Abstract
The LPP gene is the preferred translocation partner of the HMGIC gene in a subclass of human benign mesenchymal tumors known as lipomas. Here we have characterized the LPP gene product that shares 41% of sequence identity with the focal adhesion protein zyxin. LPP localizes in focal adhesions as well as in cell-to-cell contacts, and it binds VASP, a protein implicated in the control of actin organization. In addition, LPP accumulates in the nucleus of cells upon treatment with leptomycin B, an inhibitor of the export factor CRM1. The nuclear export of LPP depends on an N-terminally located leucine-rich sequence that shares sequence homology with well-defined nuclear export signals. Moreover, LPP displays transcriptional activation capacity, as measured by GAL4-based assays. Altogether, these results show that the LPP protein has multifunctional domains and may serve as a scaffold upon which distinct protein complexes are assembled in the cytoplasm and in the nucleus.
INTRODUCTION
Attachment sites to the extracellular matrix and cell-to-cell contacts play an important role in the control of cell motility and shape. In addition to these functions, these sites regulate the assembly of protein complexes that are implicated in signaling events. These dual functions require proteins at cell adhesion sites that interact, via multiple binding motifs, with components of the actin cytoskeleton and of signaling pathways that might lead to modulation of gene expression. In addition, increasing evidence suggests that proteins that shuttle from cell adhesion sites to the nucleus may form protein complexes that regulate transcription.
We have previously identified the LPP (LIM-containing lipoma-preferred partner) gene as being rearranged in a subgroup of lipomas, which are benign tumors of adipose tissue (Petit et al., 1996). The LPP gene encodes a 612-amino acid protein composed of a proline-rich N-terminal region and three C-terminally located LIM domains. LIM domains, which are rich in cysteine and histidine amino acids, form two zinc fingers capable of mediating protein-protein interactions (for reviews, see Beckerle, 1997; Dawid et al., 1998). These domains are found in a large variety of proteins, including transcription factors and proteins present at cell-substratum attachment sites. In a similar manner, the proteins zyxin (Macalma et al., 1996; Zumbrunn and Trueb, 1996) and TRIP6 (Yi and Beckerle, 1998a) are composed of a proline-rich N-terminal domain and three C-terminal LIM domains and share 41 and 53% overall identity, respectively, with LPP (Figure 1A). TRIP6 was identified as a thyroid receptor–interacting protein in a yeast two-hybrid screening (Lee et al., 1995). Zyxin (for review, see Beckerle, 1997) is present at sites of cell adhesion to the extracellular matrix and cell-to-cell contacts (Beckerle, 1986) as well as at the leading edge of migrating cells (Golsteyn et al., 1997a). Moreover, it has been shown that chicken zyxin shuttles between the cytoplasm and the nucleus (Nix and Beckerle, 1997). The N-terminal portion of LPP contains two proline-rich stretches (DFLPPPPPPLD and NFPPPPPLD) similar to the “FPPPP” motifs that are found in zyxin, in vinculin, a focal adhesion–associated protein (Price et al., 1989), and in ActA, a surface protein of the bacterium Listeria monocytogenes, which is essential for its actin-based intracellular movement (Domann et al., 1992; Kocks et al., 1992). These motifs are required for interaction with proteins of the Ena/VASP family (Brindle et al., 1996; Gertler et al., 1996; Reinhard et al., 1996; Niebuhr et al., 1997; Fedorov et al., 1999; Prehoda et al., 1999). VASP and related proteins are present in focal adhesions and at the leading edge of cells and are believed to be involved in the organization of the actin cytoskeleton by virtue of their interaction with profilin and possibly actin (Reinhard et al., 1992, 1995a; Gertler et al., 1996; Laurent et al., 1999; for reviews, see Theriot and Mitchison, 1993; Pollard, 1995).
