Abstract
We sequenced 2.2 Mb representing triplicated genome segments of Brassica oleracea, which are each paralogous with one another and homologous with a segmentally duplicated region of the Arabidopsis thaliana genome. Sequence annotation identified 177 conserved collinear genes in the B. oleracea genome segments. Analysis of synonymous base substitution rates indicated that the triplicated Brassica genome segments diverged from a common ancestor soon after divergence of the Arabidopsis and Brassica lineages. This conclusion was corroborated by phylogenetic analysis of protein families. Using A. thaliana as an outgroup, 35% of the genes inferred to be present when genome triplication occurred in the Brassica lineage have been lost, most likely via a deletion mechanism, in an interspersed pattern. Genes encoding proteins involved in signal transduction or transcription were not found to be significantly more extensively retained than those encoding proteins classified with other functions, but putative proteins predicted in the A. thaliana genome were underrepresented in B. oleracea. We identified one example of gene loss from the Arabidopsis lineage. We found evidence for the frequent insertion of gene fragments of nuclear genomic origin and identified four apparently intact genes in noncollinear positions in the B. oleracea and A. thaliana genomes.
INTRODUCTION
Polyploidy has occurred extensively in angiosperms (Leitch and Bennett, 1997) and is recognized as a key factor in the evolution of plants and their genomes (Wendel, 2000). Even the exceptionally small genome of the model species Arabidopsis thaliana has a polyploid origin, although there has been extensive rearrangement and loss of genes from the duplicated genome segments (Arabidopsis Genome Initiative, 2000; Blanc et al., 2000; Ku et al., 2000; Mayer et al., 2001). Thus, we regard genome evolution in plants as occurring by a cyclical process of polyploidy followed by diploidization, by which stable meiosis is restored (a rapid process) and the gene number reduces toward that of the diploid ancestor (a slow process). Understanding the mechanisms involved in the structural and functional evolution of genomes during this cycle is of major importance to plant biology. As well as helping to understand the processes that differentiate the genomes of closely related species, it may help us identify and understand the limited conservation of genome microstructure that can be detected between distantly related species, such as A. thaliana and tomato (Solanum lycopersicum) (Ku et al., 2000) or rice (Oryza sativa) (Mayer et al., 2001), the lineages of which diverged from that of A. thaliana ∼150 and 200 million years ago (MYA), respectively (Wolfe et al., 1989; Yang et al., 1999).
The Brassiceae tribe is a monophyletic group within the Brassicaceae family (Cruciferae) (Warwick and Black, 1997). It includes the cultivated Brassica species, which are the group of crops most closely related to A. thaliana. The Brassica and Arabidopsis lineages diverged ∼20 MYA (Yang et al., 1999). Phylogenetic analysis groups the Brassica species into the nigra and rapa/oleracea lineages (Warwick and Black, 1991), which diverged ∼8 MYA (Lysak et al., 2005). Early comparative studies conducted at the level of genetic linkage maps revealed extensive duplication within Brassica genomes (Lagercrantz and Lydiate, 1996) and tracts of collinearity disrupted by multiple rearrangements between the genomes of B. nigra and A. thaliana (Lagercrantz, 1998). Subsequent comparative analyses between B. oleracea linkage maps and the A. thaliana genome identified numerous one-to-one segmental relationships and apparent genome duplications, in addition to genome triplications (Lan et al., 2000; Babula et al., 2003; Lukens et al., 2003). Investigations of regions of the genome of B. oleracea containing specific genes of interest also revealed various numbers of related genome segments (Quiros et al., 2001; Suzuki et al., 2003). Sequence analysis of a region of the B. oleracea genome containing a family of oxoglutarate-dependent dioxygenase genes revealed extensive rearrangement compared with the homologous region of the genome of A. thaliana (Gao et al., 2004). However, the high frequency of rearrangements within tandem arrays of genes (Michelmore and Meyers, 1998; Noel et al., 1999) means that such a region is unlikely to be representative of the evolution of the genome as a whole.
A number of alternative hypotheses have been developed to explain how Brassica genome structure evolved, including an ancient tetraploidy and/or numerous segmental duplications (Lukens et al., 2004). The most likely, however, is that the diploid Brassica species, including B. oleracea, are paleohexaploids. Although there was already strong evidence for this hypothesis (Schmidt et al., 2001; Parkin et al., 2003), decisive support came in 2005 from two reports. In the first, an 8.7-Mb BAC contig of the A. thaliana genome was used to trace homologous chromosome regions in 21 species of the family Brassicaceae (Lysak et al., 2005). The results revealed that a distinctive feature of Brassiceae tribe species is that they contain triplicated genomes, indicative of being derived from a hexaploid ancestor with basic genomes similar to that of A. thaliana. In the second report, a genome-wide analysis was conducted using a linkage map of B. napus (a recent allotetraploid, formed by hybridization of B. oleracea and B. rapa) that consisted of >1000 linked restriction fragment length polymorphism loci that were mapped to homologous positions in the A. thaliana genome based on sequence similarity (Parkin et al., 2005). Twenty-one segments of the genome of A. thaliana, representing almost its entirety, could be duplicated and rearranged to generate the structure of the B. napus genome. The majority of the A. thaliana genome (11 segments) could be aligned to six segments of the B. napus genome, indicative of triplication in the genomes of both progenitor species. However, three segments of the A. thaliana genome aligned to seven segments of the B. napus genome, indicative of additional segmental duplications, and five segments of the A. thaliana genome aligned to only four or five segments of the B. napus genome, indicative of segmental loss from the hexaploid.
The results of comparative studies of genome microstructure, using physical mapping techniques, in targeted regions of the genomes of B. oleracea, B. rapa, and B. napus, relative to the genome of A. thaliana, are consistent with the fundamentally triplicated nature of the diploid Brassica genome (O'Neill and Bancroft, 2000; Rana et al., 2004; Park et al., 2005). Three paralogous segments were identified in each Brassica genome. Although synteny of genes within these collinear segments was found to be largely well conserved, there was evidence of extensive gene loss, which occurred in an interspersed pattern. The results also showed that the genomes analyzed could be differentiated by a few differences in gene content and small-scale rearrangements.
Brassica polyploids can be readily synthesized artificially and display genome instability that has been interpreted as indicating that a high rate of genome evolution occurs in polyploids (Song et al., 1995). However, natural Brassica polyploids appear to show relatively little change in genome structure compared with their progenitor species at the level of either macrostructure (Parkin et al., 1995) or microstructure (Rana et al., 2004). We aimed to characterize, at the sequence level, the consequences of the naturally occurring polyploidy observed in B. oleracea and to interpret the observations in relation to potential mechanisms of plant genome evolution. To do this, we analyzed a related set of segments of the genome of B. oleracea that have been analyzed previously by physical mapping techniques (O'Neill and Bancroft, 2000). These correspond to two sets of paralogous segments, one set being homologous with a segment of the A. thaliana genome on chromosome 4 and the other set being homologous with a segment of the A. thaliana genome on chromosome 5. These two regions of the A. thaliana genome are related by duplication (Bancroft, 2000), corresponding to the last, or α, whole genome duplication (Bowers et al., 2003), and provide outgroups for the analysis of the triplicated segments within the B. oleracea genome. Because we have complete sets of sequence data in some regions, we can also use an amalgamation of the genes present in the B. oleracea sequences as an outgroup, enabling us to detect evolutionary events affecting the duplicated segments within the corresponding parts of the A. thaliana genome. As the nigra and rapa/oleracea lineages of Brassica species diverged substantially more recently than when the genome triplication that characterizes the (monophyletic) Brassiceae tribe occurred (Lysak et al., 2005), we refer to the lineage leading to B. oleracea as the Brassica lineage, because the principal findings (i.e., those relating to the consequences of genome triplication) are expected to be common to all Brassica species.
