Abstract
Imprinted genes are expressed predominantly from either their paternal or their maternal allele. To date, all imprinted genes identified in plants are expressed in the endosperm. In Arabidopsis thaliana, maternal imprinting has been clearly demonstrated for the Polycomb group gene MEDEA (MEA) and for FWA. Direct repeats upstream of FWA are subject to DNA methylation. However, it is still not clear to what extent similar cis-acting elements may be part of a conserved molecular mechanism controlling maternally imprinted genes. In this work, we show that the Polycomb group gene FERTILIZATION-INDEPENDENT SEED2 (FIS2) is imprinted. Maintenance of FIS2 imprinting depends on DNA methylation, whereas loss of DNA methylation does not affect MEA imprinting. DNA methylation targets a small region upstream of FIS2 distinct from the target of DNA methylation associated with FWA. We show that FWA and FIS2 imprinting requires the maintenance of DNA methylation throughout the plant life cycle, including male gametogenesis and endosperm development. Our data thus demonstrate that parental genomic imprinting in plants depends on diverse cis-elements and mechanisms dependent or independent of DNA methylation. We propose that imprinting has evolved under constraints linked to the evolution of plant reproduction and not by the selection of a specific molecular mechanism.
INTRODUCTION
Reproduction of flowering plants is characterized by a double fertilization event, which involves two identical sperm cells and two female gametes, the egg cell and the central cell. Fertilization of the egg cell leads to the development of the embryo, whereas the second sperm cell independently fertilizes the central cell, giving rise to the endosperm (Guignard, 1899; Maheshwari, 1950; Berger, 2003). The endosperm controls the supply of maternal nutrients to the developing embryo and does not contribute genetic material to the next generation. In Arabidopsis thaliana and in maize (Zea mays), parental imprinted expression has been detected only in the endosperm for a few genes expressed only from their paternal or their maternal allele (Berger, 2004; Gehring et al., 2004; Köhler and Grossniklaus, 2005). In Arabidopsis, imprinting was demonstrated using allele-specific RT-PCR for the paternally expressed gene PHERES1 (Köhler et al., 2005) and the maternally expressed genes FWA (Kinoshita et al., 2004) and MEDEA (MEA) (Kinoshita et al., 1999; Vielle-Calzada et al., 1999). FWA and MEA are expressed during neither vegetative development nor male gametogenesis. During female gametogenesis, FWA and MEA become transcriptionally active, and both genes are expressed specifically in the central cell (Choi et al., 2002; Kinoshita et al., 2004). After fertilization, only the maternal alleles of FWA and MEA are expressed during endosperm development, whereas their paternal alleles remain silenced.
Imprinting of a gene thus results from a combination of silencing of the paternal allele and activation of expression of the maternal allele. Silencing of the FWA paternal allele is likely correlated with methylation of cytosine residues located in the direct repeats of the 5′ region mediated by the DNA methyltransferase MET1 (Kinoshita et al., 2004). The transcriptionally active FWA maternal allele in endosperm is characterized by demethylation of the direct repeats. This demethylation may occur in the central cell and might result from the activity of the DNA glycosylase DEMETER (DME). DME creates single-strand 5′ nicks, and such DNA breaks would be recognized by the DNA repair machinery, which in turn replaces the methylated cytosine residue with unmethylated ones (Choi et al., 2002; Gehring et al., 2006). Based on the available knowledge regarding FWA imprinting, it was possible to model a cycle for plant imprints (Berger, 2004; Kinoshita et al., 2004). According to the model, FWA is silenced during the vegetative phase by MET1. DME releases FWA silencing in the central cell. After fertilization, the activated maternal FWA allele is inherited by endosperm and is transcribed, whereas the paternal allele remains silenced because it was inherited in an inactive state from the male gamete. MET1 is active during male gametogenesis (Saze et al., 2003), but its involvement in silencing of the FWA paternal allele during male gametogenesis and endosperm development has not been demonstrated. Moreover, it is not clear whether the mode of regulation of FWA imprinting could apply to other imprinted genes.
Regulation of MEA imprinting has been analyzed, but as detailed below, the mechanism controlling MEA imprinting is still unclear. DME is also essential for MEA transcriptional activation (Choi et al., 2002), and methylated CpG residues have been identified in the MEA promoter (Xiao et al., 2003; Gehring et al., 2006), suggesting that MEA imprinting is controlled by DNA methylation similar to FWA. DNA methylation could target putative CpG islands and transposons at the MEA locus (Xiao et al., 2003; Spillane et al., 2004). However, such elements are not essential for MEA imprinting, as suggested by conflicting genetic experiments, which casts doubt on the role of MET1 in this process (Vielle-Calzada et al., 1999; Luo et al., 2000; Vinkenoog et al., 2000; Xiao et al., 2003). Although MET1 might play a regulatory role in the control of imprinting of several genes involved in endosperm development (Adams et al., 2000), its direct involvement in the control of MEA imprinting remains to be tested.
Besides DNA methylation, histone methylation was shown recently to be involved in the control of MEA imprinting (Gehring et al., 2006; Jullien et al., 2006) and likely also controls the imprinting of PHERES1, encoding a MADS box transcription factor (Köhler et al., 2005). MEA imprinting depends on transcriptional repression of its paternal allele by methylation of the Lys-27 of Histone3 by several Polycomb group complexes active during the plant life cycle (Gehring et al., 2006; Jullien et al., 2006). In endosperm, the PRC2 complex contains MEA (Grossniklaus et al., 1998; Kiyosue et al., 1999; Luo et al., 1999) and a core of three additional proteins, the VEFS domain protein FERTILIZATION-INDEPENDENT SEED2 (FIS2) (Luo et al., 1999), the WD40 protein FERTILIZATION-INDEPENDENT ENDOSPERM (FIE) (Luo et al., 1999; Ohad et al., 1999), and the WD40 protein MULTICOPYSUPPRESSOR OF IRA1 (MSI1) (Köhler et al., 2003; Guitton et al., 2004). The mutants mea, fie, msi1, and fis2 exhibit similar maternal defects in endosperm, and as a consequence, FIE, MSI1, and FIS2 are anticipated to be subject to one or more of the genetic mechanisms that regulate imprinting, including the one presented for MEA (Chaudhury et al., 1997; Grossniklaus et al., 1998; Kiyosue et al., 1999). Involvement of imprinting is also supported for FIE and FIS2 by the monoallelic maternal expression of transcriptional reporters for FIE–green fluorescent protein (GFP) (Yadegari et al., 2000) and FIS2–β-glucuronidase (GUS) (Luo et al., 2000), respectively. However, this transcriptional control may affect only the corresponding transcriptional reporter. Silencing in endosperm has been shown for the paternal copy of reporter constructs inserted at several loci, leading to the hypothesis of global silencing of the paternal genome in the first days after fertilization (Vielle-Calzada et al., 2000). Alternatively, the maternal effect associated with the loss-of-function mutations could originate from abnormal development of the female gametophyte or from dosage effects. Hence, the imprinted status of FIS2, FIE, and MSI1 remains unclear.
In this work, we show that FIS2 is subject to parental genomic imprinting. We study in detail the role of DNA methylation in the transcriptional control of FIS2 and MEA and conclude that they are distinct. We identify a cis-element upstream of FIS2 that is targeted by MET1. This element is likely associated with the control of FIS2 imprinting. Using a genetic approach, we investigated the role played by MET1 during the entire life cycle for the control of FIS2 and FWA imprinting. To this end, we investigated the effect of MET1 loss of function restricted to the vegetative developmental phase (with the MET1as line) and the effect of MET1 loss of function restricted to the gametogenesis phase (with the met1-3/+ line). Based on our results, we propose a revised model for the cycle of MET1-dependent imprinting in plants.
