Abstract
During meiosis, homologous chromosomes (homologs) undergo recombinational interactions, which can yield crossovers (COs) or noncrossovers. COs exhibit interference; they are more evenly spaced along the chromosomes than would be expected if they were placed randomly. The protein complexes involved in recombination can be visualized as immunofluorescent foci. We have analyzed the distribution of such foci along meiotic prophase chromosomes of the mouse to find out when interference is imposed and whether interference manifests itself at a constant level during meiosis. We observed strong interference among MLH1 foci, which mark CO positions in pachytene. Additionally, we detected substantial interference well before this point, in late zygotene, among MSH4 foci, and similarly, among replication protein A (RPA) foci. MSH4 foci and RPA foci both mark interhomolog recombinational interactions, most of which do not yield COs in the mouse. Furthermore, this zygotene interference did not depend on SYCP1, which is a transverse filament protein of mouse synaptonemal complexes. Interference is thus not specific to COs but may occur in other situations in which the spatial distribution of events has to be controlled. Differences between the distributions of MSH4/RPA foci and MLH1 foci along synaptonemal complexes might suggest that CO interference occurs in two successive steps.
Keywords: crossing-over, immunofluorescence, meiosis
Meiosis consists of two divisions, meiosis I and II, by which a diploid cell produces four haploid daughters. Reduction in ploidy occurs at meiosis I, when homologous chromosomes (homologs) disjoin. This event is prepared during meiotic prophase, when homologs recognize each other and form stable pairs (bivalents) that can line up in the metaphase I spindle. In most eukaryotes, including mouse and yeast, both the recognition of homologs and the formation of stable bivalents depend on recombinational interactions between homologs (reviewed in ref. 1). For this process, the meiotic prophase cell actively induces DNA double-strand breaks (DSBs) and repairs them by homologous recombination, using preferably a nonsister chromatid of the homolog as template (2). In species such as yeast and mouse, most interhomolog recombinational interactions are not resolved as reciprocal exchanges [crossovers (COs)] and probably serve homolog recognition and alignment (3, 4). A small proportion, however, yields COs, which become cytologically visible as chiasmata and are essential for the stable connection of homologs. COs are not randomly distributed among and along bivalents; every bivalent forms at least one CO (obligate CO), and, if multiple COs occur, they are more evenly spaced along the bivalent than would be expected if they were randomly placed. This phenomenon was originally detected genetically by the finding that the frequency of double recombinants involving a pair of adjacent or nearby intervals was lower than the frequency expected from recombinant frequencies for each of those intervals (reviewed in refs. 5 and 6). Interference has also been analyzed cytologically, from spatial distributions of chiasmata (7, 8) or recombination complexes along chromosomes during meiotic prophase, when recombination is in progress (9). How interference is imposed is not known.
Concomitantly with meiotic recombination, the sister chromatids of each chromosome form a common axis, the axial element (AE), and the AEs of homologs align. Then, numerous transverse filaments connect the AEs of homologs, and a zipper-like structure, the synaptonemal complex (SC), is formed between the homologs (1). Protein complexes that mediate, and mark the sites of, recombination have been localized to AEs or SCs by both EM and immunocytology (reviewed in refs. 10 and 11). These studies (9, 12), together with molecular genetic analyses (13, 14), have elicited several specific questions regarding the imposition of interference: At which step in meiotic recombination is interference first detectable? Is the level of interference the same among recombination complexes representing early and late steps in meiotic recombination? Does the SC contribute to interference? We have analyzed these questions in the mouse by examining how protein complexes that are thought to mark intermediate and late events in meiotic recombination are distributed along SCs in two stages of meiotic prophase.
In mouse, many recombination-related proteins have been identified, and the meiotic time courses of immunofluorescent foci containing these proteins have been described (15, 16). The mouse transverse filament protein SYCP1 is also known (17, 18), and SYCP1-deficient mice have been constructed (19). We have analyzed the distributions of four types of foci along mouse SCs or AEs in wild-type and/or Sycp1−/− strains: (i) MLH1 foci, which occur during pachytene and specifically mark the sites of COs (9, 20); (ii and iii) MSH4 and replication protein A (RPA) foci, which appear earlier, during zygotene, and were analyzed here at late zygotene. In mouse, these foci outnumber the prospective COs. However, a subset of them likely matures into MLH1 foci and then into COs, because early MLH1 foci colocalize with MSH4 (16, 21) but then lose MSH4 at later stages; (iv) because Sycp1−/− strains do not form MLH1 foci (19), we analyzed γH2AX signals in Sycp1−/− pachytene spermatocytes. In wild-type meiosis, γH2AX signals occur from leptotene until pachytene (22). Based on their timing and other evidence (reviewed in refs. 13 and 23), MSH4 and RPA foci likely mark early intermediate stages of recombination involving strand exchange, whereas MLH1 foci likely mark the latest stages, e.g., conversion of double Holliday junctions to COs. γH2AX signals mark various DNA lesions, including DSBs (24); in Sycp1−/− pachytene, they probably represent (perhaps diverse) unresolved recombination intermediates (19).
For the detection of genetic interference, the coefficient of coincidence (CC) is often used. However, CC is problematic as a measure for the level of interference because it is not based on the precise positions of genetic exchanges but instead is based on the frequencies of recombinants for genetic markers that delimit two adjacent or nearby chromosomal intervals. Besides the strength of interference between exchanges in adjacent/nearby intervals, the size of the analyzed intervals thus codetermines the value of CC (see, which are published as supporting information on the PNAS web site; for the mouse, cf. 25). This effect precludes a CC-based comparison of the strength of interference among two types of foci with widely different densities. Additionally, in microscopic studies, the (cytological) interference among foci will be overestimated if the size of the intervals to which foci are assigned is close to the resolution limit of the light microscope (see Supporting Text, which is published as supporting information on the PNAS web site). Assignment of foci to intervals will also result in loss of information. In short, if the positions of genetic exchanges/chiasmata/foci are precisely known, models dealing with the exact positions of events are preferable for estimating the strength of interference.