RESULTS
Generation of Brassica Sequence Contigs
A seed BAC, located approximately in the middle of each of the seven BAC contigs described by O'Neill and Bancroft (2000), was chosen for sequencing. In addition, all clones in the seven BAC contigs they reported were end-sequenced. Upon completion of the sequencing of each seed BAC, BAC end sequence data were used to determine the overlapping BAC that would produce the maximum amount of additional sequence. In some cases, BAC end sequence data were ambiguous and BACs were fingerprinted to minimize the possibility of choosing a BAC from the wrong contig. Most BACs were sequenced to GenBank phase 3 finished standards, although there were some unsequenceable regions. After the completion of sequencing, the BACs from each contig were assembled to produce a single sequence assembly, as summarized in Table 1. In all cases, these assemblies showed that the original assignment of the BACs to their respective contigs on the basis of physical mapping techniques was correct.
Table 1.
BAC Clones Sequenced for the Assembly of the Sequence Contigs
| Contig | Constituent Clones from the JBo BAC Library (Individual Length) (bp) | Length (bp) | GenBank Accession Number | ||
|---|---|---|---|---|---|
| A | 029O13 (100,534) | 061N03a (159,491) | 019I13 (123,462) | 356,505 | AC183495 |
| B | 026I24 (131,143) | 076G14a (161,694) | 284,024 | AC183493 | |
| C | 002D23 (133,880) | 039B15a (125,930) | 046B11 (179,111) | 285,752 | AC183494 |
| D | 077G15 (190,238) | 08K11a (158,552) | 041N20 (154,149) | 354,288 | AC183498 |
| E | 087H24 (159,860) | 058E11a (125,936) | 070A17 (140,046) | 385,314 | AC183496 |
| F | 041I3 (149,738) | 084C3a (162,651) | 014I23 (149,738) | 335,918 | AC183497 |
| G | 078G6a (137,351) | 044I17 (125,280) | 236,640 | AC183492 | |
Seed BAC.
Gene Content of Brassica Sequence Contigs
Annotation of the sequence data resulted in the construction of 539 gene models. These were distributed across the seven sequence contigs, as summarized in Table 2, with the highest density in contig A (93 gene models in 356 kb) and the lowest in contig E (85 gene models in 385 kb). Statistics of gene structure and gene density are shown in Table 3. Base composition was found to be very similar in B. oleracea and A. thaliana. The mean length of a B. oleracea gene (ATG to stop codon) is only 70% of that of Arabidopsis genes. This difference appears to be attributable to shorter exons in B. oleracea (233 versus 276 bp) and, on average, one fewer exon per gene. The gene density of one per 6.6 kb (excluding sequences annotated as transposon-related) is lower than that in A. thaliana (one per 4.5 kb). This difference does not appear to be attributable to a bias caused by the inclusion of species-specific hypothetical genes in the analysis. When the same analysis was performed using only conserved orthologues from the two genomes, similar results were obtained. The average number of exons per gene was 5.4 ± 0.4 (mean ± se) for Arabidopsis and 5.0 ± 0.3 for Brassica. The average exon size was 258.3 ± 11.9 for Arabidopsis and 211.2 ± 8.6 for Brassica. The average intron size was 142.7 ± 5.4 for Arabidopsis and 131.2 ± 4.9 for Brassica. The difference in exon size is highly significant (0.001 < P < 0.002) and does not appear to be attributable to a sampling bias. Details of the annotation are available online via GBrowse displays, with the URL for each sequence contig listed in Table 4.
Table 2.
Summary of Features Identified in 2,238,441 bp of B. oleracea Genome Sequences
| Contig
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Feature | A | B | C | D | E | F | G | Total |
| Gene models in A. thaliana | 53 | 42 | 31 | 67 | 31 | 27 | 20 | 271 |
| Gene models in B. oleracea | 93 | 73 | 67 | 91 | 85 | 76 | 54 | 539 |
| Conserved collinear genes | 35 | 27 | 24 | 41 | 13 | 20 | 17 | 177 |
| Genes conserved (%) | 68 | 64 | 77 | 61 | 42 | 74 | 85 | 66 |
| Fragments of collinear genes | 3 | 2 | 0 | 2 | 1 | 2 | 0 | 10 |
| Predicted transposons | 7 | 7 | 13 | 2 | 17 | 15 | 8 | 69 |
| Putatively intact relocated genes | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 4 |
| Gene fragments within models | 16 | 11 | 18 | 9 | 16 | 14 | 11 | 95 |
| Gene fragments outside models | 11 | 8 | 8 | 7 | 12 | 9 | 5 | 60 |
Table 3.
Comparison of Overall Composition of Annotated Genes (Transposons and Pseudogenes Excluded)
| Feature | B. oleracea | A. thaliana |
|---|---|---|
| GC content overall | 36.0% | 36.0% |
| GC content exons | 45.5% | 42.8% |
| GC content introns | 33.1% | 32.5% |
| GC content intergenic | 33.0% | 32.2% |
| Exon length | 233 | 276 |
| Intron length | 166 | 164 |
| Number of exons per gene | 4.5 | 5.4 |
| Gene length | 1629 bp | 2287 bp |
| Protein length | 349 amino acids | 417 amino acids |
| Gene density (kb/gene) | 6.6 | 4.5 |
Table 4.
URLs for Annotation Displays
Chronology of Lineage Divergence
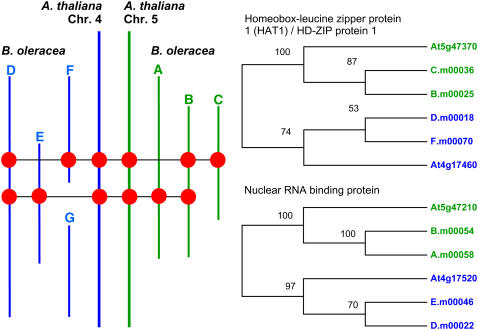
The sequence data available to us represent A. thaliana genome segments related by the α genome duplication and their sets of homologous segments in the B. oleracea genome. This provides numerous gene families for phylogenetic analysis. There are three examples of gene families in which members are single copy in both the A. thaliana chromosome 4 and 5 segments and for which there are two members in each set of B. oleracea sequence contigs. Robust phylogenies (i.e., for which trees could be constructed consistently using both neighbor-joining and maximum likelihood methods) could be constructed for two of these protein families, as shown in Figure 1. The alignments are shown in Supplemental Figure 1 online. The phylogenies confirm that the B. oleracea contigs were correctly assigned to their respective A. thaliana genome segments in the original report (O'Neill and Bancroft, 2000) and indicate that divergence of the Brassica paralogues occurred after the divergence of the Brassica and Arabidopsis lineages.