RESULTS
FIS2 Expression Is Controlled by DNA Methylation–Dependent Imprinting
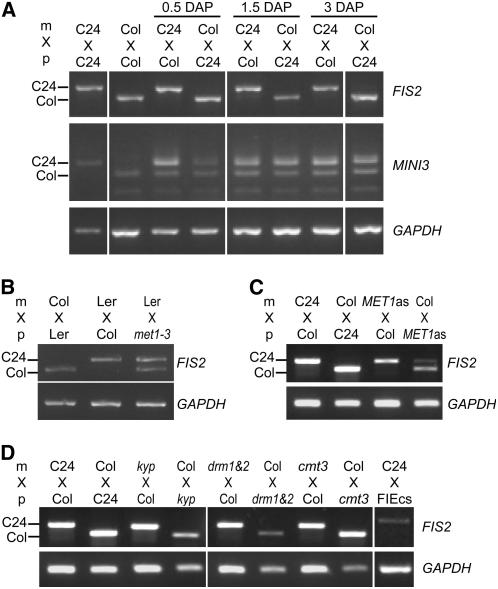
To investigate the imprinted status of FIS2, we used polymorphisms in the FIS2 coding sequence between different wild-type accessions to distinguish transcripts of each parental allele (allele-specific RT-PCR) (Figure 1A). We performed allele-specific RT-PCR on seeds originating from crosses between wild-type accessions C24 and Columbia (Col), used alternately as male or female. One would expect that if FIS2 is not imprinted, two transcripts should be detected. Contrary to this hypothesis, we observed only expression of maternal FIS2 transcripts (Figure 1A). We observed monoallelic maternal expression of FIS2 up to 5 d after pollination (DAP) (see Supplemental Figure 1 online). Maternal FIS2 transcripts stored in the ovules and inherited after fertilization could be detected in developing seeds and mimic an imprinted status. However, it is unlikely that such stored maternal transcripts could be still present at 5 DAP. It is likely that we detected transcripts from active postzygotic transcription of the FIS2 maternal allele in the endosperm. A putative general paternal silencing has been observed for reporters of endosperm-expressed genes for up to 4 DAP (Vielle-Calzada et al., 2000). It could be hypothesized that such a general mechanism also affects expression at genomic loci for genes expressed in endosperm. During all stages of development examined, we observed biparental expression of MINI3 (Figure 1A), a gene expressed only in endosperm during the first 3 DAP (Luo et al., 2005). These results and others (Berger, 2003; Kinoshita et al., 2004) indicate that there is no general paternal silencing for genes expressed in endosperm and that FIS2 monoallelic expression results from a specific silencing mechanism. In agreement with these findings, Luo et al. (2000) previously showed that the FIS2-GUS translational reporter is expressed in endosperm only from the maternal allele. We concluded that the endogenous FIS2 gene is imprinted.
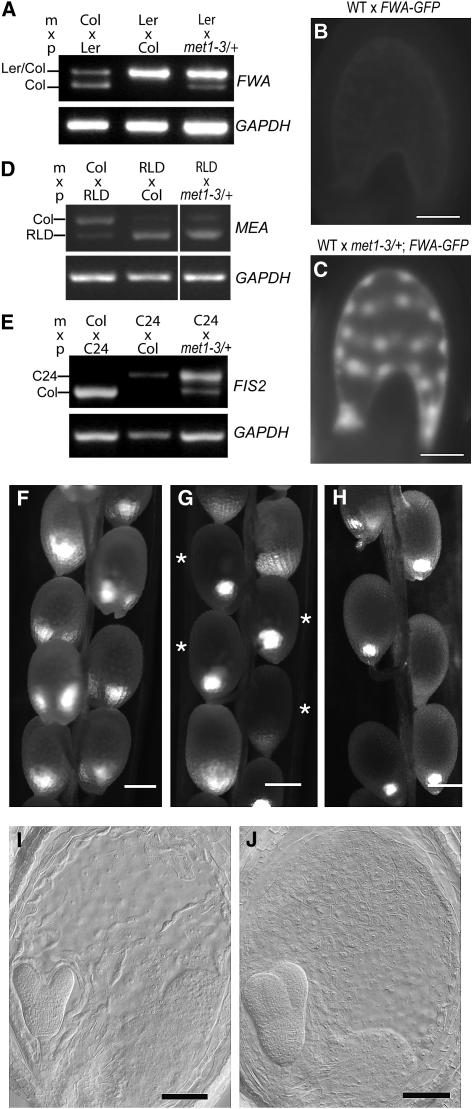
Figure 1.
FIS2 Imprinting Is Controlled by DNA Methylation.
(A) Allele-specific RT-PCR on RNAs extracted from siliques obtained at 0.5, 1.5, and 3 DAP after crosses were made between wild-type Col and wild-type C24. A 180-bp deletion in the FIS2 coding sequence of the wild-type accession Col allows the distinction of the Col FIS2 transcript from the FIS2 transcript in accession C24. Only the FIS2 maternal allele is expressed in developing seeds. By contrast, MINI3 is expressed from both parental alleles.
(B) Allele-specific RT-PCR on RNAs extracted from siliques obtained at 4 DAP after crosses between wild-type ovules and pollen from met1-3/met1-3 plants (Col) show reduced activity of the maintenance DNA MET1 during the entire life cycle. In contrast with wild-type crosses, the FIS2 paternal allele is expressed in developing seeds, which inherit a paternal allele from met1-3/met1-3 plants.
(C) Allele-specific RT-PCR on RNAs extracted from siliques obtained at 5 DAP after reciprocal crosses between wild-type plants and MET1as plants. The FIS2 paternal allele is expressed in developing seeds when pollen is provided by MET1as plants, which show reduced activity of the maintenance DNA MET1 during the vegetative developmental phase (Finnegan et al., 1996).
(D) Allele-specific RT-PCR on RNAs extracted from siliques obtained at 5 DAP after crosses made between wild-type plants and different mutants affected for histone methylation or for DNA methylation. A 180-bp deletion in the FIS2 coding sequence of the wild-type accession Col allows the distinction of the Col FIS2 transcript from the FIS2 transcript in accessions C24, Landsberg erecta (Ler), and Wassilewskija. C24 is presented as a control. Only the maternal allele of FIS2 is expressed in seeds resulting from reciprocal crosses between the wild type and homozygous mutants for other pathways controlling DNA methylation in Arabidopsis cmt3 (Lindroth et al., 2001) and drm1 and drm2 (Cao and Jacobsen, 2002a). KYP methylates Histone3 Lys-9 residues (Jackson et al., 2002; Tariq et al., 2003), whereas the Polycomb group complex containing FIE methylates Histone3 Lys-27 residues (Bastow et al., 2004).
The paternal (p) and maternal (m) alleles are indicated. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. No-RT control assays were performed, but the data are not shown.
We investigated the potential role played by MET1 in FIS2 imprinting. To this end, we used the loss-of-function allele met1-3 in the Col background (Figure 1B). To avoid nonspecific cumulative effects of inbred met1 loss-of-function lines (Saze et al., 2003; Xiao et al., 2006), we isolated met1-3/met1-3 plants from a segregating population of selfed met1-3/+ plants. Loss of MET1 activity caused expression of the paternal allele of FIS2 in crosses between wild-type female ovules and met1-3 pollen (Figure 1B). To evaluate the impact of MET1 activity during vegetative development, we used MET1as plants, which produce antisense MET1 mRNAs in vegetative tissues, leading to a loss of 70% of DNA methylation on symmetric CpG sites (Finnegan et al., 1996). We observed that loss of MET1 activity during vegetative development caused expression of the paternal allele of FIS2 in crosses between wild-type female ovules and MET1as pollen (Figure 1C). We concluded that imprinting of FIS2 depends directly or indirectly on MET1. DNA methylation is propagated through replication in a semiconservative manner (Finnegan et al., 2000). Because in the MET1as line MET1 antisense RNAs are produced only during vegetative development, we propose that demethylation of FIS2 during vegetative development is carried over to the endosperm, resulting in loss of imprinting. DNA methylation likely interacts with mechanisms that control other epigenetic marks (Tariq et al., 2003; Chan et al., 2005; Takeda and Paszkowski, 2006; Vire et al., 2006), and the effect of met1-3 and MET1as could be indirect. Thus, we tested whether other epigenetic mechanisms might be involved, including DNA methylation and histone methylation. The DOMAIN-REARRANGED METHYLTRANSFERASEs DRM1 and DRM2 control de novo methylation (Cao and Jacobsen, 2002a), and CHROMOMETHYLASE3 (CMT3), which is unique to plants, maintains methylation at asymmetric sites (Lindroth et al., 2001; Cao and Jacobsen, 2002b). Loss-of-function mutants in these genes do not influence FIS2 imprinting (Figure 1D). Transcriptional gene silencing is also regulated by methylation of histone H3. In Arabidopsis, Lys residues Lys-9 and Lys-27 of histone H3 are methylated by KRYPTONITE (KYP) (Jackson et al., 2002) and by Polycomb group complexes (Bastow et al., 2004; Jullien et al., 2006), respectively. FIS2 imprinting was not altered in crosses between wild-type ovules and pollen from mutant plants carrying loss-of-function kyp or from FIE cosuppressed plants (Katz et al., 2004) (Figure 1D). Because we could not find any effect of loss of epigenetic marks likely linked with the maintenance of DNA methylation, we conclude that MET1 directly silences FIS2.