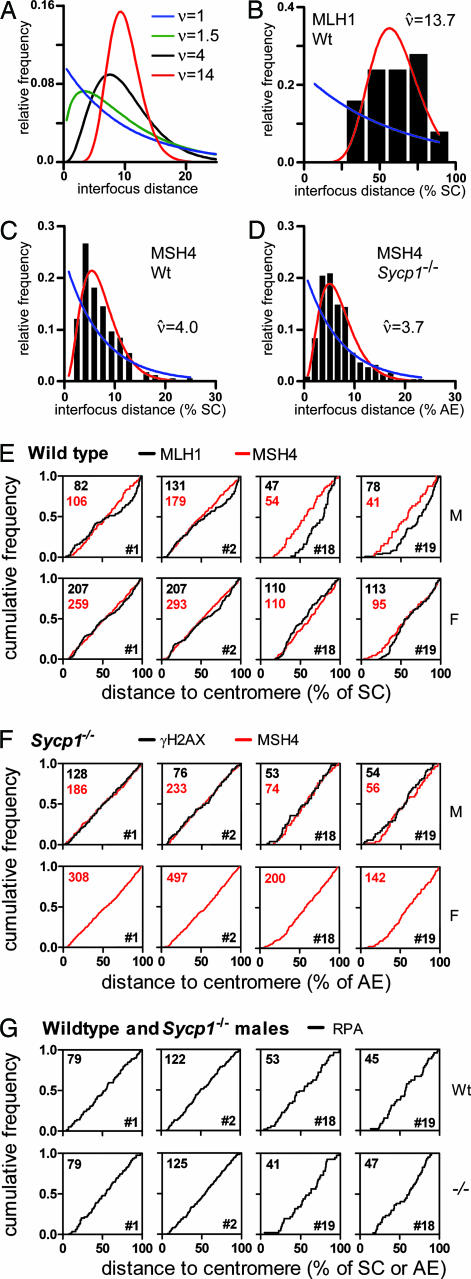
Several point process models have been considered for estimating the strength of interference (ref. 26 and references therein), and the gamma distribution has repeatedly emerged as most useful (26–29). The gamma distribution is commonly used for the analysis of distances between events along a linear axis (see Supporting Text); it describes the frequency distribution of interfocus distances that one would get if (imaginary or real) focus precursors were randomly placed along the SC, but only every nth precursor would yield a focus. Fig. 1A shows gamma distributions for various values of n (or ν, see below). The gamma distribution thus stands for a family of distributions, because each n value yields a distribution with another shape. One can determine for which n value the observed frequency distribution of interfocus distances fits best to a gamma distribution. If the best fit is obtained for n = 1, then there is no interference among foci. Fig. 1A furthermore shows that the distribution is narrower for higher n values: for a given average interfocus distance, the variance of interfocus distances decreases with increasing n. In other words, the higher the n value, the more evenly the foci are spaced and the stronger interference is. n is therefore called the interference parameter of the gamma model. Note that n is not a measure for the average interfocus distance. If one assumes that the biological mechanism of interference conforms to the gamma model (i.e., there is a mechanism that counts focus precursors; e.g., refs. 27 and 28), then n can only be a positive integer. Because it is not our purpose in this study to assume or test a specific biological interference mechanism but to instead use the gamma model as a device to estimate the strength of interference, we do not assume that n is an integer, and we will further denote the interference parameter of the gamma model as ν, which represents positive but not necessarily integer values, as distinct from n, which represents integer values only (see also Supporting Text).
Fig. 1.
Analysis of foci along bivalents. (A) Shape of gamma distributions for different ν values. The average interfocus distance equals 10 for all distributions shown. As ν increases, the very short and very long distances become sparser, and the distributions become narrower and more symmetrical. (B–D) Examples of histograms of observed interfocus distances in spermatocytes (black bars), the best fit of the observed distances to the gamma distribution (red curves), the ν value for which the best fit was obtained (ν̂), and the distributions expected if there were no interference (i.e., ν = 1; blue curves). The observed interfocus distances were binned for representation only; the best fits to the gamma distribution are based on the exact, unbinned distances. show histograms of all data sets. (E–G) Distribution of foci along bivalents. Shown are the cumulative frequencies of foci as a function of the distance to the centromeric end of the SC (wild type) or AE (Sycp1−/−). The distances are expressed as percentage of the length of the SC/AE on which the focus was located. The numbers of foci on which the curves are based are shown in the upper left corners, and the chromosome numbers are shown in the lower right corners of the graphs. A uniform distribution of foci would yield a straight line from the lower left to the upper right corner of the graph. M, male; F, female; Wt, wild type; −/−, Sycp1−/−.
The primary finding of this study is that cytological interference occurs among RPA or MSH4 foci in late zygotene, whereas interference among MLH1 foci in midpachytene was much stronger. Mouse SYCP1 was not required for cytological interference among RPA or MSH4 foci. However, our data do not allow us to decide whether SYCP1 is required for the high level of interference among MLH1 foci in wild type.
Results
Methodology.