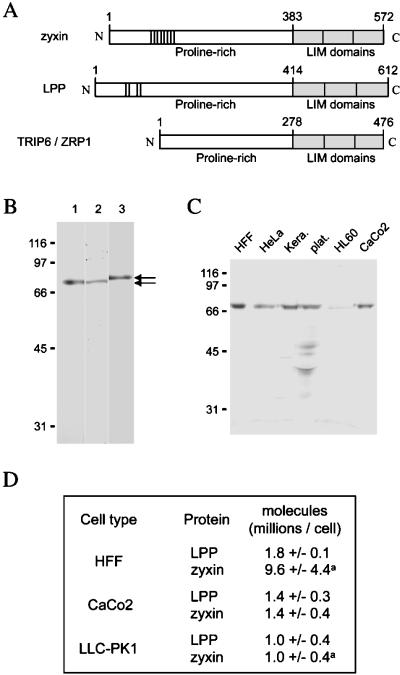
Figure 1.
Production of LPP in cells. (A) Scheme of the subdomain structure of the human zyxin, LPP, and TRIP6 proteins. All three proteins are composed of an N-terminal proline-rich domain (white box) and a C-terminal cysteine-rich portion composed of three LIM domains (gray boxes). LPP and zyxin contain FPPPP motifs in their N-terminal proline-rich domain (small boxes). (B) Specificity of anti-LPP antibodies. Total HeLa cell extracts were analyzed by SDS-PAGE and Western blotting with MP2, a serum directed against a synthetic LPP-derived peptide not present in zyxin or TRIP6 (amino acids 318–332; lane 1), with LPP2, an affinity-purified polyclonal antibody directed against a recombinant fusion protein containing amino acids 3–414 of LPP (GST-LPP3–414; lane 2), or with a monoclonal anti-zyxin antibody (lane 3). Note that zyxin migrates slower than LPP, confirming that LPP-specific antibodies do not recognize zyxin. The positions of molecular mass markers (kilodaltons) are shown on the left. (C) Expression of LPP in different human cells. Cell extracts were prepared from the following human cells and cell lines: foreskin fibroblasts (HFF), cervix carcinoma (HeLa), keratinocytes (Kera.), platelets (plat.), lymphocytes (HL60), and colon carcinoma (CaCo2). Approximately 15 μg of protein from each extract was analyzed by SDS-PAGE and Western blotting with MP2. The positions of molecular mass markers (kilodaltons) are shown on the left. (D) Quantitation of LPP and zyxin proteins in extracts from HFF, CaCo2, and LLC-PK1 cell lines. Values were obtained by quantifying LPP- or zyxin-specific protein bands with the use of a Cy5-based detection method (see MATERIALS AND METHODS). aThese values have been published elsewhere (Fradelizi et al., 1999) but are shown here for comparison.
In lipomas, the LPP gene, which is normally located at chromosome region 3q27-q28, is the preferred translocation partner of the HMGIC gene (Petit et al., 1996). The HMGIC gene is consistently altered in lipomas and a variety of other benign mesenchymal tumors characterized by aberrations of chromosome 12 at band q15 (Ashar et al., 1995; Schoenmakers et al., 1995). HMGIC is a member of the high-mobility group I (HMG) protein family (for review, see Goodwin, 1998). It comprises three short basic DNA-binding domains named AT hooks (Aravind and Landsman, 1998), which preferentially bind to AT-rich DNA sequences, and an acidic C-terminal tail. By modulating DNA conformation, HMGIC is thought to provide the correct framework for a transcriptional complex (for review, see Wolffe, 1994). In lipomas, most 12q15 translocations occur in the large third intron of the HMGIC gene, separating the three DNA-binding domains from the acidic C-terminal tail. The HMGIC/LPP fusion transcripts that have been detected encode the DNA-binding domains of HMGIC followed by the LIM domains of LPP (Petit et al., 1996). Although a causal relationship between the expression of these chimeras and tumorigenesis remains to be confirmed, it can be hypothesized that the LIM domains of LPP, targeted to specific DNA sequences via the DNA-binding domains of HMGIC, may recruit factors that alter gene transcription.
