manufacturer's protocol.
Dye incorporation was quantified by spectrophotometry (Nanodrop ND-100). Prior to hybridization, probes (50 pmol of each dye) were dried and resuspended in 12 μL of water, denatured for 2 min at 95°C, and placed on ice. Hybridization was performed as described above.
Data Analysis
Hybridized arrays were scanned on a Gen III scanner (Molecular Dynamics) with constant photomultiplicator voltage (700 V). Measurements for each fluorophore were collected separately. Raw data were extracted with Array Vision 7.0 and subtracted from the local background. Data analysis followed methods outlined in Dudoit et al. (2000) and Yang et al. (2002). Raw data consisted of eight measurements per gene per probe. Due to the dye swap, one-half of the data correspond to the Cy3 fluorescent signal and the other half to the Cy5 fluorescent signal, four for each biological duplicate. To compensate for the intensity difference between the two fluorophores, dye swap normalization was performed first. Within a swap, the global intensity of one population was equalized for the two fluorophores. By doing this it was assumed that there is no major difference between the two populations and that only a small percentage of genes varies. For each time point, an ANOVA, using the two biological repetitions (eight data points per probe per gene), was performed using Spotfire 7.0 (Spotfire), and genes were selected with a P value < 0.05 and on the basis of an expression ratio larger than 2.
Assignment of Genes to Functional Category
The functional category of genes was determined on MIPS annotation (http://mips.gsf.de/proj/thal/db/index/html). Genes were categorized into 12 major classes, and some genes contain multiple functional categories.
Chromatin Immunoprecipitation
Histones were cross linked to DNA by adding 1% formaldehyde to culture medium. Pelleted cells were resuspended in lysis buffer (1% SDS, 50 mm Tris-HCl, pH 8, 10 mm EDTA) and sonicated to shear DNA to lengths between 200 and 1,000 bp. The anti-acetyl-histone H4 antibody (06-866, Upstate) was added to the supernatant and incubated overnight with rotation. After adding salmon sperm DNA/protein A agarose slurry (Upstate, 16-157C), pelleted agarose was washed and the precipitated DNA was quantified by binding to Hoechst 33258 dye and recording fluorescence at an emission wavelength of 458 nm and an excitation of 356 nm on a Cary Eclipse Fluorescence spectrophotometer (Varian).
Supplementary Material
Acknowledgments
We thank Audrey Creff for expert technical assistance during the early part of this work, Christian Triantaphylidés and Marie-Héléne Montané for access to Q-RT-PCR equipment, Michel Delseny for support, Laboratoire de génétique et biophysique des Plantes and Biogemma lab members for useful discussions, and Keith Dudley for corrections on the manuscript.
This work was supported by the French Ministry of Industry key technologies post-genome program.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: C. Robaglia (robaglia@luminy.univ-mrs.fr).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.106.079418.