We studied the positions of foci on two long (1 and 2) and two short (18 and 19) chromosomes. All mouse chromosomes are telocentric, and the centromeric ends are marked by intense DAPI staining (see Fig. 4, which is published as supporting information on the PNAS web site). We measured the distance of each focus to the centromeric end of the SC/AE. For estimating the strength of interference we expressed interfocus distances as percentages of the length of the relevant SC/AE to account for variation in SC/AE length.
It was crucial in this study that every individual focus could be unambiguously recognized, because both confusion of background signals with foci and failure to detect foci would affect the apparent frequency distribution of interfocus distances. Of the early meiotic foci, RPA and MSH4 foci were most suitable for our study because they displayed fairly uniform immunofluorescence intensity, which was well above background, whereas their close association with SCs/AEs further facilitated the distinction of foci from background signals (19). We refrained from analyzing the still earlier RAD51 or DMC1 foci because these were less closely associated with AEs and too heterogeneous to distinguish them reliably from background. RPA and MSH4 mark nearly the same population of foci, RPA being slightly earlier than MSH4 (15). We concentrated on MSH4 foci in this study, whereas RPA foci served as a methodological control. We analyzed the positions of MSH4 and RPA foci in late zygotene cells with at least 80% synapsis (wild type) or alignment (Sycp1−/−); this represents a brief stage, which can be reliably determined both in wild type and Sycp1−/− meiosis (19). MLH1 foci, which were analyzed in midpachytene, can also be distinguished easily from background (e.g., ref. 9). Because Sycp1−/− spermatocytes do not assemble MLH1 foci (19), we analyzed γH2AX signals in Sycp1−/− pachytene (as recognized by H1t expression; cf. 19). In Sycp1−/− meiosis, these signals probably represent unresolved recombination intermediates and are also easily distinguished from background (19).
Distribution of MSH4 and RPA Foci Along Bivalents.
The density (foci per μm) of MSH4 foci was similar along the four analyzed chromosomes and was not influenced by the SYCP1 disruption; this was also found for RPA foci (Table 1). In female meiosis, SCs tended to have fewer MSH4 foci per μm of SC than in male meiosis, but because SCs were on average longer (cf. 30), the number of MSH4 foci along a given SC was similar in oocytes and spermatocytes (Table 1). On all four analyzed chromosomes, MSH4 foci were lacking from the paracentromeric region and uniformly distributed along the remainder of the AEs, i.e., the density of MSH4 foci was the same for all positions along the chromosome outside the paracentromeric region; this uniform distribution manifests itself as straight cumulative curves in Fig. 1E. These patterns are also seen for RPA foci (Fig. 1G) and were maintained in Sycp1−/− mice (Table 1 and Fig. 1 F and G). Thus, the SYCP1 disruption did not affect occurrence or positioning of MSH4 or RPA foci.
Table 1.
Density of foci on AEs/SCs in wild-type (+/+) and SYCP1-deficient (−/−) mice
| Chromosome no. | No. of foci per | Male |
Female |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| RPA* |
MSH4* |
MLH1† |
γH2AX† |
MSH4* |
MLH† |
|||||
| +/+ | −/− | +/+ | −/− | +/+ | −/− | +/+ | −/− | +/+ | ||
| 1 | Bivalent | 9.9 | 9.9 | 11.8 | 11.6 | 1.54 | 9.14 | 13.0 | 14.0 | 1.85 |
| μm of SC (AE) | 0.98 | 0.93 | 1.17 | 1.14 | 0.15 | 0.94 | 0.91 | 0.91 | 0.14 | |
| 2 | Bivalent | 12.2 | 12.5 | 12.9 | 12.9 | 1.67 | 10.70 | 12.7 | 15.1 | 1.78 |
| μm of SC (AE) | 1.14 | 1.12 | 1.15 | 1.19 | 0.15 | 1.04 | 0.92 | 0.99 | 0.13 | |
| 18 | Bivalent | 6.3 | 5.9 | 6.0 | 6.7 | 1.00 | 4.42 | 5.8 | 8.7 | 1.05 |
| μm of SC (AE) | 1.18 | 0.99 | 1.24 | 1.23 | 0.20 | 0.87 | 0.73 | 0.99 | 0.14 | |
| 19 | Bivalent | 4.5 | 3.7 | 5.1 | 6.2 | 0.96 | 3.86 | 5.3 | 6.5 | 1.00 |
| μm of SC (AE) | 1.03 | 0.84 | 1.24 | 1.32 | 0.22 | 0.85 | 0.88 | 0.99 | 0.18 | |
*Late zygotene.
†Midpachytene.
SYCP1-Independent Interference Among MSH4 and RPA Foci.
We estimated the strength of interference among early meiotic foci by fitting the frequency distribution of interfocus distances to the gamma distribution. As judged by the P values in Table 2, the fit to this model was generally good. Strikingly, there was already a significant level of interference among MSH4 foci in wild-type late zygotene, as is evident from the shapes of the frequency distributions of distances among MSH4 foci (Fig. 1C and, which are published as supporting information on the PNAS web site). In wild-type males, the estimates of the interference parameter ν for MSH4 foci were between 4.0 and 7.7 (ν̂ values in Table 2). Similar levels of interference occurred among RPA foci in wild-type male meiosis (Table 2). Interference among MSH4 foci in female meiosis tended to be slightly weaker than in male meiosis, but the ν̂ values were still well above 1 (Table 2). Importantly, MSH4 and RPA foci displayed similar levels of interference in Sycp1−/− mice as in wild type, in both males and (regarding MSH4 foci) females (Fig. 1 C and D,, and Table 2). SYCP1 is thus not required for cytological interference among these foci.
Table 2.