DNA Methylation Targets a Specific CpG-Rich Domain Upstream of the FIS2 Gene
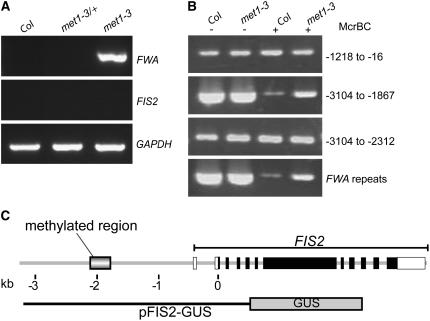
Loss of MET1 activity caused ectopic expression of FWA, as reported previously (Saze et al., 2003; Kinoshita et al., 2004), but surprisingly, it did not result in ectopic expression of FIS2 in vegetative tissues (Figure 2A). The absence of endosperm-specific transcriptional activators could explain the absence of ectopic expression of FIS2 in vegetative met1 tissues. Nevertheless, we hypothesized that loss of function of MET1 causes a loss of DNA methylation marks in FIS2 genomic sequences. To test this hypothesis, we analyzed DNA methylation in the 5′ region of FIS2 in leaf tissues. Using the methylation-sensitive restriction enzyme McrBC, we identified a region affected by DNA methylation (Figure 2B). Moreover, bisulfite sequencing analyses identified a region at ∼2 kb upstream of the start codon of FIS2 consisting of a 200-bp domain where most CpG sites were methylated (30 of 38) (Figure 2C; see Supplemental Figure 2 online). Methylation of CpG residues in this region was severely reduced in met1-3 homozygous mutant leaves (see Supplemental Figure 2 online). We conclude that MET1 targets a specific element in the 5′ region that might control FIS2 silencing in leaves as well as in pollen and endosperm.
Figure 2.
MET1 Targets a CpG-Enriched Region at the FIS2 Locus.
(A) RT-PCR analysis shows that imprinted genes FWA and FIS2 are not expressed in vegetative tissues of the wild-type accession Col. Reduced methylation of DNA in null met1-3 homozygous mutant plants causes ectopic expression of FWA but not of FIS2 in leaves. met1-3/+ plants, which show no ectopic expression of FWA, were used in the experiments presented in Figure 5A. GAPDH was used as a loading control.
(B) Analysis of DNA methylation using the methylation-sensitive restriction enzyme McrBC in wild-type Col leaves identifies a methylated region spanning −3104 to −1867 in the 5′ region of FIS2. The sensitivity to McrBC is decreased in the met1/met1 background, which indicates that MET1 controls DNA methylation in the −3104 to −1867 region. FWA repeats, which are methylated by MET1, were used as controls (Kinoshita et al., 2004).
(C) Bisulfite sequencing of the FIS2 5′ region, which contains a 200-bp domain enriched in methylated cytosine residues predominantly on CpGs (see Supplemental Figure 2 online for the full sequence). Analysis was done with genomic DNA from rosette leaves. The structure of the FIS2 genomic sequence is depicted, with the start codon indicated by 0. Exons are indicated by black boxes, and the 5′ and 3′ untranslated regions are indicated by white boxes. The extent of the partial translational fusion FIS2-GUS is indicated below the schematic genomic structure.
Activation of the FIS2 Maternal Allele by DME in the Central Cell
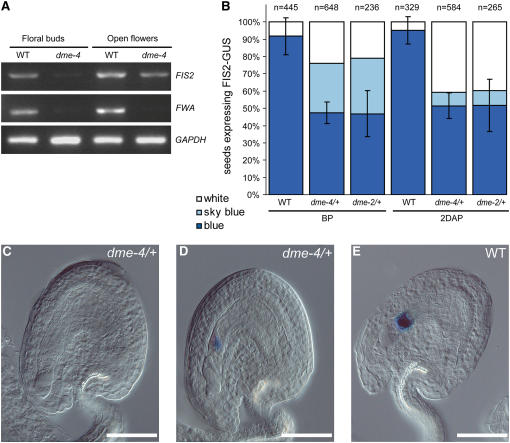
To investigate the role of DME on FIS2 expression, we tested the effect of dme loss of function on FIS2 expression. To this end, RT-PCR experiments were performed with mRNA extracted from buds and flowers, where DME was shown to be expressed (Choi et al., 2002). We used FWA as a control for the action of the dme-4 allele, which was isolated independently from other dme alleles (Guitton et al., 2004). FWA is expressed in the wild type before and after fertilization (Kinoshita et al., 2004), whereas in dme-4, a strong reduction of FWA transcription was observed (Figure 3A), as shown previously with dme-1 (Kinoshita et al., 2004). Similarly, dme-4 caused a reduction of FIS2 expression (Figure 3A), but the level of FIS2 expression was only partially reduced. The limited reduction of FIS2 expression by dme-4 was confirmed by analysis of the expression of the FIS2-GUS reporter, which includes most of the FIS2 locus and is only maternally expressed in wild-type endosperm (Figure 2C) (Luo et al., 2000). In self-pollinated FIS2-GUS/FIS2-GUS, dme-4/DME plants, all ovules inherit FIS2-GUS and 50% inherit the dme-4 mutation. Accordingly, we observed a significant reduction of FIS2-GUS expression in 50% of the ovules in which dme-4 was expected to be present (Figures 3B to 3D) compared with wild-type ovules (Figures 3B and 3E). However, half of dme-4 ovules still expressed the reporter at lower levels than wild-type ovules (Figures 3B and 3D). After fertilization, we observed a similar reduction of FIS2-GUS expression in endosperm, which inherits a maternal dme-4 (Figure 3B). Homozygous dme-4/dme-4 mutants are viable (Guitton et al., 2004), whereas dme-2/dme-2 causes seed developmental arrest (Choi et al., 2002). Hence, partial reduction of FIS2 expression in the dme-4 background could result from incomplete penetrance of the dme-4 mutation. However, a similar partial reduction of FIS2-GUS expression was obtained with the strong allele dme-2 (Choi et al., 2002) (Figure 3B). We conclude that DME is required for the expression of FIS2 in the central cell as well as in the endosperm. Persisting FIS2 expression in a fraction of dme ovules could be accounted for by either redundant functions for DME or by additional activation mechanisms sufficient to overcome the repressive effect of DNA methylation.
Figure 3.
Maternal Control of FIS2 Expression by DME.
(A) RT-PCR analyses performed on mRNA from buds (stages 11 and 12) and flowers (stage 13) (Smyth et al., 1990) show a lack of FWA expression and reduced FIS2 expression in the dme-4 homozygous mutant compared with the wild-type C24 accession. GAPDH was used as a control.
(B) to (E) Effects of dme on FIS2-GUS expression.
(B) Percentage of ovules expressing the FIS2-GUS reporter construct in dme-2/+ and dme-4/+ before pollination (BP) and at 2 DAP. Three different classes are observed: no expression (white), little expression (light blue), and wild-type expression (dark blue). Error bars correspond to the sd associated with each set of measurements.
(C) to (E) Micrographs showing the three categories of FIS2-GUS expression in the dme-4/+ mutant background: no expression (white; [C]), little expression (light blue; [D]), and wild-type expression (dark blue; [E]). Bars = 20 μm.