Figure 1.
Phylogenetic Relationships of Conserved Genes.
Left, overview of the positions of the members of the analyzed gene family across the sequence contigs. Right, phylogenies illustrated as maximum likelihood cladograms. Bootstrap support, calculated from 1000 replicates, is shown at each branch as a percentage.
To determine the relative divergence of the respective lineages, we calculated distances for every possible paralogous/homologous gene pair. The results, which are summarized in Table 5, show that there is no significant difference between the mean synonymous base substitution rate (Ks) value for any comparison between sets of Brassica genes and their Arabidopsis homologues. The Ks value for all Brassica–Arabidopsis comparisons calculated from the pooled data is 0.53 ± 0.19 (n = 179). Similarly, none of the mean Ks values for any pairwise comparison between sets of Brassica paralogues is significantly different from any other. The Ks value for all Brassica–Brassica comparisons calculated from the pooled data is 0.44 ± 0.19 (n = 64), a value that is significantly different from the Brassica–Arabidopsis mean Ks (P = 0.001). Thus, the triplication of the Brassica genome occurred some time after the divergence of the Brassica and Arabidopsis lineages from a common ancestor. However, it is not possible to determine whether the triplication in Brassica occurred as a single event or as successive events.
Table 5.
Ks Values for the Estimation of Time since the Divergence of Genome Segments
| At5 | A | B | At4 | D | E | |||
|---|---|---|---|---|---|---|---|---|
| No. genesa | At4 | 33 | ||||||
| Ksb | 0.99 ± 0.41 | |||||||
| ID nucleotides (%)c | 71.8 ± 13.6 | |||||||
| ID amino acids (%)d | 70.2 ± 15.9 | |||||||
| No. genes | A | 34 | D | 37 | ||||
| Ks | 0.50 ± 0.17 | 0.53 ± 0.19 | ||||||
| ID nucleotides (%) | 84.3 ± 5.5 | 84.4 ± 5.5 | ||||||
| ID amino acids (%) | 83.1 ± 9.7 | 84.3 ± 9.1 | ||||||
| No. genes | B | 23 | 15 | E | 12 | 5 | ||
| Ks | 0.50 ± 0.14 | 0.40 ± 0.13 | 0.46 ± 0.12 | 0.37 ± 0.11 | ||||
| ID nucleotides (%) | 84.2 ± 5.0 | 86.1 ± 4.9 | 84.9 ± 4.1 | 86.5 ± 3.6 | ||||
| ID amino acids (%) | 82.6 ± 9.0 | 84.6 ± 8.4 | 83.1 ± 7.5 | 85.3 ± 6.1 | ||||
| No. genes | C | 22 | 13 | 9 | F + G | 51 | 20 | 2 |
| Ks | 0.49 ± 0.15 | 0.38 ± 0.14 | 0.45 ± 0.12 | 0.58 ± 0.23 | 0.49 ± 0.27 | 0.57 ± 0.35 | ||
| ID nucleotides (%) | 84.5 ± 4.5 | 85.6 ± 8.2 | 82.9 ± 5.9 | 83.0 ± 5.5 | 83.6 ± 7.9 | 79.0 ± 7.4 | ||
| ID amino acids (%) | 83.7 ± 7.6 | 84.5 ± 13.1 | 79.7 ± 10.4 | 82.2 ± 9.4 | 81.2 ± 11.4 | 71.8 ± 5.7 |
Number of genes analyzed in this pairwise comparison of paralogues or homologues.
Ks for this set of genes ± sd.
Percentage nucleotide identity for this set of genes ± sd.
Percentage amino acid identity for this set of genes ± sd.
Using the commonly adopted estimate of mutational rate of 1.5 × 10−8 synonymous substitutions per site per year (Koch et al., 2000), we estimated the times of lineage divergence. Although mutation rate estimates are likely to be imprecise, the approach does provide a robust analysis of the order of events, providing that the same methodology is used. We estimated the Brassica and Arabidopsis lineages to have diverged ∼35 MYA, with the subsequent divergence of the three Brassica paralogous segments from a common Brassica ancestor ∼29 MYA (i.e., distinctly after divergence from the Arabidopsis lineage). Using the same methodology and the sequences of the A. thaliana genome segments, we estimate the α genome duplication to have occurred ∼66 MYA. Therefore, the triplicated subgenomes of Brassica species have been diverging for approximately half the length of time that the α genome duplicated pairs, as represented in A. thaliana, have been evolving.
Gene Loss
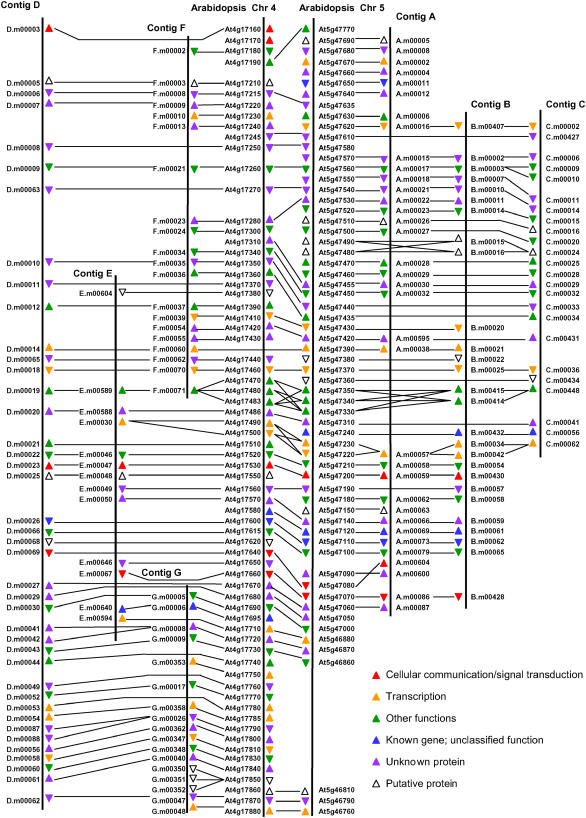
In total, 177 of the genes identified in the B. oleracea sequences correspond to collinear homologues of genes in the Arabidopsis genome segments, as summarized schematically in Figure 2. These genes represent 66% of the ∼271 genes inferred to be present in these genome segments upon genome triplication (i.e., the sum of the number of genes in the corresponding segments of the A. thaliana genome). There is only one exception to conservation of synteny and strand orientation: the order of the homologues in contig A of At5g47090 and At5g47080 (i.e., A.m00604 and A.m00600) is reversed. As the strand orientation of At5g47090 and A.m00600 is conserved, whereas that of At5g17080 and A.m000604 is reversed, the rearrangement appears to have involved the translocation and inversion of the ancestor of At5g47080 or A.m00604 in one lineage.
Figure 2.
Alignment of Conserved Genes.