The structural relationship of LPP with zyxin, which is associated with the actin cytoskeleton, and with proteins that are involved in the regulation of gene transcription suggests that LPP may play a role in the communication between the cellular cortex and the nucleus. To identify this role, we characterized the intracellular distribution and binding partners of LPP. We show that LPP is localized in focal adhesions and cell-to-cell contacts. We demonstrate that LPP interacts with VASP family members, supporting its role in the spatial control of actin organization. In addition, we find that LPP is transiently located in the nucleus. We identified and characterized a nuclear export sequence (NES) localized in the N-terminal part of LPP. Strikingly, we show that LPP activates transcription in a GAL4-based transactivation assay and that this activity can be increased when the NES sequence is removed, causing an accumulation of LPP in the nucleus. We find two regions (the LIM domains and a sequence comprising residues 248–413) that are important for this activity. Altogether, our results suggest that LPP can act as a link between the actin cytoskeleton and gene regulation.
MATERIALS AND METHODS
Reagents
Leptomycin B was provided by B. Wolff-Winiski (Novartis, Basel, Switzerland) and by M. Yoshida (The University of Tokyo, Tokyo, Japan). Cy2-linked goat anti-rabbit, Texas red–coupled goat anti-mouse, and Cy5-coupled donkey anti-rabbit immunoglobulin G antibodies were purchased from Jackson Immunoresearch Laboratories (West Grove, PA). Alkaline phosphatase–coupled goat anti-rabbit or anti-mouse immunoglobulin G antibodies were purchased from Promega (Madison, WI).
Cell Lines and Cells
The human cervix carcinoma HeLa cell line (American Type Culture Collection [ATCC; Rockville, MD] CCL-2), African green monkey kidney Vero cell line (ATCC CCL-81), porcine proximal kidney epithelial LLC-PK1 cell line (ATCC CL-101), human colon adenocarcinoma CaCo2 cell line (ATCC HTB-37), human penis carcinoma keratinocytes (Rogel-Gaillard et al., 1992; Robine et al., 1993), and human foreskin fibroblasts (HFF) (Dr. A. Rochat, Ecole Normale Superieure, Paris, France) were grown in DMEM supplemented with 10% FCS (complete medium). The HL60 (ATCC CCL-240) (a gift from Dr. B. Bauvois, Institut Curie, Paris, France) cell line was cultured in RPMI 1640 medium supplemented with 2 mM glutamine and 10% FCS. The human embryonic kidney epithelial 293 cell line (ATCC CRL-1573) and the mouse embryonic fibroblast 3T3-L1 cell line (ATCC CL-173; previously known as ATCC CCL-92.1) were grown in DMEM/F12 (1:1) supplemented with 10% FCS. All cell lines were cultured at 37°C and 5% CO2. Human platelets were received from Dr. C. Gachet (Institut National pour la Santé et la Recherche Medicale, U311, Strasbourg, France).