Imprinting of FWA and FIS2 in Endosperm Requires the Maintenance of Silencing by MET1 during Male Gametogenesis
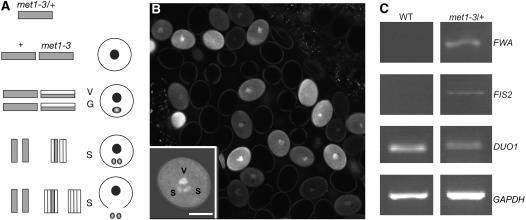
In contrast with female gametogenesis, male gametogenesis does not promote the expression of FWA, FIS2, and MEA (Luo et al., 2000; Kinoshita et al., 2004). We have investigated how silencing of FWA and FIS2 is maintained during male gametogenesis. Expression of MET1 has been detected in pollen, by transcriptome analysis (Honys and Twell, 2004), and there is evidence for MET1 function in the silencing of exogenous transcriptional reporters during male gametogenesis (Saze et al., 2003). Thus, we tested whether the loss of MET1 function affected the expression of FWA and FIS2 paternal alleles in pollen and later in endosperm. To this end, we used heterozygous met1-3/+ plants, which exhibit reduced MET1 activity only during gametogenesis. In met1-3/+ plants, the mutant met1 allele is inherited by half of the microspores, which develop in pollen grains after two cell divisions (McCormick, 2004). The generative cell is expected to inherit a hemimethylated copy and divides further to produce two male gametes, in which the genomic DNA is demethylated (Figure 4A). Accordingly, we observed ectopic expression of the transcriptional reporter FWA-GFP in segregating pollen from met1-3/+, FWA-GFP/FWA-GFP plants (Figure 4B) (47.4% of pollen bearing FWA-GFP fluorescence, sd = 6.7, n = 1372). Expression of FWA-GFP was observed in the sperm cells, which indicates that met1 pollen grains are able to provide a transcriptionally active FWA-GFP copy (Figure 4B, inset).
Figure 4.
Maintenance of FIS2 and FWA Silencing during Male Gametogenesis.
(A) During pollen development in met1-3/+ plants, half of haploid microspores produced by meiosis inherit the met1-3 mutation, and the other half inherit the wild-type MET1 allele. All microspores inherit a methylated copy of the genome (gray rectangles). DNA replication in met1-3 microspores produces hemimethylated DNA from the methylated template (gray/white rectangles). Hemimethylated DNA is inherited after mitosis by the vegetative nucleus (V) and the nucleus of the generative cell (G), which divides a second time and produces the two sperm cells (S). After this division, one sperm cell inherits a fully demethylated DNA copy (white rectangles) and the other sperm cell inherits hemimethylated DNA. The vegetative nucleus does not participate in fertilization. During sperm cell maturation, DNA replicates (Durbarry et al., 2005), and at fertilization, all sperm from met1-3 microspores is expected to contain at least one hemimethylated copy.
(B) Segregating population of pollen grains from FWA-GFP/FWA-GFP, met1-3/+ plants (confocal section). The inset represents a confocal section of a pollen grain expressing FWA-GFP in the vegetative cell (V) and in the two sperm cells (s). Bar = 5 μm.
(C) Ectopic expression of FWA and FIS2 observed by RT-PCR in RNA purified from stamens. GAPDH was used as a control to assess equal amounts of RNA loaded. DUO1 is expressed specifically in pollen (Rotman et al., 2005) and was used as a quality control for pollen mRNA extraction.
Our cytological observations were supported by RT-PCR experiments showing ectopic expression of FWA in mRNA populations extracted from stamens of met1-3/+ plants (Figure 4C). Thus, we used met1-3/+ plants to test whether loss of function of MET1 during male gametogenesis was sufficient to allow the expression of FWA paternal alleles in endosperm. In seeds resulting from crosses between wild-type ovules and met1-3/+ pollen, FWA transcripts were detected from both the paternal and maternal alleles (Figure 5A; note that polymorphism for Col is represented by a double band). Accordingly, the paternal copy of FWA-GFP is expressed in endosperm of seeds produced by crosses between wild-type ovules and pollen from met1-3/+, FWA-GFP/FWA-GFP plants (Figure 5C) (46.7%, sd = 5.2, n = 358). By contrast, no expression was observed when FWA-GFP was provided from a paternal wild-type background (Figure 5B; n = 126) (Kinoshita et al., 2004). We conclude that during gametogenesis, MET1 maintains FWA silencing and that FWA imprinting in endosperm requires MET1 activity during male gametogenesis. The MEA paternal allele is imprinted in endosperm (Kinoshita et al., 1999; Vielle-Calzada et al., 1999) and is predominantly expressed from its maternal allele during the first days after pollination (Jullien et al., 2006). We tested the effect of MET1 on MEA paternal allele expression in endosperm. MEA paternal allele expression was examined in seeds resulting from crosses between wild-type ovules and met1-3/+ pollen. In such crosses, we detected a predominance of maternal MEA allele expression, which is similar to the result obtained when crosses were made with wild-type male accessions. We concluded that MEA imprinting was not affected by loss of DNA methylation during male gametogenesis (Figure 5D). A similar result was recently obtained independently (Gehring et al., 2006). Thus, MET1 does not appear to play a major role in MEA silencing comparable to its role in FWA and FIS2 imprinting.
Figure 5.
Imprinting of FWA and FIS2 in Endosperm Relies on the Maintenance of Their Silencing during Male Gametogenesis.
The imprinting status of FWA, MEA, and FIS2 was analyzed in developing seeds resulting from crosses between wild-type ovules and pollen from met1-3/+ plants.
(A) Allele-specific RT-PCR detecting expression of the paternal and maternal alleles of FWA in seeds resulting from crosses between wild-type ovules (Ler accession) and pollen from met1-3/+ plants. (For the Col accession, two bands were obtained because digestion of the restriction polymorphism was partial; the lowest band was Col-specific [3 DAP].) We used met1-3/+ plants, which did not show expression of FWA in vegetative tissues (see Figure 2A). Although crosses between wild-type Ler and Col showed only expression of the maternal FWA allele, the FWA paternal allele was expressed in crosses with met1-3/+ pollen.
(B) and (C) Accordingly, at 2 DAP, the FWA-GFP reporter was expressed paternally in endosperm when provided by pollen from met1-3/+ plants (C) but not from wild-type plants (B).
(D) Allele-specific RT-PCR performed on crosses between wild-type accessions RLD and Col detected expression of the maternal MEA allele, typical of MEA imprinting (1 DAP). MEA imprinting was not altered if met1-3/+ plants provided the MEA paternal allele.
(E) Crosses between wild-type accessions C24 and Col showed expression only from the maternal FIS2 allele typical of FIS2 imprinted status in wild-type endosperm. By contrast, allele-specific RT-PCR showed expression of the paternal and maternal alleles of FIS2 in seeds resulting from crosses between ovules from wild-type C24 and pollen from met1-3/+ plants (Col accession; 3 DAP).
(F) to (J) Rescue of endosperm development upon removal of the FIS2 paternal allele imprint in met1-3 pollen.
(F) and (H) GFP fluorescence of the enhancer trap marker KS117 was increased uniformly in endosperm of the fis2 homozygous mutant (F) compared with the wild type (H), in which it was restricted to the endosperm posterior pole at 5 DAP.
(G) After pollination of fis2/fis2, KS117/KS117 plants by pollen from met1-3/+ plants, half of the seeds showed a pattern of KS117 expression similar to the KS117 pattern in wild-type endosperm (asterisks). Such seeds also showed a rescue of cytological alteration in fis2 endosperm ([I] and [J]).
(I) The phenotype conferred by fis2 is characterized by the absence of cellularization of the syncytial endosperm and the enlargement of the posterior pole opposite the embryo (Guitton et al., 2004).
(J) Rescue of such phenotypic traits was obtained in 19% of seeds resulting from crosses between fis2/fis2 plants and met1-3/+ pollen.
Bars = 50 μm ([B], [C], [I], and [J]) and 200 μm ([F] to [H]). GAPDH was used as a control for the RT-PCR analyses ([A], [D], and [E]).
Similarly, we tested the impact of met1 loss of function during male gametogenesis on FIS2 silencing in pollen and imprinting in endosperm. FIS2 was ectopically expressed in stamens from met1-3/+ plants (Figure 4C). Pollination of wild-type ovules with met1-3/+ plants caused expression of the FIS2 paternal allele in endosperm (Figure 5E), which indicates that FIS2 silencing by MET1 during male gametogenesis is required for FIS2 imprinting. Previously, it was shown that maternal inheritance of fis2 loss-of-function alleles causes major defects in endosperm development, including overexpression of the KS117 marker (Sørensen et al., 2001). In addition, endosperm produced by fis2 mothers fails to cellularize during the syncytial stage (Chaudhury et al., 1997). In crosses between fis2 ovules and met1 pollen, the wild-type FIS2 paternal allele is activated by met1 and is expected to rescue the maternal loss-of-function fis2 allele. We next tested whether in met1-3/+ plants the demethylated expressed paternal allele of FIS2 is sufficient to rescue the inactive mutant fis2 allele provided maternally to the developing endosperm. Heterozygous met1-3/+ plants produce 50% met1 pollen. The KS117 expression pattern was rescued in 49% of seeds produced by crosses with pollen from met1-3/+ plants and fis2/fis2, KS117/KS117 plants used as females (sd = 5.3, n = 459) (Figure 5G). In these crosses, 19% of seeds showed cellular endosperm similar to wild-type seeds (sd = 5.1, n = 365) (Figure 5J). Hence, met1 loss of function during pollen development is sufficient to remove the repressive epigenetic marks from the FIS2 paternal allele, which remains active in endosperm.