Vertical lines denote sequence contigs, and horizontal lines join members of homologous/paralogous gene families. The triangle by each gene model name indicates the coding stand of the gene and is color-coded to indicate the Munich Information Center for Protein Sequences FunCat functional classification of the predicted protein, as shown in the key at bottom.
Despite the highly conserved synteny of genes represented in both genomes, there is extensive breakdown of the conservation of genome microstructure in that many genes expected to be present in the B. oleracea sequence contigs were not predicted by the annotation pipeline. An analysis of the sequence data using BLASTN alignment revealed that, in most cases, there is no trace of sequence homology with the missing genes. However, fragments of 10 genes were detected in collinear positions. These were homologues of At5g47370, At5g47380, and At5g47190 in contig A, homologues of At5g47360 and At5g47080 in contig B, homologues of At4g17430 and At4g1766 in contig D, a homologue of At4g17390 in contig E, and homologues of At4g17270 and At4g17330 in contig F. In all cases, only part of the gene is present, but the sequences of this part were well conserved. As an example, the alignment between the coding sequence of At5g47080 and the sequences present in B. oleracea contig B is shown in Supplemental Figure 2 online.
Previous studies have indicated that genes involved in signal transduction and transcription may be preferentially retained after duplication (Blanc and Wolfe, 2004). We classified the conserved genes using the Munich Information Center for Protein Sequences functional category of the proteins (FunCat) that they are predicted to encode, as indicated in Figure 2. Collectively, the A. thaliana chromosome 4 and 5 segments contain 175 gene models. For the 29 classified as cellular communication/signal transduction or transcription, there are 38 orthologues in the B. oleracea sequences, and for the 43 classified with other functions, there are 50 orthologues in the B. oleracea sequences. These proportions do not differ from the null hypothesis of equal retention irrespective of functional classification (χ2 = 0.32). The only underrepresented category was that described by FunCat as putative proteins, which correspond to a subset of the proteins annotated as hypothetical protein by The Institute for Genomic Research (TIGR) pipeline. For the 28 classified as putative proteins in A. thaliana, there are 18 orthologues in B. oleracea (χ2 = 4.46, P < 0.05).
We can identify one example of gene loss having occurred from the A. thaliana lineage. Genes A.m00038 and B.m00021 are orthologous with At5g47390. They are also closely related to genes D.m00014 and F.m00060, which occur in collinear positions in the B. oleracea contigs related to the A. thaliana chromosome 4 segment. However, there is no homologue in the corresponding position in the A. thaliana genome (or anywhere else in the genome). The occurrence of homologous genes in two A. thaliana chromosome 4–related B. oleracea contigs (D and F), and in the corresponding A. thaliana chromosome 5 segments, indicates that there was a copy of the gene, between the ancestors of At4g17430 and At4g17440, at the time of divergence of the Arabidopsis and Brassica lineages that has subsequently been lost from the Arabidopsis lineage.
Analysis of Tandem Arrays
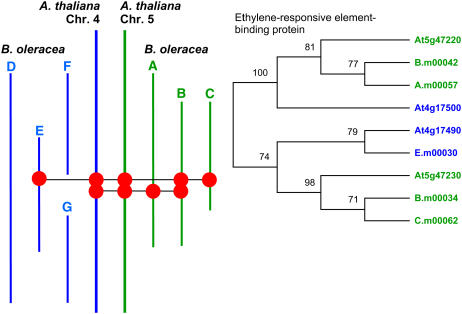
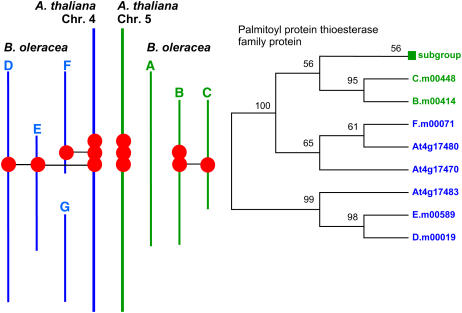
One of the unexpected features of plant genome structure is the frequent occurrence of tandem arrays of genes. The Brassica sequence contigs contain numerous examples of tandem arrays. In four cases, these represent amplification of genes present in single copy per locus in A. thaliana. These were D.m00041/D.m00042 related to At4g17720, D.m0087/D.m0088 related to At4g17790, G.m00350/G.m00351/G.m00352 related to At4g17850, and C.m00009/C.m00010 related to At5g47560. The A. thaliana chromosome 4 and chromosome 5 segments contain one common duplicate array (At4g17490/At4g17500 related to At5g47220/At5g47230) and one common triplicate array (At4g17470/At4g17480/At4g17483 related to At5g47330/At5g47340/At5g47350). The representation of these gene families is generally reduced in the B. oleracea genome segments, as shown in Figure 2. However, we can use the sequences of the protein families to identify orthology relationships and to assess how stable the arrays observed in A. thaliana might be. We constructed phylogenies for the protein families encoded by these sets of genes. Phylogenetic analysis of the protein family related to the tandem duplication resulted in consistent trees using both neighbor-joining and maximum likelihood methods, as illustrated in Figure 3. The alignments are shown in Supplemental Figure 3 online. The results show that B. oleracea genes B.m00034 and C.m00062 are orthologous with At5g47230, A.m00057 and B.m00042 are orthologous with At5g47220, E.m00030 is orthologous with At4g17490, and the sequenced regions contain no orthologue of At4g17500. The phylogeny also indicates that the tandem duplication is of ancient origin, predating the α genome duplication. Phylogenetic analyses were conducted for the protein family related to the tandem triplication. Part of the tree was consistently constructed using either the neighbor-joining or maximum likelihood method, as illustrated in Figure 4. The alignments are shown in Supplemental Figure 4 online. The results show that B. oleracea genes D.m00019 and E.m00589 are orthologous with At4g17483, F.m00071 is orthologous with At4g17480, and the sequenced regions contain no orthologue of At4g17470. However, we were unable to construct a robust phylogeny for At5g47330, At5g47340, At5g47350, and B.m00415 and were unable to define the orthology relationship of B. oleracea genes B.m00414 and C.m00448. We postulate that these results are the consequence of the loss of members of the gene family from the ancestor of the A. thaliana chromosome 5 segment (including the orthologue of B.m00414 and C.m0448) and of amplification, to three copies, of the orthologue of B.m00415.
Figure 3.
Phylogenic Analysis of an Inverted Duplicate Array Present in Both A. thaliana Daughter Segments of the α Genome Duplication.
Left, overview of the positions of the members of the analyzed gene family across the sequence contigs. Right, phylogeny illustrated as a maximum likelihood cladogram. Bootstrap support, calculated from 1000 replicates, is shown at each branch as a percentage.
Figure 4.
Phylogenic Analysis of a Parallel Triplicate Array Present in Both A. thaliana Daughter Segments of the α Genome Duplication.
Left, overview of the positions of the members of the analyzed gene family across the sequence contigs. Right, phylogeny illustrated as a maximum likelihood cladogram. Bootstrap support, calculated from 1000 replicates, is shown at each branch as a percentage. The subgroup represents a cluster of sequences (At5g47350, B.m00415, At5g47340, and At5g47330) for which a robust phylogeny could not be constructed.