Interference among RPA and MSH4 foci in wild-type (+/+) and SYCP1-deficient (−/−) mice*
| Sex; focus type | Chromosome no. | +/+ |
−/− |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Intervals | ν̂† (SE) | P‡ | Corr. ν̂§ | Intervals | ν̂† (SE) | P‡ | Corr. ν̂§ | ||
| Male; RPA | 1 | 71 | 3.2 (0.5) | 0.6 | 2.7 | 71 | 4.1 (0.7) | 0.2 | 3.8 |
| 2 | 112 | 3.0 (0.4) | 0.08 | 2.3 | 115 | 3.1 (0.4) | 0.5 | 2.4 | |
| 18 | 48 | 4.6 (0.9) | 0.2 | 4.1 | 39 | 4.0 (0.9) | 0.8 | 3.5 | |
| 19 | 35 | 4.6 (1.1) | 0.4 | 4.1 | 30 | 4.2 (0.1) | 0.5 | 3.8 | |
| Male; MSH4 | 1 | 97 | 5.3 (0.7) | 0.6 | 5.0 | 170 | 3.3 (0.3) | 0.2 | 2.6 |
| 2 | 165 | 4.0 (0.4) | 0.2 | 3.4 | 215 | 3.7 (0.3) | 0.005 | 3.1 | |
| 18 | 45 | 7.7 (1.6) | 0.2 | 7.4 | 63 | 4.5 (0.8) | 0.3 | 3.9 | |
| 19 | 33 | 5.8 (1.4) | 0.2 | 5.4 | 47 | 3.7 (0.7) | 0.04 | 2.7 | |
| Female; MSH4 | 1 | 252 | 3.5 (0.3) | 0.1 | 3.1 | 273 | 3.0 (0.2) | 0.007 | 2.5 |
| 2 | 270 | 3.2 (0.3) | 0.0002 | 2.8 | 461 | 3.5 (0.2) | 0.0001 | 3.1 | |
| 18 | 91 | 3.4 (0.5) | 0.5 | 3.1 | 177 | 3.4 (0.3) | 0.08 | 2.9 | |
| 19 | 77 | 3.2 (0.5) | 0.2 | 2.7 | 120 | 4.2 (0.5) | 0.3 | 3.8 | |
*Late zygotene.
†Maximum likelihood estimate of the interference parameter ν in the gamma model (with estimated SE).
‡Estimated P value; P is the probability of finding an as-bad or worse fit of the observations to the gamma distribution due to sampling error. The estimate of P was based on the deviance of the observations from the values expected based on the gamma equation with parameter ν̂.
§ν̂ corrected (Corr.) for the limited range of observable interfocus distances.
Distribution of MLH1 Foci and γH2AX Signals Along SCs/AEs.
Like MSH4 and RPA foci, MLH1 foci were lacking from the paracentromeric region during pachytene of wild-type meiosis (Fig. 1E). However, in contrast to MSH4 and RPA foci, MLH1 foci were not uniformly distributed along the SCs. In male meiosis, the density of MLH1 foci was high in the centromere-distal subtelomeric region of all four analyzed chromosomes (cf. 9), and the middle part of chromosomes 1 and 2 had a lower MLH1 focus density than average for those chromosomes. This nonuniform distribution of MLH1 foci does not ensue from a corresponding distribution of MSH4 or RPA foci (described above; Fig. 1 E–G). In female meiosis, these deviations from a uniform distribution of MLH1 foci on chromosomes 1 and 2 were less pronounced (Fig. 1E). Along chromosome 18 and 19, the distribution of MLH1 foci deviated somewhat from uniform in female meiosis but did not display the high focus density seen in the centromere-distal subtelomeric regions in male meiosis.
The short chromosomes had more MLH1 foci per micrometer of SC than the long chromosomes (Table 1). This finding has also been reported for MLH1 foci in male meiosis (9), genetic exchanges in female meiosis (29), and chiasmata in male and female meiosis (8) of the mouse and probably reflects the phenomenon of obligate COs. The higher density of MLH1 foci on short chromosomes does not ensue from a higher density of RPA and MSH4 foci (Table 1). Thus, there are two indications that the positions of MLH1 foci do not ensue in a simple way from the positions of RPA and MSH4 foci: first, the density of MLH1 foci is higher on short than on long chromosomes, whereas the density of MSH4 and RPA foci is similar on long and short chromosomes; and second, particularly in male meiosis, the density of MLH1 foci, but not of RPA and MSH4 foci, is higher than average in the centromere-distal subtelomeric regions. Nevertheless, MLH1 foci most likely arise from MSH4 foci, because some MLH1 foci colocalize with MSH4 in early to midpachytene (16, 21).
The γH2AX signals in Sycp1−/− pachytene spermatocytes do not share these features with MLH1. γH2AX signals are lacking in paracentromeric regions, like all other analyzed foci, but do not display a higher-than-average density in the subtelomeric regions (Fig. 1F). Further, the density of γH2AX signals is not higher on short than on long chromosomes (Table 1). Apparently, mere progression from zygotene to pachytene at the cellular level does not bring along the specific features seen for MLH1 positioning in wild-type pachytene spermatocytes.
Interference in Pachytene of Wild-Type Mice.
The ν̂ values for MLH1 in pachytene were 2- to 4-fold higher than the ν̂ values for MSH4 or RPA in late zygotene; therefore, interference among MLH1 foci is much stronger than among MSH4 or RPA foci (compare the shapes of the frequency distributions in Fig. 1 B and C and compare Tables 2 and 3). The density of MLH1 foci was much lower than for MSH4 or RPA foci, as expected (Table 1). As noted above, ν̂ represents the strength of interference (evenness of spacing), not the focus density.