We conclude that MET1 activity is essential to maintain the silencing of FIS2 and FWA during male gametogenesis. If MET1 activity is absent during pollen development, the sperm cells deliver a transcriptionally active paternal allele to endosperm and imprinting of FIS2 and FWA in endosperm is compromised. Regulation of MEA imprinting appears to depend on a distinct regulatory mechanism independent of MET1. Instead, recent evidence supports the notion that MEA silencing relies on histone methylation by Polycomb group complexes (Gehring et al., 2006; Jullien et al., 2006).
MET1 Is Required for Silencing the FWA Paternal Allele in Endosperm
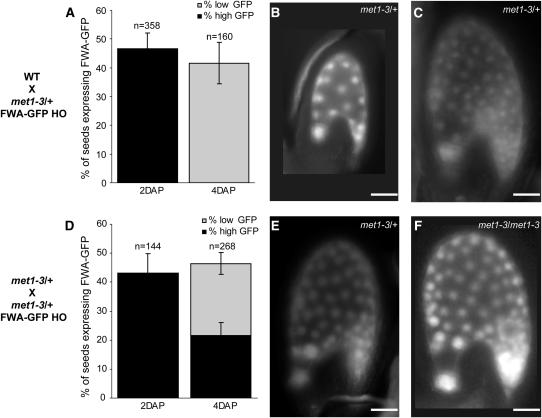
We have shown that FIS2 and FWA imprinted status in endosperm requires MET1 activity before fertilization. However, the requirement of MET1 after fertilization and during endosperm development has not yet been reported. Although the paternal FWA-GFP copy provided by met1 pollen becomes expressed in endosperm, its expression gradually decreases in signal intensity between 2 and 4 DAP (Figures 6A to 6C) and becomes gradually undetectable after 6 DAP (data not shown). We hypothesized that this resulted from progressive silencing of the paternal FWA-GFP copy by maternally provided MET1 activity in met1/+ endosperm. According to this hypothesis, a loss of function of MET1 in met1/met1 endosperm is expected to sustain the expression of a paternally provided copy of FWA-GFP. Transmission of met1-3 through male and female gametes follows Mendelian proportions (see Supplemental Table 1 online), and we expected production of 25% met1/met1 seeds in crosses between met1/+ plants. We performed crosses between ovules of met1-3/+ plants and pollen from met1/+, FWA-GFP/FWA-GFP plants, leading to 25% met1/met1 seeds and 25% met1/+ seeds with a paternal FWA-GFP copy from met1 pollen. We observed two distinct classes of seeds expressing FWA-GFP either in met1-3/+ or in met1/met1 endosperm. One-quarter of seeds showed low expression levels comparable to those of seeds with met1/+ endosperm (Figures 6D and 6E), and another one-quarter of seeds showed high levels of FWA-GFP expression at 4 DAP (Figures 6D and 6F). Given the Mendelian proportion of each seed class, the latter class presumably corresponds to met1/met1 endosperm with no MET1 activity, in which progressive silencing of a demethylated FWA-GFP paternal copy does not occur. These results suggest that MET1 is continuously required for silencing of the paternal FWA allele during endosperm development to maintain FWA imprinted status.
Figure 6.
Maintenance of FWA Imprinting in Endosperm Depends on MET1.
(A) At 2 DAP, after fertilization of wild-type ovules with pollen from met1-3/+, FWA-GFP/FWA-GFP plants, expression of the paternally provided allele of FWA-GFP is observed in endosperm in half of the seeds.
(B) and (C) The intensity of FWA-GFP expression decreases during endosperm development from 2 DAP (B) to 4 DAP (C).
(D) In contrast with the gradual silencing of FWA-GFP paternal expression in met1-3/+ endosperm, one-quarter of the seeds show expression of the FWA-GFP paternal allele in crosses between ovules from met1-3/+ plants and pollen carrying met1-3/+ and FWA-GFP/FWA-GFP.
(E) and (F) One-quarter of the seeds express low levels of FWA-GFP (E) and are presumed to be heterozygous for met1-3. Another one-quarter of the seeds show higher sustained expression of FWA-GFP (F) and are presumably homozygous for met1-3 in endosperm.
Bars = 80 μm ([B], [C], [E], and [F]). The number of seeds observed for each cross is indicated above each bar. sd is indicated for each measurement. HO, homozygous.
DISCUSSION
Imprinting of FIS2 and FWA Relies on MET1 Activity during Male Gametogenesis
We have shown that FIS2 undergoes monoallelic maternal expression in endosperm. A gene is considered to be imprinted when it is expressed after fertilization and primarily from one of its two parental alleles (Vielle-Calzada et al., 1999). We detected FIS2 maternal transcripts up to 5 d after pollination and showed that the FIS2 paternal allele can be expressed in endosperm during met1 loss of function in gametogenesis. Thus, FIS2 is actively transcribed after fertilization and is expressed only from its maternal allele. We conclude that FIS2 is a maternally expressed imprinted gene. MET1 activity is necessary during endosperm development for sustained silencing of the paternal copy of FWA. We anticipate a similar dependence of the FIS2 paternal allele silencing on MET1 activity in endosperm. In conclusion, when MET1 activity is removed, as in met1-3 mutants, the expression of MET1-dependent imprinted genes is maintained beyond the wild-type schedule for both parental alleles.
Continuous DNA methylation by MET1 at specific sites during vegetative development maintains the silencing of both parental FWA and FIS2 alleles. The imprinted genes are expressed during female gamete maturation, whereas they remain silent during male gametogenesis. This imbalance of gene expression in gametes prefigures imprinting in endosperm after fertilization. In the central cell, silencing of imprinted genes expressed in endosperm would be removed via DNA demethylation mediated by the glycosylase DME. This was shown for FWA (Kinoshita et al., 2004), and our data indicate that DME also acts on FIS2 together with other unknown activators, because FIS2 expression is only partially suppressed in dme ovules. We have shown that MET1 is required during male gametogenesis to maintain FWA and FIS2 silencing. This provides an example for the role played by MET1 during male gametogenesis, which was originally demonstrated with an exogenous reporter construct (Saze et al., 2003). Furthermore, we have shown that sperm cells deficient for met1 provide activated FIS2 paternal alleles, which are able to rescue endosperm defects caused by the maternally provided fis2 null alleles. In conclusion, imprinting results from the maintenance of DNA methylation in sperm cells, whereas DNA methylation is removed in the female gamete (Figure 7). Our model thus extends and complements the model proposed previously (Berger, 2004; Kinoshita et al., 2004).
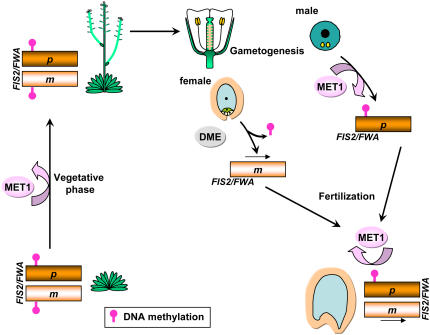
Figure 7.
Model for the Dual Control of Parental Genomic Imprinting by DNA Methylation in Plants.
The genes FIS2 and FWA are imprinted in endosperm and silenced by the continuous action of MET1, which targets methylation (pink circles) to an element in the promoter specific for each gene. MET1-dependent silencing of FIS2 and FWA is required during the vegetative phase, male gametogenesis, and endosperm development, when it maintains silencing of the paternal (p) allele. FIS2 and FWA expression is activated during female gametogenesis by the DNA glycosylase DME, leading to the expression of their maternal (m) allele in endosperm after fertilization. In endosperm, the paternal copy remains silenced through the continuous action of MET1, whereas the maternal copy is active, leading to an imprinted expression.