References
- Aubert S, Gout E, Bligny R, Marty-Mazars D, Barrieu F, Alabouvette J, Marty F, Douce R (1996) Ultrastructural and biochemical characterization of autophagy in higher plant cells subjected to carbon deprivation: control by the supply of mitochondria with respiratory substrates. J Cell Biol 133: 1251–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelos M, Curie C, Mazzolini L, Bardet C, Lescure B (1992) A protocol for transient gene expression in Arabidopsis thaliana protoplast isolated cell suspension. Plant Physiol Biochem 30: 123–128 [Google Scholar]
- Bassham DC (2002) Golgi-independent trafficking of macromolecules to the plant vacuole. Adv Bot Res 38: 65–92 [Google Scholar]
- Branco-Price C, Kawaguchi R, Ferreira RB, Bailey-Serres J (2005) Genome-wide analysis of transcript abundance and translation in Arabidopsis seedlings subjected to oxygen deprivation. Ann Bot (Lond) 96: 647–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouquisse R, James F, Raymond P, Pradet A (1991) Study of glucose starvation in excised maize root tips. Plant Physiol 96: 619–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan MT, Yu S-M (1998) The 3′ untranslated region of a rice alpha-amylase gene functions as a sugar-dependent mRNA stability determinant. Proc Natl Acad Sci USA 95: 6543–6547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M-H, Liu L-F, Chen Y-R, Wu HK, Yu S-M (1994) Expression of α-amylases, carbohydrate metabolism, and autophagy in cultured rice cells is coordinately regulated by sugar nutrient. Plant J 6: 626–636 [DOI] [PubMed] [Google Scholar]
- Cheng WH, Taliercio EW, Chourey PS (1999) Sugars modulate an unusual mode of control of the cell-wall invertase gene (Incw1) through its 3′ untranslated region in a cell suspension culture of maize. Proc Natl Acad Sci USA 96: 10512–10517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contento AL, Kim SJ, Bassham DC (2004) Transcriptome profiling of the response of Arabidopsis suspension culture cells to Suc starvation. Plant Physiol 135: 2330–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coruzzi G, Bush DR (2001) Nitrogen and carbon nutrient and metabolite signalling in plants. Plant Physiol 125: 61–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coruzzi G, Zhou L (2001) Carbon and nitrogen sensing and signalling in plants: emerging matrix effect. Curr Opin Plant Biol 4: 247–253 [DOI] [PubMed] [Google Scholar]
- de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB (2003) Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J 15: 737–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudoit P, Yang YH, Callow MJ, Speed TP (2000) Statistical methods for identifying differentially expressed genes in replicated cDNA microarray experiments. Technical report no. 578. Department of Statistics, University of California, Berkeley, CA
- Finkelstein RR, Gibson SI (2002) ABA and sugar interactions regulating development: cross-talk or voices in a crowd? Curr Opin Plant Biol 5: 26–32 [DOI] [PubMed] [Google Scholar]
- Fujiki Y, Ito M, Nishida I, Watanebe A (2001) Leucine and its keto acid enhance the coordinated expression of genes for branched-chain amino acid catabolism in Arabidopsis under sugar starvation. FEBS Lett 449: 161–165 [DOI] [PubMed] [Google Scholar]
- Gazzarrini S, McCourt P (2001) Genetics interactions between ABA, ethylene and sugar signalling pathways. Curr Opin Plant Biol 4: 387–391 [DOI] [PubMed] [Google Scholar]
- Ho S-L, Chao Y-C, Tong W-F, Yu S-M (2001) Sugar coordinately and differentially regulates growth and stress-related gene expression via a complex signal transduction network and multiple control mechanisms. Plant Physiol 125: 877–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journet EP, Bligny R, Douce R (1986) Biochemical changes during sucrose deprivation in higher plant cells. J Biol Chem 261: 3193–3199 [PubMed] [Google Scholar]
- Kim T-H, Kim B-H, Yahalom A, Chamovitz DA, Von Arnim AG (2004) Translational regulation via 5′ mRNA leader sequences revealed by mutational analysis of the Arabidopsis translation initiation factor subunit elF3h. Plant Cell 16: 3341–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch KE (1996) Carbohydrate-modulated gene expression in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 509–540 [DOI] [PubMed] [Google Scholar]
- Lloyd JC, Zakhleniuk OV (2004) Responses of primary and secondary metabolism to sugar accumulation revealed by microarray expression analysis of the Arabidopsis mutant, pho3. J Exp Bot 55: 1221–1230 [DOI] [PubMed] [Google Scholar]