Table 3.
Interference among MLH1 foci in wild type (WT) and among γH2AX foci in SYCP1-deficient mice*
| Mouse type; focus type | Chromosome no. | Intervals | ν̂† (SE) | P‡ | Corr. ν̂§ |
|---|---|---|---|---|---|
| WT male; MLH1 | 1 | 27 | 14.4 (3.9) | 0.3 | 11.5 |
| 2 | 50 | 13.7 (2.7) | 0.02 | 11.8 | |
| 18 | 0 | - | - | - | |
| 19 | 0 | - | - | - | |
| WT female; MLH1 | 1 | 95 | 8.9 (1.3) | 0.3 | 7.6 |
| 2 | 88 | 11.7 (1.7) | 0.3 | 10.1 | |
| 18 | 7 | 14.3 (7.5) | - | - | |
| 19 | 3 | - | - | - | |
| Sycp 1−/− male; γH2AX | 1 | 114 | 5.0 (0.7) | 0.07 | 4.8 |
| 2 | 96 | 4.8 (0.7) | 0.2 | 4.6 | |
| 18 | 41 | 5.1 (1.1) | 0.06 | 4.8 | |
| 19 | 40 | 7.9 (1.7) | 0.09 | 7.7 |
*Midpachytene.
†Maximum likelihood estimate of the interference parameter ν in the gamma model (with SE).
‡Estimated P value.
§ν̂ corrected (Corr.) for the limited range of observable interfocus distances.
Interference Among γH2AX Signals in SYCP1-Deficient Mice.
Interference among γH2AX signals in Sycp1−/− midpachytene (Table 3) was slightly stronger than interference among MSH4 foci in Sycp1−/− late zygotene (Table 2) but far weaker than interference among MLH1 foci in wild-type midpachytene. Thus, mere progression from late zygotene to midpachytene at the cellular level does not bring about the strong interference seen by MLH1 foci. Our results therefore do not provide a clear answer as to the role of the SC in the strong interference seen for COs/MLH1 foci.
Discussion
This study shows that cytological interference is already detectable in late zygotene among MSH4 or RPA foci but that interference occurs at a much higher level in pachytene among MLH1 foci. Furthermore, it shows that interference among MSH4 or RPA foci does not depend on transverse filament protein SYCP1. The presented data do not allow us to decide whether SYCP1 is required for the strong interference among MLH1 foci.
Distribution of Foci Along SCs.
In male mouse meiosis, the density of MLH1 foci in the centromere-distal subtelomeric region is higher than average for the entire SC, and this is not due to a corresponding higher-than-average density of MSH4 or RPA foci (Fig. 1 E–G). The short chromosomes 18 (5.0 μm) and 19 (4.4 μm) display a similar distribution of MLH1 foci as the distal half of the long chromosomes 1 (10.2 μm) and 2 (11.0 μm) (Fig. 1E), which suggests that the distance to the distal telomere codetermines the position of the most distal MLH1 focus. Perhaps certain sequence elements enhance MLH1 focus formation in the distal subtelomeric region (31). In the context of the tension model for interference (32), clamping of the telomeres in spermatocytes could explain the observed MLH1 focus patterns in these cells.
Use of the Gamma Model for Estimating the Strength of Interference.
As judged by the P values in Tables 2 and 3, our data fit reasonably well to the gamma distribution in most cases, but the fit tends to be less good as more interfocus distances are available for analysis; this indicates that (some of) the distributions of interfocus distances resemble but are not identical to gamma distributions. Possibly, the mechanism(s) of interference conform to the gamma model, but other factors, such as the limited range of observable interfocus distances or local chromatin properties, have additional, minor influences. Alternatively, the mechanism(s) of interference do not conform to the gamma model (e.g., ref. 32) but coincidentally yield frequency distributions of interfocus distances that resemble gamma distributions (further discussion below).
Regardless, the gamma model remains a useful tool for comparing interference levels (26, 29). Broman et al. (29) found a fairly good fit of genetic distances in the female mouse to the gamma model. For chromosomes 1 and 2, their estimates of ν were 10.2 and 9.6, which fits well with the ν̂ values based on MLH1 focus positions (8.9 and 11.7; Table 3). This correspondence renders the large-scale discrepancies between the recombination maps and MLH1 focus maps of these chromosomes unlikely and virtually rules out that cytological interference among MLH1 foci is a merely spatial phenomenon without any consequence at the genetic level. For MSH4 and RPA foci, however, this possibility cannot be excluded.
Two Levels of Interference.
In late zygotene, we found cytological interference among MSH4 foci in all analyzed situations: in males and females, in wild-type and Sycp1−/− mice, and on long and short chromosomes. The independently analyzed RPA foci displayed similar levels of interference as the MSH4 foci, which confirms that interference among foci of recombination-related proteins is already detectable in late zygotene and does not depend on SYCP1. Two obvious questions emerge: First, when during the recombination process does interference first arise? If the interference mechanism conforms to the gamma model (e.g., 27, 28), then randomly distributed focus precursors (e.g., DSBs or pre-DSB complexes) must exist in early meiosis, on which that mechanism acts. On the other hand, it is conceivable that a different type of process that coincidentally yields frequency distributions of interfocus distances resembling gamma distributions causes interference; then there is no reason to suppose an underlying population of randomly distributed focus precursors, and the earliest recombination precursors might already display interference. Second, are the weak interference among MSH4 foci in late zygotene and the strong interference among MLH1 foci in pachytene due to a single, progressive mechanism that causes increasingly stronger interference among recombination-related protein complexes as meiotic prophase proceeds, are they imposed independently by two different mechanisms, or are they imposed in two steps? For wild type, a single, progressive interference mechanism is unlikely, because MLH1 and MSH4 foci are distributed differently along the SCs, particularly in male meiosis (Fig. 1E). This difference indicates that the factors determining MLH1 focus positions along wild-type SCs differ from those determining MSH4 focus positions. We therefore propose that interference is imposed in at least two temporally or functionally distinguishable ways.