Similarly, in mammals, loss of function of de novo DNA methylation during gametogenesis caused a lack of imprints (epigenetic marks responsible for silencing) establishment in mice (Monk, 1990; Kaneda et al., 2004). Thus, during gametogenesis, crucial steps are made necessary for imprinting genes in both mammals and plants, but the establishment of imprints occurs through distinct DNA methylation mechanisms. In contrast with plants, extensive demethylation occurs in the mammalian germ line and erases parental imprints (Delaval and Feil, 2004). Imprints are reestablished in the male and female germ lines by a specific de novo DNA methyltransferase (Kaneda et al., 2004) and not by the maintenance DNA methyltransferase, as in flowering plants.
MET1 Controls Imprinting in Arabidopsis via Diverse cis-Elements
In spite of the similarities between the expression patterns of maternally imprinted genes identified in Arabidopsis, we show that imprinting mechanisms of different genes are distinct. Our results demonstrate that MET1 plays a major role in FIS2 and FWA imprinting, but some aspects of MEA imprinting do not appear to depend critically on MET1. Silencing of the MEA paternal allele is not mediated by MET1 but rather depends on the continuous action of histone methylation mediated by Polycomb group complexes (Gehring et al., 2006; Jullien et al., 2006). Nevertheless, activation of the maternal allele of MEA depends on the removal of MET1-dependent DNA methylation (Gehring et al., 2006). It is not clear, however, whether DME also leads to the removal of Polycomb group–dependent histone methylation marks. The regulation of MEA imprinting also could involve DDM1, a SWI2/SNF2 factor, which interferes with MEA function (Vielle-Calzada et al., 1999) and interacts with DNA methyl binding proteins (Zemach et al., 2005).
MET1 was shown to target tandem repeats in the promoter of FWA and to regulate its imprinting (Kinoshita et al., 2004), although a direct connection between these two events remains to be demonstrated. Additional silencing mechanisms are involved in the silencing of FWA (Chan et al., 2004), but it is not clear whether they control FWA imprinting. Upstream of the FIS2 coding sequence, we could detect neither repetitive sequence nor transposons with bioinformatics analysis, which successfully detected such elements in the MEA promoter (Spillane et al., 2004). Rather, we show that methylation is directed to a 200-bp region upstream of the FIS2 coding sequence. A putative MET1-targeted control element has been detected upstream of one of the maize homologues of FIE, which is also imprinted and expressed in endosperm (Danilevskaya et al., 2003). This element consists of CpG-enriched sequences different from the cis-elements identified in Arabidopsis for FIS2. Based on the limited data available, we are able to conclude that imprinting in flowering plants relies on several types of cis-elements and several silencing mechanisms. It is also becoming clear that imprinting in mammals does not rely on a single type of epigenetic transcriptional regulation. Both maintenance and de novo DNA methyltransferase are involved, as well as Polycomb group activities.
DNA methylation targets several types of elements in imprinting control regions, which can be related to altered transposable elements and CpG-enriched regions (Constancia et al., 2004; Delaval and Feil, 2004). The diversity of cis control elements and trans-acting mechanisms involved in imprinting found in flowering plants and mammals contrasts with the relative similarity of functions for some targets of imprinting. Most imprinted genes identified in flowering plants play an essential role in endosperm development and thus indirectly control embryo nutrition in the developing seed. In animals, several imprinted genes were found to have an essential function in reproduction and to affect interactions between the mother and the developing embryo or infant (Constancia et al., 2004). It is thus likely that the evolution of imprinting did not depend on selection of a molecular epigenetic mechanism but more likely stemmed from biological constraints linked to reproductive innovations. Selective mechanisms might have used available silencing machineries and targeted potential cis-elements already present at the loci of some genes important for reproduction. This evolutionary process eventually led to the actual state of diverse cis and trans control elements associated with imprinting.
METHODS
Plant Materials and Growth Conditions
The MET1 antisense line of Arabidopsis thaliana was provided by J. Finnegan (Finnegan et al., 1996). This line is characterized by a loss of 70% CpG methylation and by a dominant effect of the MET1 antisense in vegetative diploid tissues. As the line is maintained by selfing, it likely accumulates epimutations of unknown nature, which may interfere with our experiments. Hence, for most experiments, we used the met1-3 (Col) line provided by J. Paszkowski (Saze et al., 2003) and took care to propagate the met1-3 line as a heterozygote. Homozygous mutants cmt3, drm1, drm2, and kyp (Ler) came from S.E. Jacobsen's laboratory (Lindroth et al., 2001; Cao and Jacobsen, 2002a; Jackson et al., 2002). The mutant dme-2 (Col) was provided by R. Fischer (Choi et al., 2002). The mutants dme-4 and fis2-6 (C24) were characterized previously in the laboratory of F.B. (Guitton et al., 2004). FIEcs (Col) plants were characterized previously by N.O.'s group (Katz et al., 2004). We used met1-3/+ plants in which neither FIS2 nor FWA transcripts could be detected in vegetative tissues (Figure 2A), which indicated that the parent plants were not affected by the mutation during the vegetative phase but only during the production of male gametes.
The KS117 line was identified after a screen in Jim Haseloff's enhancer trap line collection (Haseloff, 1999). The transgenic reporter line FIS2-GUS was kindly provided by A. Chaudhury (Chaudhury et al., 1997; Luo et al., 2000). The FWA-GFP fusion was described previously (Kinoshita et al., 2004).
After 3 d at 4°C in the dark, seeds were germinated and grown on soil. Plants were cultured in a growth chamber under short days (8 h of light at 20°C/16 h of dark at 16°C; 60 to 70% RH) until rosettes were formed. Plants were transferred to long days at 20°C (14 h of light/10 h of dark) to induce flowering and grown until seeds were harvested. Crosses were performed by emasculating flowers and pollinating them manually. Siliques were harvested at various times after pollination, as indicated in figure legends and in the text.
RT-PCR Analyses
Sample tissues were collected from Arabidopsis plants and immediately frozen in liquid nitrogen. Tissues were ground, and total RNA was prepared using the RNeasy mini kit (Qiagen). DNase treatment was done on 2 μg of total RNA using the DNase free kit (Ambion).
For reverse transcription, 500 ng of total RNA was incubated for 1 h at 42°C with 200 units of RevertAid Moloney murine leukemia virus reverse transcriptase (Fermentas) in a 20-μL reaction mixture containing 4 μM oligo(dT) 12 primer, appropriate reaction buffer, 1 mM deoxynucleotide triphosphate, and 40 units of recombinant RNasin ribonuclease inhibitor (Promega). The reaction was stopped by incubation at 70°C for 10 min. Equal amounts of RT products were used to perform subsequent PCRs. Primers used to amplify FIS2 were Fis2-R5018 (5′-CCTGCATTGTTTGGGAGTGATAGAA-3′) and Fis2-F3412 (5′-GGATGATGTAGGAAATCCCCAATTGAGCCCTTTG-3′). Allele-specific RT-PCR of FWA was performed as described (Kinoshita et al., 2004). Allele-specific RT-PCR of MEA was performed as described (Kinoshita et al., 1999). Primers used to amplify FWA were FWA-RTf2 (5′-GTTACATGGATTGAACAAGCGG-3′) and FWA-RTr2 (5′-ACCTTGAATGAGTGCAGCAGTTG-3′). Primers used to amplify the control, GAPDH RNA, were GAPDH3′ (5′-GTAGCCCCACTCGTTGTCGTA-3′) and GAPDH5′ (5′-AGGGTGGTGCCAAGAAGGTTG-3′). A negative control without RT was used for each RT-PCR experiment. We designed primers spanning introns, such that genome-specific and cDNA-specific PCR products could easily be distinguished by size differences.
Microscopy
FIS2-GUS expression was analyzed as described by Jullien et al. (2006). KS117 GFP fluorescence was analyzed in 5-DAP seeds with a stereomicroscope (MZ16FA; Leica). GUS-stained developing seeds or pistils cleared with a derivative of Hoyer's medium were observed with differential interference contrast optics. In developing seeds mounted in 50% glycerol solution, FWA-GFP fluorescence was analyzed with a GFP-specific filter set with a ×20 planapo objective (DM6000 B; Leica). Images were acquired with a DXM1200F digital camera (Nikon) and processed using Metamorph (version 6.2; Universal Imaging). The settings for observation were saved and applied to all seeds observed to allow comparison of fluorescence intensities at 2 and 4 DAP. Pollen was prepared as described (Rotman et al., 2005). In pollen, FWA-GFP fluorescence was analyzed by confocal microscopy (LSM 510) with a ×40 water-immersion objective, and images were further processed with Adobe Photoshop and Adobe Illustrator.