Plasmid Constructs
DNA manipulations were performed as described by Sambrook et al. (1989). Human LPP cDNA (GenBank accession number U49957) was subcloned into the NdeI site of the pGEM5Zf(+) vector (Promega). For expression in Escherichia coli, sequences encoding domains of ActA, zyxin, or LPP were inserted into the pGEX-2T vector (Pharmacia, Piscataway, NJ). For expression in eukaryotic cells, sequences encoding full-length or partial LPP were inserted into the pEGFP-C1 (Clontech, Palo Alto, CA) or pM (Sadowski et al., 1992) expression vectors. To generate GFP-LPP, a BamHI site was inserted into the NsiI site of the LPPpGEM construct with the use of a linker (TGAGGATCCATGCA). To reconstitute full-length LPP, SalI/BamHI and BamHI/BamHI cDNA fragments encoding the N-terminal and C-terminal portions of LPP, respectively, were inserted into the SalI/BamHI pEGFP-C1 vector. To generate GST-LPP3–414, a cDNA encoding the proline-rich domain of LPP with 5′ NcoI and 3′ PmlI flanking sites was generated by PCR with the use of pMP50 cDNA, which encodes full-length LPP, as a template. This fragment was subcloned into a modified pUHD vector 5′ to a sequence encoding the 9E10 myc epitope tag (Evan et al., 1985). From this, the coding sequence of LPP3–414 and the myc epitope were subcloned into a modified pGEX2 vector to produce GST-LPP3–414. To produce a coding sequence that included a 6xHis tag, oligonucleotides encoding six histidines were inserted 3′ to the myc epitope sequence. The resulting plasmid encodes a protein of 668 amino acids composed of a GST domain, followed by amino acids 3–414 of LPP (the proline-rich domain), and a 27-amino acid sequence that contains the 9E10 myc epitope and the 6xHis tag (642-HVAACNMEQKLISEEDLNMNFHHHHHH-668). To construct GFP-LPP1–414-M, a DNA fragment encoding amino acids 1–414 of LPP was generated by PCR with the use of oligonucleotides with add-on sequences that allowed cloning of this fragment into the pIM20-M vector, in frame with a 3′ sequence encoding the membrane anchor of ActA (LILAMLAIGVFSLGAFIKIIQLRKNN; a gift of P. Cossart, Institut Pasteur). An ApaI/BamHI fragment of this pIM20-LPP1–414-M construct encoding a portion of the LPP sequence (amino acids 183–414) fused to the ActA membrane anchor sequence was inserted into the GFP-LPP construct, replacing a DNA sequence that encoded amino acids 183–612 of LPP, yielding GFP-LPP1–414-M. The constructs for the expression of GFP-LPPΔ387–397 and GFP-LPPΔ117–128 were generated by PCR with the use of the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the supplier's instructions. Constructs for the expression of the chimeric proteins GFP-HMGIC/LPP-long and GFP-HMGIC/LPP-short were generated by subcloning cDNAs encoding these fusions into the PstI and the PstI/ApaI sites, respectively, of the pEGFP-C1 vector (Clontech). Constructs for the expression of GAL4 chimeric proteins were made by subcloning the cDNAs into one of the pM vectors (Sa-dowski et al., 1992). The construct encoding GAL4-LPP1–612 was constructed by subcloning the full-length LPP coding region into the SmaI site of the pM1 vector. The LPP deletion construct encoding GAL4-LPP1–413 was generated by PCR with the use of the QuickChange site-directed mutagenesis kit introducing a TAG stop codon between the T and G at positions 1239 and 1240 of the LPP coding region. All other deletion constructs of LPP were generated by digestion and ligation with the use of the appropriate unique restriction sites (XbaI, SacI, ApaI, and MscI at positions 202, 429, 739, and 1116, respectively, of the LPP coding region). Constructs pMP53 and pMP54 for the expression of the chimeric proteins GAL4-HMGIC/LPP-short and GAL4-HMGIC/LPP-long, respectively, were generated by subcloning the appropriate HMGIC/LPP fusion cDNAs into the PstI and XbaI sites of the pM2 vector or in the EcoRI and SmaI sites of the pM3 vector, respectively.
Production of GST Fusion Proteins in Bacteria
The GST-LPP3–414 fusion protein comprising a myc tag and a 6xHis tag at its C terminus was produced in bacteria and purified from extracts by two-step affinity chromatography. The fusion protein was first purified on a Ni nitrilo triacetic acid resin (Qiagen, Chatsworth, CA) according to the manufacturer's instructions and then on a glutathione–Sepharose 4B column as described by Smith and Johnson (1988). GST-Zyx21–41 (Fradelizi et al., 1999) and GST-ActA235–584 (Golsteyn et al., 1997a) were produced and purified as described previously. Recombinant proteins were quantified by the method described by Bradford (1976).