If the factors positioning MSH4 foci differ from those positioning MLH1 foci, then an ensuing question is whether MLH1 foci are placed entirely independently of MSH4 foci or whether the positions of MSH4 foci are determined first, whereupon, in a second step, MLH1 foci are recruited from the MSH4 foci in such a way that MLH1 foci display stronger interference than MSH4 foci and, particularly in males, are distributed differently along the SCs than MSH4 foci. The colocalization of MSH4 and MLH1 in some early pachytene foci (16, 21) is consistent with positioning of MLH1 foci in two steps, with MLH1 foci being recruited from weakly interfering MSH4 foci. However, it does not rule out positioning of MLH1 foci in one step, because it is conceivable that those MSH4 foci that will develop into MLH1 foci are positioned independently from all other MSH4 foci. Our data do not allow distinguishing between these two possible sequences of events, because the expected outcomes in terms of inter-MSH4 focus distances differ only slightly (simulations not shown).
How the two levels of interference are imposed cannot be decided from our data, either, and it cannot be excluded that interference among MSH4 foci arises by another mechanism than interference among MLH1 foci. However, one recent chromosome model explicitly suggests the possibility of interference phenomena in more than one stage of meiosis by the same mechanism (32). The model is based on the observation that chromatin globally expands and contracts a number of times during the mitotic cycle and during meiosis, with three expansion/contraction cycles during meiotic prophase. Chromatin expansion would be globally withstood by structural chromatin components, which would generate mechanical stress. Mechanical stress might then promote (some of) the covalent DNA changes during the recombination process, and if such a change were to occur, it would be accompanied by local stress relief around the involved recombination complex, for instance, by local detachment of chromatin from constraining structures. This would reduce the probability of a similar event nearby and thus bring about interference. Because there is more than one expansion/contraction cycle during meiotic prophase, interference phenomena might occur in more than one meiotic prophase stage. The model furthermore implies that the expansion/contraction cycles serve to coordinate events and control the spacing of events within the nucleus (32). Interference among MSH4 foci in late zygotene would thus reflect control of the spacing of early strand-exchange intermediates, which in turn might or might not reflect imposition of interference at an earlier stage (e.g., formation of axis-associated DSBs). Obviously, such control is essential at various steps in the recombination process: DSBs should not be too closely spaced, and on another distance scale the same is true for COs (33).
Comparison with Other Species.
In tomato, ultrastructural maps of early recombination nodules (34), which roughly correspond to Rad51/Dmc1 foci (35), and late recombination nodules, which correspond to COs (36), have been constructed. We have found that both early and late recombination nodules display interference (C.H., unpublished work); therefore, the phenomenon of two levels of interference is not unique to meiosis in the mouse.
Cytological interference among foci other than MLH1 has been analyzed previously only in yeast (12). The comparison of cytological interference in yeast and mouse is complicated for several reasons. First, Mlh1 foci have not been studied in yeast. Second, part of the COs in yeast belong to a noninterfering category (37). Third, the number of Msh4 foci in yeast corresponds to the number of interfering COs, whereas in mouse, MSH4 foci in late zygotene far outnumber the prospective COs. This difference may imply differences in detectability, turnover rates, real differences in function, or other variables. Yeast Msh4 foci display cytological interference in both wild-type strains and strains lacking the yeast SYCP1 analogue, Zip1 (12, 38); this finding resembles the result obtained for mouse MSH4 foci. However, as most yeast Msh4 foci yield COs, it appears as though the mechanism that causes strong interference among Mlh1 foci does not operate in yeast. Alternatively, both the weak (MSH4-linked) and the strong (MLH1-linked) interference mechanisms detected in mouse are active in yeast but act on the same distance scale (see figure 3 in ref. 39). That interpretation might explain why yeast zip1 cells display cytological interference among Zip2 foci, which largely colocalize with Msh4 foci (38), but no CO interference: yeast zip1 cells might be capable of imposing cytological interference among Zip2 (and Msh4) foci but be incapable of converting these foci into COs, with the COs actually analyzed for interference not reflecting the normal process. The same interpretation might also explain why yeast mlh1Δ mutants still display a reduced level of CO interference (40, 41), because the mechanism for interference among Msh4 foci might still function, and part of the Msh4 foci might still yield a CO in the absence of Mlh1 (reviewed in ref. 42).
Materials and Methods
Cytological Techniques.
All antibodies used have been described (19). Testis cell suspensions (43) were spread by the dry-down procedure (44). We collected ovaries (45), incubated them for 15 min in hypotonic buffer (44), isolated the oocytes from the ovaries, and spread them on microscope slides in 2% paraformaldehyde/0.15% Triton X-100 as described (45). Slides were incubated for immunofluorescence labeling, stained with DAPI, and micrographed as described (43, 46, 47). Then we removed the coverslips, subjected the slides to FISH (48) using FITC-labeled probes for chromosomes 1 and 19 and biotin-labeled probes for chromosomes 2 and 18 (STARFISH probes; Cambio, Cambridge, U.K.), visualized binding of the biotin-labeled probes by using Texas red-labeled avidin according to the supplier’s instructions, collected the FISH images, and combined the images with the corresponding immunofluorescence and DAPI images by using the adobe photoshop software package (Fig. 2). We measured the lengths of SCs and AEs and the positions of foci on SCs/AEs using the public-domain program object-image (available at http://simon.bio.uva.nl), which is an extended version of nih image (developed at the National Institutes of Health, Bethesda) by N. Vischer (University of Amsterdam).