- Martin DE, Hall MN (2005) The expanding TOR signalling network. Curr Opin Cell Biol 17: 159–166 [DOI] [PubMed] [Google Scholar]
- Menand B, Desnos T, Nussaume L, Berger F, Bouchez D, Meyer C, Robaglia C (2002) Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc Natl Acad Sci USA 99: 6422–6427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menand B, Robaglia C (2004) Plant cell growth. In MN Hall, G Thomas, eds, Cell Growth: Control of Cell Size, Vol 42. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 625–637
- Menges M, Murray JA (2002) Synchronous Arabidopsis suspension cultures for analysis of cell-cycle gene activity. Plant J 30: 203–212 [DOI] [PubMed] [Google Scholar]
- Moore B, Zhou L, Rolland F, Hall Q, Cheng WH, Liu YX, Hwang I, Jones T, Sheen J (2003) Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300: 332–336 [DOI] [PubMed] [Google Scholar]
- Moriyasu Y, Ohsumi Y (1996) Autophagy in tobacco suspension-cultured cell in response to sucrose starvation. Plant Physiol 111: 1233–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss T, Baron-Benhamou J, Ansorge W, Hentze MW (2003) Homodirectional changes in transcriptome composition and mRNA translation induced by rapamycin and heat shock. Nat Struct Biol 10: 1039–1047 [DOI] [PubMed] [Google Scholar]
- Price J, Laxmi A, St Martin SK, Jang J-C (2004) Global transcription profiling reveals multiple sugar signal transduction mechanisms in Arabidopsis. Plant Cell 16: 2128–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riou-Khamlichi C, Menges M, Healy JM, Murray JA (2000) Sugar control of the plant cell cycle: differential regulation of Arabidopsis D-type cyclin gene expression. Mol Cell Biol 20: 4513–4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roitsch T (1999) Source-sink regulation by sugar and stress. Curr Opin Plant Biol 2: 198–206 [DOI] [PubMed] [Google Scholar]
- Rolland F, Moore B, Sheen J (2002) Sugar sensing and signalling in plants. Plant Cell 14: 185–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook F, Gerrits N, Kortstee A, van Kampen M, Borrias M, Weisbeek P, Smeekens S (1998) Sucrose-specific signalling represses translation of the Arabidopsis ATB2 bZIP transcription factor gene. Plant J 15: 253–263 [DOI] [PubMed] [Google Scholar]
- Sheen J, Zhou L, Jang JC (1999) Sugars as signalling molecules. Curr Opin Plant Biol 2: 410–418 [DOI] [PubMed] [Google Scholar]
- Singh KB, Foley RC, Onate-Sanchez L (2002) Transcription factors in plant defense and stress responses. Curr Opin Plant Biol 5: 430–436 [DOI] [PubMed] [Google Scholar]
- Smeekens S (2000) Sugar-induced signal transduction in plants. Annu Rev Plant Physiol 51: 49–81 [DOI] [PubMed] [Google Scholar]
- Smirnova JB, Selley JN, Sanchez-Cabo F, Carroll K, Eddy AA, McCarthy JE, Hubbard SJ, Pavitt GD, Grant CM, Ashe MP (2005) Global gene expression profiling reveals widespread yet distinctive translational responses to different eukaryotic translation initiation factor 2B-targeting stress pathways. Mol Cell Biol 25: 9340–9349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M (1999) Nitrate regulation of metabolism and growth. Curr Opin Plant Biol 2: 178–186 [DOI] [PubMed] [Google Scholar]
- Thiel G, Lietz M, Hohl M (2004) How mammalian transcriptional repressors work. Eur J Biochem 271: 2855–2862 [DOI] [PubMed] [Google Scholar]
- Tian L, Chen ZJ (2001) Blocking histone deacetylation in Arabidopsis induces pleiotropic effects on plant gene regulation and development. Proc Natl Acad Sci USA 98: 200–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Fong MP, Wang JJ, Wei NE, Jiangs H, Doerge R, Chen ZJ (2005) Reversible histone acetylation and deacetylation mediate genome-wide promoter-dependent and locus-specific changes in gene expression during plant development. Genetics 169: 337–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Wang J, Fong MP, Chen M, Cao H, Gelvin SB, Chen ZJ (2003) Genetic control of developmental changes induced by disruption of Arabidopsis histone deacetylase 1 (AtHD1) expression. Genetics 165: 399–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese A, Elzinga N, Wobbes B, Smeekens S (2004) A conserved upstream open reading frame mediates sucrose-induced repression of translation. Plant Cell 16: 1717–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP (2002) Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res 30: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu SM (1999) Cellular and genetic responses of plants to sugar starvation. Plant Physiol 121: 687–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.