For each measurement of FWA-GFP and FIS2-GUS in developing seeds, counting was established on individual siliques. Measurements of FWA-GFP expression in pollen were performed by placing several anthers on a slide. Fifteen individual fields were observed, each containing 20 to 50 pollen grains, which were scored, thus establishing the data set for determining the percentage of positive expressing pollen. Standard deviation was calculated for each genetic background used based on the average obtained per silique or per field of pollen grains observed.
McrBC Analysis
DNA methylation status in the FIS2 promoter was analyzed by PCR amplification of DNA that had been pretreated with McrBC endonuclease, which digests methylated DNA (New England Biolabs). One hundred nanograms of genomic DNA from rosette leaves of met1-3 and Col-0 plants was digested overnight with 10 units of McrBC, according to the manufacturer's instructions. Ten nanograms of template DNA was then amplified by PCR with ExTaq polymerase (Takara) using the following primers: F3697Tf (5′-AAAGAGTTATGGGYYGAAG-3′) and F3697Tr (5′-GGGCAGAAACATGGTCCA-3′), FIS2.-3104BisBf (5′-ACAARTCACAACCAAAACCTTAA-3′) and FIS2.-2312r (5′-TGCCAAATAGCACAATGAGGA-3′), FIS2.-3104BisBf and FIS2.-1867r (5′-ATGTTGCGCCTTCACCACTT-3′), and FIS2.-1218BisBf (5′-TCCARTCCACTATTCTTTACTCTT-3′) and FIS2.-16r (5′-GTAGTTGAATCTTATTTTCCCACCTGA-3′).
Bisulfite Sequencing
Bisulfite sequencing was performed as described (Paulin et al., 1998). DNA was purified from rosette leaves. After chemical bisulfite reaction, the top strand of the FIS2 promoter from −2185 to −1768 (relative to the translation start) was amplified with primers FIS2.-2185BisTf (5′-AGGTYYAATYGYATATTTATTTAGGGTTTYGGGT-3′) and FIS2.-1768BisTr (5′-TCCTACATTTTAATAAAATATTACTRAATCTAARCA-3′), and the bottom strand from −2110 to −1670 was amplified with primers FIS2.-2110BisBf (5′-TACCAAACCCRAARAARAAAAATTTACAA-3′) and FIS2.-1670BisBr (5′-TGATGGYAGTAGAGATTATAAGAAAAGA-3′). The amplified PCR fragments were gel-purified and cloned into pT7Blue plasmid (Novagen), and then six to nine independent clones were sequenced. The ASA1 gene was used as a positive control for the bisulfite chemical reaction (Jeddeloh et al., 1998). We used met1-3 homozygous mutants segregated from met1-3/+ heterozygotes because these plants do not accumulate epimutations, in contrast with MET1as selfed plants.
Accession Numbers
Arabidopsis Genome Initiative numbers for the genes used in this study are At1g02580 (MEA), At2g35670 (FIS2), At4g25530 (FWA), and At5g04560 (DME).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Analysis of FIS2 Imprinting at 3 and 5 DAP.
Supplemental Figure 2. Bisulfite Sequencing of the FIS2 5′ Region Contains a 200-bp Domain Enriched in Methylated Cytosine Residues Predominantly on CpGs.
Supplemental Table 1. Resistance to BASTA Is Associated with met1-3.
Supplementary Material
Acknowledgments
We thank all contributors of materials cited in Methods and the Nottingham Arabidopsis Resource Centre and ABRC. P.E.J. and F.B. were supported by the Temasek Lifesciences Laboratory and by the National University of Singapore. N.O. was supported by the Israel Science Foundation (Grant 574-04) and by the U.S.–Israel Binational Agricultural Research and Development Fund (Grant IS-3604-04c).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Frédéric Berger (fred@tll.org.sg).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.106.041178.
References
- Adams, S., Vinkenoog, R., Spielman, M., Dickinson, H.G., and Scott, R.J. (2000). Parent-of-origin effects on seed development in Arabidopsis thaliana require DNA methylation. Development 127 2493–2502. [DOI] [PubMed] [Google Scholar]
- Bastow, R., Mylne, J.S., Lister, C., Lippman, Z., Martienssen, R.A., and Dean, C. (2004). Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427 164–167. [DOI] [PubMed] [Google Scholar]
- Berger, F. (2003). Endosperm: The crossroad of seed development. Curr. Opin. Plant Biol. 6 42–50. [DOI] [PubMed] [Google Scholar]
- Berger, F. (2004). Plant sciences. Imprinting—A green variation. Science 303 483–485. [DOI] [PubMed] [Google Scholar]
- Cao, X., and Jacobsen, S.E. (2002. a). Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr. Biol. 12 1138–1144. [DOI] [PubMed] [Google Scholar]
- Cao, X., and Jacobsen, S.E. (2002. b). Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc. Natl. Acad. Sci. USA 99 (suppl. 4), 16491–16498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, S.W., Henderson, I.R., and Jacobsen, S.E. (2005). Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat. Rev. Genet. 6 351–360. [DOI] [PubMed] [Google Scholar]
- Chan, S.W., Zilberman, D., Xie, Z., Johansen, L.K., Carrington, J.C., and Jacobsen, S.E. (2004). RNA silencing genes control de novo DNA methylation. Science 303 1336. [DOI] [PubMed] [Google Scholar]
- Chaudhury, A.M., Ming, L., Miller, C., Craig, S., Dennis, E.S., and Peacock, W.J. (1997). Fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 94 4223–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, Y., Gehring, M., Johnson, L., Hannon, M., Harada, J.J., Goldberg, R.B., Jacobsen, S.E., and Fischer, R.L. (2002). DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell 110 33–42. [DOI] [PubMed] [Google Scholar]
- Constancia, M., Kelsey, G., and Reik, W. (2004). Resourceful imprinting. Nature 432 53–57. [DOI] [PubMed] [Google Scholar]
- Danilevskaya, O.N., Hermon, P., Hantke, S., Muszynski, M.G., Kollipara, K., and Ananiev, E.V. (2003). Duplicated fie genes in maize: Expression pattern and imprinting suggest distinct functions. Plant Cell 15 425–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaval, K., and Feil, R. (2004). Epigenetic regulation of mammalian genomic imprinting. Curr. Opin. Genet. Dev. 14 188–195. [DOI] [PubMed] [Google Scholar]
- Durbarry, A., Vizir, I., and Twell, D. (2005). Male germ line development in Arabidopsis. duo pollen mutants reveal gametophytic regulators of generative cell cycle progression. Plant Physiol. 137 297–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan, E.J., Peacock, W.J., and Dennis, E.S. (1996). Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc. Natl. Acad. Sci. USA 93 8449–8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan, E.J., Peacock, W.J., and Dennis, E.S. (2000). DNA methylation, a key regulator of plant development and other processes. Curr. Opin. Genet. Dev. 10 217–223. [DOI] [PubMed] [Google Scholar]
- Gehring, M., Choi, Y., and Fischer, R.L. (2004). Imprinting and seed development. Plant Cell 16 (suppl.), S203–S213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring, M., Huh, J.H., Hsieh, T.F., Penterman, J., Choi, Y., Harada, J.J., Goldberg, R.B., and Fischer, R.L. (2006). DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell 124 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus, U., Vielle-Calzada, J.P., Hoeppner, M.A., and Gagliano, W.B. (1998). Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 280 446–450. [DOI] [PubMed] [Google Scholar]
- Guignard, M.L. (1899). Sur les anthérozoïdes et la double copulation sexuelle chez les végétaux angiospermes. Rev. Gen. Bot. 11 129–135. [DOI] [PubMed] [Google Scholar]
- Guitton, A.E., Page, D.R., Chambrier, P., Lionnet, C., Faure, J.E., Grossniklaus, U., and Berger, F. (2004). Identification of new members of Fertilisation Independent Seed polycomb group pathway involved in the control of seed development in Arabidopsis thaliana. Development 131 2971–2981. [DOI] [PubMed] [Google Scholar]
- Haseloff, J. (1999). GFP variants for multispectral imaging of living cells. Methods Cell Biol. 58 139–151. [DOI] [PubMed] [Google Scholar]
- Honys, D., and Twell, D. (2004). Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol. 5 R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, J.P., Lindroth, A.M., Cao, X., and Jacobsen, S.E. (2002). Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416 556–560. [DOI] [PubMed] [Google Scholar]