Estimating the Interference Parameter ν.
We obtained a first estimate of ν by fitting the observed frequency distribution of interfocus distances to the gamma distribution by the maximum likelihood method using the genstat software package (VSN International, Hemel Hempstead, U.K.). Then we applied a correction for the limited range of interfocus distances that we can observe. The upper limit is the SC length, and the lower limit is the smallest interfocus distance that we can distinguish in the immunofluorescence images (0.2 μm). For each of the four analyzed SCs, we determined the effect of these limits on the estimate of ν by simulating the focus positions along at least 5,000 SCs for the observed average number of foci per SC and integer values of ν close to the first estimate. For each of these “input” values of ν, we fitted those simulated interfocus distances that fell within the above-mentioned limits to the gamma model to obtain “output” values of ν. The first estimate of ν was then corrected based on the comparison of the input and output values of ν in the simulations.
Supplementary Material
Acknowledgments
We thank N. Kleckner for valuable input into the text, N. Vischer for providing us with the object-image program, F. Lhuissier (Wageningen University) for adapting the program, C. Her (Washington State University, Pullman) for anti-MSH4 antibodies, the animal facilities at the Leiden University and Wageningen University for expert technical support, and H. Offenberg and F. Lhuissier for several useful comments. The Netherlands Society for Scientific Research (NWO) Grant 901-01-097 financially supported this work.
Abbreviations
- AE
axial element
- CO
crossover
- SC
synaptonemal complex
- RPA
replication protein A
- DSB
double-strand break.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Zickler D., Kleckner N. Annu. Rev. Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]
- 2.Schwacha A., Kleckner N. Cell. 1997;90:1123–1135. doi: 10.1016/s0092-8674(00)80378-5. [DOI] [PubMed] [Google Scholar]
- 3.Carpenter A. T. C. BioEssays. 1987;6:232–236. doi: 10.1002/bies.950060510. [DOI] [PubMed] [Google Scholar]
- 4.Tessé S., Storlazzi A., Kleckner N., Gargano S., Zickler D. Proc. Natl. Acad. Sci. USA. 2003;100:12865–12870. doi: 10.1073/pnas.2034282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hillers K. J. Curr. Biol. 2004;14:R1036–R1037. doi: 10.1016/j.cub.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 6.Muller H. J. Am. Nat. 1916;50:193–221. [Google Scholar]
- 7.Jones G. H. Symp. Soc. Exp. Biol. 1984;38:293–320. [PubMed] [Google Scholar]
- 8.Lawrie N. M., Tease C., Hultén M. A. Chromosoma. 1995;104:308–314. doi: 10.1007/BF00352262. [DOI] [PubMed] [Google Scholar]
- 9.Froenicke L., Anderson L. K., Wienberg J., Ashley T. Am. J. Hum. Genet. 2002;71:1353–1368. doi: 10.1086/344714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson L. K., Stack S. M. Cytogenet. Genome Res. 2005;109:198–204. doi: 10.1159/000082400. [DOI] [PubMed] [Google Scholar]
- 11.Ashley T., Plug A. W. Curr. Top. Dev. Biol. 1998;37:201–239. doi: 10.1016/s0070-2153(08)60175-1. [DOI] [PubMed] [Google Scholar]
- 12.Fung J. C., Rockmill B., Odell M., Roeder G. S. Cell. 2004;116:795–802. doi: 10.1016/s0092-8674(04)00249-1. [DOI] [PubMed] [Google Scholar]
- 13.Börner G. V., Kleckner N., Hunter N. Cell. 2004;117:29–45. doi: 10.1016/s0092-8674(04)00292-2. [DOI] [PubMed] [Google Scholar]
- 14.Storlazzi A., Xu L., Schwacha A., Kleckner N. Proc. Natl. Acad. Sci. USA. 1996;93:9043–9048. doi: 10.1073/pnas.93.17.9043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moens P. B., Kolas N. K., Tarsounas M., Marcon E., Cohen P. E., Spyropoulos B. J. Cell Sci. 2002;115:1611–1622. doi: 10.1242/jcs.115.8.1611. [DOI] [PubMed] [Google Scholar]
- 16.Kolas N. K., Svetlanov A., Lenzi M. L., Macaluso F. P., Lipkin S. M., Liskay R. M., Greally J., Edelmann W., Cohen P. E. J. Cell Biol. 2005;171:447–458. doi: 10.1083/jcb.200506170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meuwissen R. L. J., Offenberg H. H., Dietrich A. J. J., Riesewijk A., van Iersel M., Heyting C. EMBO J. 1992;11:5091–5100. doi: 10.1002/j.1460-2075.1992.tb05616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sage J., Martin L., Meuwissen R., Heyting C., Cuzin F., Rassoulzadegan M. Mech. Dev. 1999;80:29–39. doi: 10.1016/s0925-4773(98)00191-9. [DOI] [PubMed] [Google Scholar]
- 19.de Vries F. A. T., de Boer E., van den Bosch M., Baarends W. M., Ooms M., Yuan L., Liu J.-G., Heyting C., Pastink A. Genes Dev. 2005;19:1376–1389. doi: 10.1101/gad.329705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baker S. M., Plug A. W., Prolla T. A., Bronner C. E., Harris A. C., Yao X., Christie D.-M., Monell C., Arnheim N., Bradley A., et al. Nat. Genet. 1996;13:336–342. doi: 10.1038/ng0796-336. [DOI] [PubMed] [Google Scholar]