Generation of LPP-specific Antibodies
The polyclonal rabbit antiserum MP2 was prepared against peptide G318QQGHPNTWKREPGY332 and synthesized as a multiple-antigen peptide conjugate (Research Genetics, Huntsville, AL), according to the procedure described by van de Loo et al. (1997). The polyclonal antiserum LPP2 was prepared against the GST-LPP3–414 fusion protein by immunization of rabbits (Harlow and Lane, 1988). MP2 and LPP2 antibodies were purified by GST-LPP3–414 affinity chromatography as described by Harlow and Lane (1988).
Transfection
HeLa cells were transfected with the use of a calcium phosphate DNA precipitation method described previously (Matthias et al., 1983). 293 and 3T3-L1 cells were transfected with the use of FuGene 6 transfection reagent (Boehringer Mannheim, Indianapolis, IN), according to the supplier's instructions. Cells were analyzed 24 h after the addition of DNA.
Preparation of Cell Extracts and Western Blotting
Eukaryotic cell extracts were prepared as described previously (Fradelizi et al., 1999). Proteins from cell lysates were separated by SDS-PAGE under reducing conditions (Laemmli, 1970). Transfer to nitrocellulose and antibody incubation were performed according to the method described by Burnette (1981). For analytical Western blotting, proteins were revealed by alkaline phosphatase detection. For quantitative Western blotting, extracts from a known number of cells were analyzed. Standard curves were established with the use of dilution series of GST-LPP3–414 and GST-Zyx 21–41. B38 anti-zyxin (Macalma et al., 1996) and MP2 anti-LPP sera were used as primary antibodies. Cy5-coupled secondary antibodies were detected by a PhosphorImager and analyzed by ImageQuant software (Molecular Dynamics, Sunnyvale, CA) (Fradelizi et al., 1999).
Analysis of Cells by Epifluorescence and Confocal Laser Scanning Microscopy
Cells were treated as described in the figure legends or in RESULTS and fixed with 3% paraformaldehyde. For detection of GFP fluorescence, coverslips were mounted and analyzed directly. For immunofluorescence labeling, cells were detergent permeabilized for 5 min with 0.4% Triton X-100 and labeled as described previously (Friederich et al., 1995). For detection of vinculin and zyxin, tissue culture supernatants of hybridoma cells 7F9 (Glukhova et al., 1990) (a gift of M. Glukhova, Institut Curie, Paris, France) and 164D4 (Krause and Wehland, unpublished data), respectively, were used. The 164D4 antibody is directed against an epitope not present in the LPP protein. A monoclonal anti-VASP antibody (C43) was purchased from Transduction Laboratories (Lexington, KY). Labeled cells were analyzed by epifluorescence or by confocal laser scanning microscopy (Leica, Wetzlar, Germany; TCS4D). For confocal scanning, typically, images were collected within the linear range of detection, eight scans per image and 8–12 images, to encompass the cell volume from the basolateral to the apical surfaces. Images were processed with Leica SCANware.
Overlay Detection of ActA- and Zyxin-interacting Proteins
Vero cell extracts enriched by profilin affinity chromatography were prepared as described previously (Golsteyn et al., 1997a). Samples containing proteins retained by profilin-Sepharose were separated by SDS-PAGE and transferred to nitrocellulose. A final concentration of 5 μg/ml GST-LPP3–414 or 1 μg/ml GST-ActA235–584 was added to the blocking solution and incubated for 30 min with the nitrocellulose membrane. The membrane was then processed as described for Western blotting with the antibodies specified in the figure legends.