- Jeddeloh, J.A., Bender, J., and Richards, E.J. (1998). The DNA methylation locus DDM1 is required for maintenance of gene silencing in Arabidopsis. Genes Dev. 12 1714–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien, P.E., Katz, A., Oliva, M., Ohad, N., and Berger, F. (2006). Polycomb group complexes self-regulate imprinting of the polycomb group gene MEDEA in Arabidopsis. Curr. Biol. 16 486–492. [DOI] [PubMed] [Google Scholar]
- Kaneda, M., Okano, M., Hata, K., Sado, T., Tsujimoto, N., Li, E., and Sasaki, H. (2004). Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 429 900–903. [DOI] [PubMed] [Google Scholar]
- Katz, A., Oliva, M., Mosquna, A., Hakim, O., and Ohad, N. (2004). FIE and CURLY LEAF polycomb proteins interact in the regulation of homeobox gene expression during sporophyte development. Plant J. 37 707–719. [DOI] [PubMed] [Google Scholar]
- Kinoshita, T., Miura, A., Choi, Y., Kinoshita, Y., Cao, X., Jacobsen, S.E., Fischer, R.L., and Kakutani, T. (2004). One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science 303 521–523. [DOI] [PubMed] [Google Scholar]
- Kinoshita, T., Yadegari, R., Harada, J.J., Goldberg, R.B., and Fischer, R.L. (1999). Imprinting of the MEDEA polycomb gene in the Arabidopsis endosperm. Plant Cell 11 1945–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosue, T., Ohad, N., Yadegari, R., Hannon, M., Dinneny, J., Wells, D., Katz, A., Margossian, L., Harada, J.J., Goldberg, R.B., and Fischer, R.L. (1999). Control of fertilization-independent endosperm development by the MEDEA polycomb gene in Arabidopsis. Proc. Natl. Acad. Sci. USA 96 4186–4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler, C., and Grossniklaus, U. (2005). Seed development and genomic imprinting in plants. Prog. Mol. Subcell. Biol. 38 237–262. [DOI] [PubMed] [Google Scholar]
- Köhler, C., Hennig, L., Bouveret, R., Gheyselinck, J., Grossniklaus, U., and Gruissem, W. (2003). Arabidopsis MSI1 is a component of the MEA/FIE polycomb group complex and required for seed development. EMBO J. 22 4804–4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler, C., Page, D.R., Gagliardini, V., and Grossniklaus, U. (2005). The Arabidopsis thaliana MEDEA polycomb group protein controls expression of PHERES1 by parental imprinting. Nat. Genet. 37 28–30. [DOI] [PubMed] [Google Scholar]
- Lindroth, A.M., Cao, X., Jackson, J.P., Zilberman, D., McCallum, C.M., Henikoff, S., and Jacobsen, S.E. (2001). Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 292 2077–2080. [DOI] [PubMed] [Google Scholar]
- Luo, M., Bilodeau, P., Dennis, E.S., Peacock, W.J., and Chaudhury, A. (2000). Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc. Natl. Acad. Sci. USA 97 10637–10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, M., Bilodeau, P., Koltunow, A., Dennis, E.S., Peacock, W.J., and Chaudhury, A.M. (1999). Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96 296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, M., Dennis, E.S., Berger, F., Peacock, W.J., and Chaudhury, A. (2005). MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) kinase gene, are regulators of seed size in Arabidopsis. Proc. Natl. Acad. Sci. USA 102 17531–17536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshwari, P. (1950). An Introduction to the Embryology of Angiosperms. (New York: McGraw-Hill).
- McCormick, S. (2004). Control of male gametophyte development. Plant Cell 16 (suppl.), S142–S153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk, M. (1990). Changes in DNA methylation during mouse embryonic development in relation to X-chromosome activity and imprinting. Philos. Trans. R. Soc. Lond. B Biol. Sci. 326 299–312. [DOI] [PubMed] [Google Scholar]
- Ohad, N., Yadegari, R., Margossian, L., Hannon, M., Michaeli, D., Harada, J.J., Goldberg, R.B., and Fischer, R.L. (1999). Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell 11 407–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulin, R., Grigg, G.W., Davey, M.W., and Piper, A.A. (1998). Urea improves efficiency of bisulphite-mediated sequencing of 5′-methylcytosine in genomic DNA. Nucleic Acids Res. 26 5009–5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotman, N., Durbarry, A., Wardle, A., Yang, W.C., Chaboud, A., Faure, J.E., Berger, F., and Twell, D. (2005). A novel class of MYB factors controls sperm-cell formation in plants. Curr. Biol. 15 244–248. [DOI] [PubMed] [Google Scholar]
- Saze, H., Scheid, O.M., and Paszkowski, J. (2003). Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat. Genet. 34 65–69. [DOI] [PubMed] [Google Scholar]
- Smyth, D.R., Bowman, J.L., and Meyerowitz, E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen, M.B., Chaudhury, A.M., Robert, H., Bancharel, E., and Berger, F. (2001). Polycomb group genes control pattern formation in plant seed. Curr. Biol. 11 277–281. [DOI] [PubMed] [Google Scholar]
- Spillane, C., Baroux, C., Escobar-Restrepo, J.M., Page, D.R., Laoueille, S., and Grossniklaus, U. (2004). Transposons and tandem repeats are not involved in the control of genomic imprinting at the MEDEA locus in Arabidopsis. Cold Spring Harb. Symp. Quant. Biol. 69 465–475. [DOI] [PubMed] [Google Scholar]
- Takeda, S., and Paszkowski, J. (2006). DNA methylation and epigenetic inheritance during plant gametogenesis. Chromosoma 115 27–35. [DOI] [PubMed] [Google Scholar]
- Tariq, M., Saze, H., Probst, A.V., Lichota, J., Habu, Y., and Paszkowski, J. (2003). Erasure of CpG methylation in Arabidopsis alters patterns of histone H3 methylation in heterochromatin. Proc. Natl. Acad. Sci. USA 100 8823–8827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielle-Calzada, J.P., Baskar, R., and Grossniklaus, U. (2000). Delayed activation of the paternal genome during seed development. Nature 404 91–94. [DOI] [PubMed] [Google Scholar]
- Vielle-Calzada, J.P., Thomas, J., Spillane, C., Coluccio, A., Hoeppner, M.A., and Grossniklaus, U. (1999). Maintenance of genomic imprinting at the Arabidopsis medea locus requires zygotic DDM1 activity. Genes Dev. 13 2971–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinkenoog, R., Spielman, M., Adams, S., Fischer, R.L., Dickinson, H.G., and Scott, R.J. (2000). Hypomethylation promotes autonomous endosperm development and rescues postfertilization lethality in fie mutants. Plant Cell 12 2271–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vire, E., et al. (2006). The polycomb group protein EZH2 directly controls DNA methylation. Nature 439 871–874. [DOI] [PubMed] [Google Scholar]
- Xiao, W., Custard, K.D., Brown, R.C., Lemmon, B.E., Harada, J.J., Goldberg, R.B., and Fischer, R.L. (2006). DNA methylation is critical for Arabidopsis embryogenesis and seed viability. Plant Cell 18 805–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, W., Gehring, M., Choi, Y., Margossian, L., Pu, H., Harada, J.J., Goldberg, R.B., Pennell, R.I., and Fischer, R.L. (2003). Imprinting of the MEA polycomb gene is controlled by antagonism between MET1 methyltransferase and DME glycosylase. Dev. Cell 5 891–901. [DOI] [PubMed] [Google Scholar]
- Yadegari, R., Kinoshita, T., Lotan, O., Cohen, G., Katz, A., Choi, Y., Nakashima, K., Harada, J.J., Goldberg, R.B., Fischer, R.L., and Ohad, N. (2000). Mutations in the FIE and MEA genes that encode interacting polycomb proteins cause parent-of-origin effects on seed development by distinct mechanisms. Plant Cell 12 2367–2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach, A., Li, Y., Wayburn, B., Ben-Meir, H., Kiss, V., Avivi, Y., Kalchenko, V., Jacobsen, S.E., and Grafi, G. (2005). DDM1 binds Arabidopsis methyl-CpG binding domain proteins and affects their subnuclear localization. Plant Cell 17 1549–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.