- 21.Santucci-Darmanin S., Walpita D., Lespinasse F., Desnuelle C., Ashley T., Paquis-Flucklinger V. FASEB J. 2000;14:1539–1547. doi: 10.1096/fj.14.11.1539. [DOI] [PubMed] [Google Scholar]
- 22.Mahadevaiah S. K., Turner J. M. A., Baudat F., Rogakou E. P., de Boer P., Blanco-Rodriguez J., Jasin M., Keeney S., Bonner W. M., Burgoyne P. S. Nat. Genet. 2001;27:271–276. doi: 10.1038/85830. [DOI] [PubMed] [Google Scholar]
- 23.Whitby M. C. Biochem. Soc. Trans. 2005;33:1451–1455. doi: 10.1042/BST0331451. [DOI] [PubMed] [Google Scholar]
- 24.Rogakou E. P., Boon C., Redon C., Bonner W. M. J. Cell Biol. 1999;146:905–915. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Broman K. W. Genetics. 2005;169:1133–1146. doi: 10.1534/genetics.104.035212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McPeek M. S., Speed T. P. Genetics. 1995;139:1031–1044. doi: 10.1093/genetics/139.2.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stam P. Genetics. 1979;92:573–594. doi: 10.1093/genetics/92.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Foss E., Lande R., Stahl F. W., Steinberg C. M. Genetics. 1993;133:681–691. doi: 10.1093/genetics/133.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Broman K. W., Rowe L. B., Churchill G. A., Paigen K. Genetics. 2002;160:1123–1131. doi: 10.1093/genetics/160.3.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lynn A., Koehler K. E., Judis L., Chan E. R., Cherry J. P., Schwartz S., Seftel A., Hunt P. A., Hassold T. J. Science. 2002;296:2222–2225. doi: 10.1126/science.1071220. [DOI] [PubMed] [Google Scholar]
- 31.Starling J. A., Maule J., Hastie N. D., Allshire R. C. Nucleic Acids Res. 1990;18:6881–6888. doi: 10.1093/nar/18.23.6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kleckner N., Zickler D., Jones G. H., Dekker J., Padmore R., Henle J., Hutchinson J. Proc. Natl. Acad. Sci. USA. 2004;101:12592–12597. doi: 10.1073/pnas.0402724101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maguire M. P. Heredity. 1980;45:127–131. doi: 10.1038/hdy.1980.56. [DOI] [PubMed] [Google Scholar]
- 34.Anderson L. K., Hooker K. D., Stack S. M. Genetics. 2001;159:1259–1269. doi: 10.1093/genetics/159.3.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anderson L. K., Offenberg H. H., Verkuijlen W. C., Heyting C. Proc. Natl. Acad. Sci. USA. 1997;94:6868–6873. doi: 10.1073/pnas.94.13.6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sherman J. D., Stack S. M. Genetics. 1995;141:683–708. doi: 10.1093/genetics/141.2.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de los Santos T., Hunter N., Lee C., Larkin B., Loidl J., Hollingsworth N. M. Genetics. 2003;164:81–94. doi: 10.1093/genetics/164.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Novak J. E., Ross-Macdonald P. B., Roeder G. S. Genetics. 2001;158:1013–1025. doi: 10.1093/genetics/158.3.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Boer E., Heyting C. Chromosoma. 2006;115:220–234. doi: 10.1007/s00412-006-0057-5. [DOI] [PubMed] [Google Scholar]
- 40.Argueso J. L., Kijas A. W., Sarin S., Heck J., Waase M., Alani E. Mol. Cell. Biol. 2003;23:873–886. doi: 10.1128/MCB.23.3.873-886.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Argueso J. L., Wanat J., Gemici Z., Alani E. Genetics. 2004;168:1805–1816. doi: 10.1534/genetics.104.032912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoffmann E. R., Borts R. H. Cytogenet. Genome Res. 2004;107:232–248. doi: 10.1159/000080601. [DOI] [PubMed] [Google Scholar]
- 43.Heyting C., Dietrich A. J. Methods Cell Biol. 1991;35:177–202. doi: 10.1016/s0091-679x(08)60573-7. [DOI] [PubMed] [Google Scholar]
- 44.Peters A. H. F. M., Plug A. W., van Vugt M. J., de Boer P. Chromosome Res. 1997;5:66–71. doi: 10.1023/a:1018445520117. [DOI] [PubMed] [Google Scholar]
- 45.Dietrich A. J. J., Mulder R. J. P. Chromosoma. 1983;88:377–385. doi: 10.1007/BF00285860. [DOI] [PubMed] [Google Scholar]
- 46.Eijpe M., Offenberg H., Jessberger R., Revenkova E., Heyting C. J. Cell Biol. 2003;160:657–670. doi: 10.1083/jcb.200212080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Revenkova E., Eijpe M., Heyting C., Hodges C., Hunt P. A., Liebe B., Scherthan H., Jessberger R. Nat. Cell Biol. 2004;6:555–562. doi: 10.1038/ncb1135. [DOI] [PubMed] [Google Scholar]
- 48.Turner J. M. A., Mahadevaiah S. K., Fernandez-Capetillo O., Nussenzweig A., Xu X., Deng C. X., Burgoyne P. S. Nat. Genet. 2005;37:41–47. doi: 10.1038/ng1484. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.