Immunoprecipitation
RIPA extracts from Vero cells, previously stored at −80°C, were thawed and cleared by centrifugation at 12,000 × g for 10 min at 4°C. Nonimmune and affinity-purified MP2 antibodies were added to extracts at a final concentration of ∼1 μg/ml. The samples were incubated for 3 h at 4°C and centrifuged at 12,000 × g for 10 min at 4°C, and the immune complexes were isolated from the supernatant fraction by incubation with protein A–Sepharose (Pharmacia). The beads were collected and washed three times with RIPA buffer, transferred to new tubes, and then resuspended in SDS sample buffer. VASP was detected by Western blotting with the use of mouse anti-VASP antibodies.
Luciferase Reporter Assay
Twenty-four hours after seeding, semiconfluent 293 cells on six-well plates were transiently cotransfected with 800 ng of DNA of a construct expressing the GAL4DBD (DNA-binding domain) or GAL4DBD/LPP fusion proteins, 1 μg of a luciferase reporter construct, and 200 ng of RSV-β-galactosidase DNA (internal control for transfection efficiency). The reporter construct contains the gene encoding the firefly luciferase enzyme, which is under the control of a minimal promoter containing five consecutive GAL4-binding sequences (kindly provided by Dr. W. Schaffner and Dr. D. Escher, Institute of Molecular Biology, Zürich, Switzerland). Cell lysates were prepared 18–24 h after transfection and assayed for luciferase activity as described previously (Jansen et al., 1997), with some minor modifications. For each experiment, luciferase activity was determined in duplicate wells. Each experiment was repeated at least three times. The activation values (fold) were corrected for transfection efficiency (with the use of the RSV-β-galactosidase internal control) and are reported relative to the activity of the GAL4DBD alone. The cellular localization of the GAL4DBD chimeric proteins was determined by indirect immunofluorescence analysis with the use of a GAL4-specific antibody (Santa Cruz Biotechnology, Santa Cruz, CA), as described previously (van de Loo et al., 1997).
RESULTS
Characterization of Anti-LPP Antibodies
We prepared two anti-LPP antibodies to analyze the expression and the intracellular distribution of LPP in cultured cells. One antibody (MP2) was directed against a synthetic LPP-derived peptide whose sequence was not present in zyxin and TRIP6 (amino acids 318–332 of LPP), and the other antibody (LPP2) was directed against a recombinant N-terminal domain of LPP (GST-LPP3–414). Both antibodies recognized a protein of an apparent molecular mass of 80 kDa in HeLa cell extracts (Figure 1B, lanes 1 and 2) migrating slower in SDS gels than would be expected from the theoretically calculated molecular mass of LPP (72.7 kDa). Aberrant migration behavior is also observed for the LPP-related protein zyxin and may be due to the high proline content (Macalma et al., 1996). LPP2 (Figure 3E, lane 2) and MP2 (our unpublished results) antibodies reacted also with a GFP-LPP fusion produced in cultured cells transfected with the corresponding DNA. Anti-LPP serum did not recognize zyxin, which migrates with an apparent molecular mass of 84 kDa, as shown by the specific anti-zyxin antibodies (Figure 1B, lane 3). Note that LPP antiserum did not recognize TRIP6, which migrates at 50 kDa (Yi and Beckerle, 1998b).
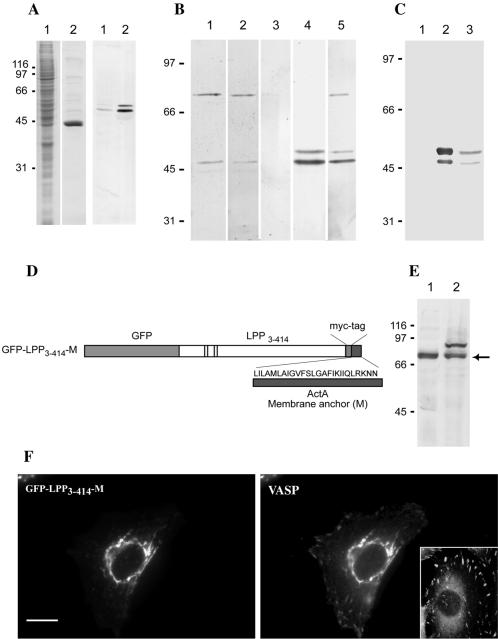
Figure 3.