Gene Insertion
We observed 13 examples of gene models in A. thaliana for which we had sequences for all three corresponding segments of the genome of B. oleracea, but none of them contained homologues. These could be indicative of insertions in the A. thaliana genome segments since the divergence of the Arabidopsis and Brassica lineages. Six of them (At4g17700, At5g47400, At5g47410, At5g47580, At5g47590, and At5g47600) are classified as hypothetical or unknown proteins, so they may not correspond to genuine genes. One (At5g47320) encodes 40S ribosomal protein S19 and is irrefutably a genuine gene. Almost adjacent to this is a block of six genes: At5g47250 to At5g47300. This block includes four genes annotated as disease resistance proteins, one annotated as a myb family transcription factor, and one annotated as an F-box family protein. This structure is characteristic of a disease resistance locus. Either all copies of all seven genes (At5g47250 to At5g47300 plus At5g47320) have been deleted from all three paralogues of the B. oleracea genome or, more likely, they have been inserted into their present location in the A. thaliana genome, probably as a single event, since the divergence of the Arabidopsis and Brassica lineages.
Analysis of the sequence homologies of modeled genes, where significant matches were identified, generally revealed high similarity to A. thaliana nuclear genes. However, a contiguous segment of sequence contig B (encompassing gene models B.m00046 to B.m00053) revealed high homology (E value < 1e−18) with genes RPOB, PSAB, PSAA, YCF3, TRNS.3, RPS4, TRNT.2, TRNL.1, TRNF, NDHJ, PSBG, ACCD, PSAI, and YCF4, which are encoded in the segment 23 to 60 kb of the A. thaliana chloroplast genome. The sequence assembly was rechecked, and the origin of the sequence data was determined to have been from an internal region of the BAC insert, ruling out chimerism. Thus, the result is indicative of the integration of plastid DNA into the nuclear genome.
There is little database evidence supporting most of the B. oleracea gene models outside the conserved collinear genes. There are, however, four models, as listed in Table 6, that appear to contain intact genes that have homologues located elsewhere in the A. thaliana genome. There are no related genes in the other B. oleracea sequence contigs, suggesting that the observed breakdown of collinearity between B. oleracea and A. thaliana is not likely to be the result of deletion of the corresponding genes from the Arabidopsis lineage since divergence. More likely, these were inserted into their present positions in the B. oleracea genome since the genome triplication in the Brassica lineage.
Table 6.
Genes Apparently Relocated since the Divergence of the Arabidopsis and Brassica Lineages
| B. oleracea Gene | A. thaliana Gene | BLASTN E Value | Brassica EST? | Predicted Function |
|---|---|---|---|---|
| A.m00037 | At4g34880 | 8.1e−90 | No | Amidase family protein |
| A.m00074 | At3g01910 | 1.4e−86 | Yes | Sulfite oxidase, putative |
| B.m00017 | At3g15310 | 1.0e−107 | No | Expressed protein |
| F.m00523 | At2g31290 | 7.9e−148 | No | Hypothetical protein |
Dispersal of Gene Fragments
One of the genomic rearrangements identified on the basis of hybridization-based mapping was the apparent translocation of a homologue of At4g17280 to a position between homologues of At4g17570 and At4g17650 in contig E (O'Neill and Bancroft, 2000). We were able to characterize the event in detail at the sequence level. Gene model E.m00054 in sequence contig E corresponds to this insertion, which occurred between homologues of At4g17570 and At4g17650. However, only 155 bp of the gene was inserted, as illustrated in Supplemental Figure 5 online. The inserted sequence represents the 3′ end of the gene and retains a high level of sequence identity (85%).
To assess how common the translocation and insertion of gene fragments might be, we analyzed all of the sequence contigs by deconstructing them to 1000-bp overlapping segments and using BLASTN to identify homology with the coding regions of genes annotated in A. thaliana. Full details of the homologies detected across the sequence contigs by this approach are presented in Supplemental Table 1 online. The result, after exclusion of transposons, collinear gene fragments, and the four noncollinear apparently intact genes, was the identification of 155 apparent gene fragments distributed across the sequence contigs, as summarized in Table 2. Twelve of these were selected, on the basis of covering a wide range of BLASTN E values, and the alignments of the sequences were characterized in greater detail. The results are summarized in Table 7. None of the sequences represented intact genes. Eight of these gene fragments corresponded to regions of genes containing introns, and in all cases low homology sequences were present in the positions expected for introns. This finding shows the gene fragments to be of genomic origin rather than pseudogenes derived from reverse transcription of mRNA. There was also a strong bias for the sequences being homologous with the 3′ half of genes, with seven being near (or including) the 3′ end, five being near the middle of the gene, and none being near the 5′ end. To assess the proportion of these fragments that may be artifacts caused by the erroneous inclusion of transposon sequences into the A. thaliana gene models, we conducted BLAST analyses of the coding sequences of these genes against the whole genome sequence and found no evidence of homology with transposons known in A. thaliana. We conclude that the 155 interstitial gene fragments are largely derived from protein-coding genes.
Table 7.
Interstitial Gene Fragments Analyzed in Detail
| B. oleracea Gene Model | A. thaliana Gene | BLAST Score | Introns Present? | Position within Gene | Predicted Function |
|---|---|---|---|---|---|
| B.m00067 | At2g45730 | 5.5e−70 | Yes | Middle | Eukaryotic initiation factor 3 γ subunit family protein |
| B.m00063 | At3g23690 | 1.2e−57 | Yes | Including 3′ end | Basic helix-loop-helix family protein |
| C.m00439 | At4g23570.2 | 1.2e−55 | Yes | Including 3′ end | Phosphatase-related |
| A.m00081 | At1g74310 | 3.7e−46 | n/aa | Middle | Heat shock protein 101 |
| A.m00600/A.m00085 | At2g36310 | 3.4e−35 | Yes | Including 3′ end | Inosine-uridine–preferring nucleoside hydrolase family protein |
| B.m00420 | At2g47170.1 | 1.0e−34 | Yes | Middle | ADP-ribosylation factor 1 |
| E.m00626/E.m00622 | At4g31130 | 9.3e−22 | n/a | Middle | Expressed protein |
| E.m00583 | At5g19850 | 8.8e−20 | Yes | Including 3′ end | Hydrolase, α/β fold family protein |
| E.m00060 | At2g46470 | 5.1e−16 | Yes | Near 3′ | OXA1 protein, putative |
| E.m00577/E.m00012 | At1g14750 | 9.6e−12 | n/a | Near 3′ | Cyclin, putative (SDS) |
| G.m0005/G.m0006 | At4g36970 | 3.3e−10 | Yes | Middle | Remorin family protein |
| D.m00065/D.m00017 | At1g18900 | 1.4e−7 | n/a | Near 3′ | Pentatricopeptide repeat–containing protein |
n/a, not applicable.
DISCUSSION
The Evolution of Tandem Arrays
Tandem arrays of amplified genes, first characterized on a genome-wide scale in A. thaliana (Arabidopsis Genome Initiative, 2000), are a common feature of plant genomes. A mechanism involving homologous recombination and unequal crossing over can account for the amplification and reduction of arrays at disease resistance loci (Michelmore and Meyers, 1998; Noel et al., 1999). Comparative analysis has enabled the identification of numerous examples of the formation/loss of tandem arrays in the Brassica and Arabidopsis lineages. In A. thaliana, we have identified a putative example of gene loss and reamplification of a three-gene tandem array. However, tandem arrays can also be very stable; for example, we identified a tandem duplication event in A. thaliana that predates the α genome duplication. The greater stability of this array may be a consequence of it being an inverted duplication rather than a parallel duplication, and thus being less susceptible to rearrangement by unequal crossing over.
Genome Evolution by Gene Loss
Interspersed gene loss from duplicated genome segments was observed in A. thaliana by the analysis of genome sequence data (Arabidopsis Genome Initiative, 2000; Bancroft, 2001) and is also extensive in the triplicated genome segments of B. oleracea. The mechanism for such gene loss cannot be discerned from the analysis of a single genome. However, Brassica and Arabidopsis are sufficiently closely related that sequence drift alone would not be expected to have completely erased the sequence homology in pseudogenes. Most of the genes inferred to have been lost show no detectable residual sequence homology in B. oleracea, indicating a deletion mechanism for gene loss, rather than sequence drift. A prediction of the deletion hypothesis is that, in some instances, only parts of genes will have been deleted and the residual fragments should show discernible homology. We observed this on 10 occasions, confirming that gene deletion is most likely the mechanism responsible for the widely observed interstitial gene loss from duplicated segments of plant genomes.
Although it is the loss of genes from the triplicated B. oleracea genome segments that is predominant, we were able to identify one example of deletion of a gene in the Arabidopsis lineage. This finding demonstrates that, although the rate of genome change in A. thaliana is much lower than that in B. oleracea, presumably because the much greater genetic redundancy in the more recent polyploid means that loss of genes is less likely to be detrimental to fitness, its genome is still undergoing the diploidization process. Therefore, the genome of A. thaliana provides a good, but imperfect, representation of the ancestral genome of the Brassica lineage.
Increase in Gene Number by Genome Duplication
Excluding transposable elements, the 2.2 Mb of the B. oleracea genome we sequenced contains at least 181 genes. Of these, 177 show conserved synteny with their homologues in the two A. thaliana genome segments, and 4 are in noncollinear positions. Assuming that the genome of the progenitor of the Brassiceae at the point of genome triplication contained three intact genome segments, each representing the Arabidopsis-like progenitor genome, a total of 274 genes would have been present (271 corresponding to the genuine genes still present in the A. thaliana genome segment, as shown in Figure 2 and listed in Table 1, plus three copies of the gene inferred to have been present in that ancestral species between the predecessors of genes modeled as At4g17430 and At4g17440). Thus, 65% of genes have been retained in B. oleracea. Extrapolation to the whole genome scale provides us with an estimation of the minimum number of genes in B. oleracea as 48,750 in collinear positions and >1000 in noncollinear positions relative to A. thaliana (assuming that the regions are representative of the genome as a whole and that A. thaliana contains 25,000 genuine genes). The genome of B. oleracea is more than four times the size of the A. thaliana genome (Arumuganthan and Earle, 1991), but our results show that only a small proportion of the increased size is attributable to additional genes.
Genome Evolution by Sequence Insertion
Transposable elements represent one source of intergenic sequences. These ranged in frequency from one per 177 kb in contig D to one per 23 kb in contig E. There were, however, other sources of inserted sequences. There was one example of integration of plastid sequences into the nuclear genome. This appears to be common in plants, with many examples identified in rice (International Rice Genome Sequencing Project, 2005). An unexpected source of new sequences is genomic fragments of genes originating from elsewhere within the genome. This has occurred extensively, with 155 potential instances having been identified. Many of these (95) coincide with gene models that do not represent conserved collinear orthologues of A. thaliana genes. Because part of the evidence for automated gene model construction is homology with known genes, it is likely that these insertions are driving extensive erroneous gene prediction. Extrapolating to the whole genome, B. oleracea could contain >40,000 interstitial insertions of gene fragments. A likely mechanism is capture and replication by transposable elements, akin to Pack-MULE (Jiang et al., 2004) or Helitron-mediated translocation of gene fragments (Lai et al., 2005), although the 3′ bias of the inserted gene fragments, which was not observed for Pack-MULE or Helitron-mediated translocation, suggests that a different transposon system is likely to be responsible.
Conclusions
We conducted an extensive sequence-based analysis of the Brassica genome triplication event across complete sets of paralogous Brassica genome segments descended from both daughters of the α genome duplication. Our studies have provided direct evidence that the interspersed loss of protein-coding genes, which has been widely observed in duplicated segments of plant genomes, is the result of a deletion mechanism rather than of sequence drift. We have also provided evidence in Brassica for the insertion of apparently intact protein-coding genes that originate from elsewhere in the genome (as opposed to originating by tandem duplication). In addition, our studies have demonstrated, in a dicot species, that the dispersal of fragments of protein-coding genes, of genomic origin, is extensive; such widespread insertions were previously reported only in grasses (Jiang et al., 2004; International Rice Genome Sequencing Project, 2005; Lai et al., 2005). We implicate such inserted fragments in the overcalling that we (and others) often encounter with automated sequence annotation systems. Thus, many of the insights from our analyses must be taken into account when using comparative approaches to gene isolation from Brassica. In particular, we can expect many exceptions to genome collinearity, even for functional genes.
METHODS
BAC Sequencing
Purified BAC DNA was sheared by nebulization, size-selected (2 to 3 or 10 to 12 kb), and ligated into a pUC-derived vector, pHOS1, using BstXI linkers. Clones were sequenced from both ends using ABI Big Dye terminator chemistry on ABI 3700 or 3730xl sequencing machines. Sequences were assembled using the TIGR assembler, and additional directed sequencing reactions were performed as necessary to complete the sequence to high quality.
Sequence Annotation
Annotation of the BAC assemblies was performed using the TIGR annotation pipeline, a collection of software known as Eukaryotic Genome Control that serves as the central data management system. Each BAC sequence was processed through a series of algorithms for predicting genes (Genscan+, Genemark.hmm, and Glimmer) (Burge and Karlin, 1997; Lukashin and Borodovsky, 1998; Salzberg et al., 1999), splice sites (Hebsgaard et al., 1996; Pertea et al., 2001), and tRNAs (Lowe and Eddy, 1997). The AAT package (Huang et al., 1997) was used for homology searches against nucleotide and protein databases, including plant-specific cDNA and EST sequences, TIGR plant gene indices (Quackenbush et al., 2000), a nonredundant amino acid database filtered from public sources, and SwissProt (Bairoch and Apweiler, 2000). Proteins encoded by gene models generated by the database searches and predictions were further searched against Markov model databases, including Pfam (Bateman et al., 2002), and automatically assigned a putative name based on domain hits or homology with previously identified proteins. Gene structures and names were manually inspected and refined as necessary. Annotated gene models were also searched against a curated database of transposon-encoded proteins (ftp://ftp.tigr.org/pub/data/TransposableElements/transposon_db.pep). The top match from each hit was used to classify the transposons, which are treated as a separate data set. Predicted proteins <100 amino acids in length with no database support were excluded from the annotation and gene counts.
Sequence Deconstruction and BLAST Analysis Tool
We developed an alignment tool that deconstructs a sequence contig into overlapping segments of 1000 bp and conducts BLASTN searches against a database of Arabidopsis thaliana coding sequences named by Arabidopsis Genome Initiative code. The tabular output permits a rapid (<5 min) visual inspection of collinearity between the genes present on the Brassica BAC and the Arabidopsis genome. This differs from existing synteny assessment tools in that it is fast and uses all of the BAC sequence, rather than restricting the analysis to the predicted proteins. Thus, the tool can detect gene fragments not incorporated into gene models. The tool is accessible as part of a publicly available analysis pipeline that is under development (http://brassica.bbsrc.ac.uk/annotate.html).
Phylogenetic Analysis
Protein sequences were aligned using the MUSCLE program (Edgar, 2004), and alignments were edited with GeneDoc (Nicholas et al., 1997). Phylogenetic analysis was performed with the PHYLIP version 3.6 software package (Felsenstein, 1989). The SEQBOOT program was used to generate 1000 bootstrap alignments. The maximum likelihood program PROML was used to generate the bootstrap trees, for each of which the sequence order was jumbled 10 times, with the global rearrangement option enabled. The final extended majority rule consensus tree was generated with the CONSENSE program. Cladograms were displayed with Tree Explorer, in the MEGA version 3.1 software (Kumar et al., 2004), and rooted with the midpoint method using RETREE in the PHYLIP version 3.6 software package (Felsenstein, 1989). To check consistency between approaches, phylogenies were also constructed as neighbor-joining trees with p-distance correction, using MEGA version 3.1 (Kumar et al., 2004).
Divergence Calculations
For every possible paralogous or homologous gene pair, we performed protein back-translated nucleotide alignments and calculated the synonymous substitution rate as well as overall sequence identity using the PAML package assuming model 0, similar to the approach described by Maere et al. (2005). Putative pairs with relatively low sequence identity (<50% at the amino acid level) generated anomalous values and were excluded.
Accession Numbers
Accession numbers for the sequence contigs are listed in Table 1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Summary of BLASTN Analysis with Contig Sequences Deconstructed to Overlapping 1-kb Segments.
Supplemental Figure 1. Multiple Alignments for Figure 1.
Supplemental Figure 2. Deletion Mechanism for Gene Loss.
Supplemental Figure 3. Multiple Alignments for Figure 3.
Supplemental Figure 4. Multiple Alignments for Figure 4.
Supplemental Figure 5. Gene Fragment Accounting for Rearrangement Detected by Hybridization-Based Mapping.
Supplementary Material
Acknowledgments
This work was funded by a U.K. Biotechnology and Biological Sciences Research Council competitive support grant to I.B. and M.T. and by U.S. National Science Foundation Cooperative Agreement DBI-9813586 to TIGR.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Ian Bancroft (ian.bancroft@bbsrc.ac.uk).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.106.041665.
References
- Arabidopsis Genome Initiative (2000). Analysis of the genome of the flowering plant Arabidopsis thaliana. Nature 408 796–815. [DOI] [PubMed] [Google Scholar]
- Arumuganthan, K., and Earle, E.D. (1991). Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9 208–218. [Google Scholar]
- Babula, D., Kaczmarek, M., Barakat, A., Delseney, M., Quiros, C.F., and Sadowski, J. (2003). Chromosomal mapping of Brassica oleracea based on ESTs from Arabidopsis thaliana: Complexity of the comparative map. Mol. Genet. Genomics 268 656–665. [DOI] [PubMed] [Google Scholar]
- Bairoch, A., and Apweiler, R. (2000). The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 28 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancroft, I. (2000). Insights into the structural and functional evolution of plant genomes afforded by the nucleotide sequences of chromosomes 2 and 4 of Arabidopsis thaliana. Yeast 17 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancroft, I. (2001). Duplicate and diverge: The evolution of plant genome microstructure. Trends Genet. 17 89–93. [DOI] [PubMed] [Google Scholar]
- Bateman, A., Birney, E., Cerruti, L., Durbin, R., Etwiller, L., Eddy, S.R., Griffiths-Jones, S., Howe, K.L., Marshall, M., and Sonnhammer, E.L. (2002). The Pfam protein families database. Nucleic Acids Res. 30 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc, G., Barakat, A., Guyot, R., Cooke, R., and Delseny, M. (2000). Extensive duplication and reshuffling in the Arabidopsis genome. Plant Cell 12 1093–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc, G., and Wolfe, K.H. (2004). Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16 1679–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers, J.E., Chapman, B.A., Rong, J., and Paterson, A.H. (2003). Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422 433–438. [DOI] [PubMed] [Google Scholar]
- Burge, C., and Karlin, S. (1997). Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268 78–94. [DOI] [PubMed] [Google Scholar]
- Edgar, R.C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein, J. (1989). PHYLIP—Phylogeny Inference Package (version 3.2). Cladistics 5 164–166. [Google Scholar]
- Gao, M.Q., Li, G.Y., Yang, B., McCombie, W.R., and Quiros, C.F. (2004). Comparative analysis of a Brassica BAC clone containing several major aliphatic glucosinolate genes with its corresponding Arabidopsis sequence. Genome 47 666–679. [DOI] [PubMed] [Google Scholar]
- Hebsgaard, S.M., Korning, P.G., Tolstrup, N., Engelbrecht, J., Rouze, P., and Brunak, S. (1996). Splice site prediction in Arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic Acids Res. 24 3439–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X., Adams, M.D., Zhou, H., and Kerlavage, A.R. (1997). A tool for analyzing and annotating genomic sequences. Genomics 46 37–45. [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005). The map-based sequence of the rice genome. Nature 436 793–800. [DOI] [PubMed] [Google Scholar]
- Jiang, N., Bao, Z., Zhang, X., Eddy, S.R., and Wessler, S.R. (2004). Pack-MULE transposable elements mediate gene evolution in plants. Nature 431 569–573. [DOI] [PubMed] [Google Scholar]
- Koch, M.A., Haubold, B., and Mitchell-Olds, T. (2000). Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol. Biol. Evol. 17 1483–1498. [DOI] [PubMed] [Google Scholar]
- Ku, H.-M., Vision, T., Liu, J., and Tanksley, S.D. (2000). Comparing sequenced segments of the tomato and Arabidopsis genomes: Large-scale duplication followed by selective gene loss creates a network of synteny. Proc. Natl. Acad. Sci. USA 97 9121–9126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S., Tamura, K., and Nei, M. (2004). MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5 150–163. [DOI] [PubMed] [Google Scholar]
- Lagercrantz, U. (1998). Comparative mapping between Arabidopsis thaliana and Brassica nigra indicates that Brassica genomes have evolved through extensive genome replication accompanied by chromosome fusions and frequent rearrangements. Genetics 150 1217–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagercrantz, U., and Lydiate, D. (1996). Comparative genome mapping in Brassica. Genetics 144 1903–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, J., Li, Y., Messing, J., and Dooner, H.K. (2005). Gene movement by Helitron transposons contributes to the haplotype variability of maize. Proc. Natl. Acad. Sci. USA 102 9068–9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan, T.H., DelMonte, T.A., Reischmann, K.P., Hyman, J., Kowalski, S., McFerson, J., Kresovich, S., and Paterson, A.H. (2000). An EST-enriched comparative map of Brassica oleracea and Arabidopsis thaliana. Genome Res. 10 776–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch, I.J., and Bennett, M.D. (1997). Polyploidy in angiosperms. Trends Plant Sci. 2 470–476. [Google Scholar]
- Lowe, T.M., and Eddy, S.R. (1997). tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukashin, A.V., and Borodovsky, M. (1998). GeneMark.hmm: New solutions for gene finding. Nucleic Acids Res. 26 1107–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukens, L., Zou, F., Lydiate, D., Parkin, I., and Osborn, T. (2003). Comparison of a Brassica oleracea genetic map with the genome of Arabidopsis thaliana. Genetics 164 359–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukens, L.N., Quijada, P.A., Udall, J., Pires, J.C., Schranz, M.E., and Osborn, T.C. (2004). Genome redundancy and plasticity within ancient and recent Brassica crop species. Biol. J. Linn. Soc. 82 665–674. [Google Scholar]
- Lysak, M.A., Koch, M.A., Pecinka, A., and Schubert, I. (2005). Chromosome triplication found across the tribe Brassiceae. Genome Res. 15 516–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere, S., De Bodt, S., Raes, J., Casneuf, T., Van Montagu, M., Kuiper, M., and Van de Peer, Y. (2005). Modeling gene and genome duplications in eukaryotes. Proc. Natl. Acad. Sci. USA 102 5454–5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, K., et al. (2001). Conservation of microstructure between a sequenced region of the genome of rice and multiple segments of the genome of Arabidopsis thaliana. Genome Res. 11 1167–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore, R.W., and Meyers, B.C. (1998). Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 8 1113–1130. [DOI] [PubMed] [Google Scholar]
- Nicholas, K.B., Nicholas, H.B., Jr., and Deerfield, D.W., II (1997). GeneDoc: Analysis and visualization of genetic variation. EMBnet.news 4 1–4. [Google Scholar]
- Noel, L., Moores, T.L., van der Biezen, E.A., Parniske, M., Daniels, M.J., Parker, J.E., and Jones, J.D.G. (1999). Pronounced intraspecific haplotype divergence at the RPP5 complex disease resistance locus of Arabidopsis. Plant Cell 11 2099–2111. [PMC free article] [PubMed] [Google Scholar]
- O'Neill, C.M., and Bancroft, I. (2000). Comparative physical mapping of segments of the genome of Brassica oleracea var alboglabra that are homoeologous to sequenced regions of the chromosomes 4 and 5 of Arabidopsis thaliana. Plant J. 23 233–243. [DOI] [PubMed] [Google Scholar]
- Park, J.Y., et al. (2005). Physical mapping and microsynteny of Brassica rapa ssp. pekinensis genome corresponding to a 222 kb gene-rich region of Arabidopsis chromosome 4 and partially duplicated on chromosome 5. Mol. Genet. Genomics 274 579–588. [DOI] [PubMed] [Google Scholar]
- Parkin, I.A.P., Gulden, S.M., Sharpe, A.G., Lukens, L., Trick, M., Osborn, T.C., and Lydiate, D.J. (2005). Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics 171 765–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin, I.A.P., Sharpe, A.G., Keith, D.J., and Lydiate, D.J. (1995). Identification of the A and C genomes of amphidiploid Brassica napus (oilseed rape). Genome 38 1122–1131. [DOI] [PubMed] [Google Scholar]
- Parkin, I.A.P., Sharpe, A.G., and Lydiate, D.J. (2003). Patterns of genome duplication within the Brassica napus genome. Genome 46 291–303. [DOI] [PubMed] [Google Scholar]
- Pertea, M., Lin, X., and Salzberg, S.L. (2001). GeneSplicer: A new computational method for splice site prediction. Nucleic Acids Res. 29 1185–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quackenbush, J., Liang, F., Holt, I., Pertea, G., and Upton, J. (2000). The TIGR gene indices: Reconstruction and representation of expressed gene sequences. Nucleic Acids Res. 28 141–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiros, C.F., Grellet, F., Sadowski, J., Suzuki, T., Li, G., and Wroblewski, T. (2001). Arabidopsis and Brassica comparative genomics: Sequence, structure and gene content in the ABI -Rps2-Ck chromosomal segment and related regions. Genetics 157 1321–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana, D., van den Boogaart, T., O'Neill, C.M., Hynes, L., Bent, E., Macpherson, L., Park, J.Y., Lim, Y.P., and Bancroft, I. (2004). Conservation of the microstructure of genome segments in Brassica napus and its diploid relatives. Plant J. 40 725–733. [DOI] [PubMed] [Google Scholar]
- Salzberg, S.L., Pertea, M., Delcher, A.L., Gardner, M.J., and Tettelin, H. (1999). Interpolated Markov models for eukaryotic gene finding. Genomics 59 24–31. [DOI] [PubMed] [Google Scholar]
- Schmidt, R., Acarkan, A., and Boivin, K. (2001). Comparative structural genomics in the Brassicaceae family. Plant Physiol. Biochem. 39 253–262. [Google Scholar]
- Song, K., Lu, P., Tang, K., and Osborn, T.C. (1995). Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc. Natl. Acad. Sci. USA 92 7719–7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, T., Grellet, F., Potter, D., Li, G., and Quiros, C.F. (2003). Structure, sequence and phylogeny of the members of the Ck1 gene family in Brassica oleracea and Arabidopsis thaliana (Brassicaceae). Plant Sci. 164 735–742. [Google Scholar]
- Warwick, S.I., and Black, L.D. (1991). Molecular systematics of Brassica and allied genera (subtribe Brassicinae, Brassiceae)—Chloroplast genome and cytodeme congruence. Theor. Appl. Genet. 82 81–92. [DOI] [PubMed] [Google Scholar]
- Warwick, S.I., and Black, L.D. (1997). Molecular phylogenies from theory to application in Brassica and allies (tribe Brassiceae, Brassicaceae). Opera Bot. 132 159–168. [Google Scholar]
- Wendel, J.F. (2000). Genome evolution in polyploids. Plant Mol. Biol. 42 225–249. [PubMed] [Google Scholar]
- Wolfe, K.H., Gouy, M., Yang, Y.W., Sharp, P.M., and Li, W.H. (1989). Date of the monocot-dicot divergence estimated from the chloroplast DNA sequence data. Proc. Natl. Acad. Sci. USA 86 6201–6205.2762323 [Google Scholar]
- Yang, Y.W., Lai, K.N., Tai, P.Y., and Li, W.H. (1999). Rates of nucleotide substitution in angiosperm mitochondrial DNA sequences and dates of divergence between Brassica and other angiosperm lineages. J. Mol. Evol. 48 597–604. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.