Characterization of LPP/VASP binding. (A) Preparation of VASP-enriched extracts. Total Vero cell extracts (lanes 1) or proteins bound to profilin-Sepharose after incubation with Vero cell extracts (lanes 2) were analyzed by SDS-PAGE and stained with Coomassie blue (left panel) or transferred onto a nitrocellulose membrane and stained with anti-VASP antibodies (right panel). The positions of molecular mass markers (kilodaltons) are shown on the left. (B) The N-terminal portion of LPP interacts with VASP in vitro. The profilin-bound proteins were separated by SDS-PAGE and transferred onto a nitrocellulose membrane. (Lanes 1–3) Identical membranes were incubated with myc-tagged GST-LPP3–414 (lane 1), with myc-tagged GST-LPP3–414 in the presence of a 100-fold molar excess of GST (lane 2), or without protein (lane 3) and then probed with anti-myc antibodies. (Lane 4) An identical membrane was probed directly with anti-VASP antibodies to indicate the positions of VASP bands. (Lane 5) A membrane was incubated with GST-ActA235–584 and then probed with anti-ActA3 (Golsteyn et al., 1997a). The positions of molecular mass markers (kilodaltons) are shown on the left. (C) VASP coimmunoprecipitates with LPP. Vero cell extracts were incubated with nonimmune (lane 1) or purified MP2 (lane 2) antibody. Immunoprecipitates were analyzed by SDS-PAGE and Western blotting with mouse anti-VASP antibody. Total cell extracts were used to indicate the positions of VASP bands (lane 3). The positions of molecular mass markers (kilodaltons) are shown on the left. (D) Scheme of GFP-LPP chimera. GFP-LPP3–414-M is the fusion of Green Fluorescent Protein (GFP), the N-terminal part of LPP (amino acids 3–414), the myc tag, and the membrane anchor of ActA protein (amino acids 613–639). This ActA sequence is able to target proteins to the outer membrane of mitochondria. (E) Production of GFP-LPP3–414-M chimera in transiently transfected HeLa cells. Total extracts of nontransfected (lane 1) or transiently transfected HeLa cells producing GFP-LPP3–414-M (lane 2) were analyzed by SDS-PAGE and Western blotting with LPP2 antiserum. The antibody recognizes a band at 90 kDa that is not present in nontransfected cells. The arrow indicates the position of endogenous LPP. The positions of molecular mass markers (kilodaltons) are shown on the left. (F) LPP recruits VASP to an ectopic mitochondrial localization. HeLa cells transiently transfected to produce GFP-LPP3–414-M were fixed, permeabilized, and stained with anti-VASP antibody. Note that in addition to structures identified as mitochondria in separate experiments, VASP also localizes to focal adhesions, as in a nontransfected cell (inset). Bar, 20 μm.
LPP Is Expressed in Cultured Cells Derived from Various Tissues
To determine the expression pattern of the LPP protein in cells, we prepared extracts from different human cell lines and analyzed them by Western blotting techniques (Figure 1C). LPP was easily detected in most of the cell lines, with the exception of HL60, a peripheral blood lymphocyte cell line, in which the expression level was relatively low. Additional lower-molecular-mass bands were detected in platelet extracts and most likely correspond to degradation products of LPP. We next calculated the amount of LPP and zyxin present in cultured cells (Figure 1D) with the use of quantitative Western blotting analysis, as reported previously (Fradelizi et al., 1999). Two human cell lines of fibroblastic (HFF) and epithelial (CaCo2) origin as well as a porcine epithelial cell line (LLC-PK1) were analyzed. The amount of LPP and zyxin in extracts prepared from a fixed number of cells was compared with a dilution series of known amounts of the standard proteins (see MATERIALS AND METHODS). By this method, we detected 1–2 million molecules of LPP per cell in all cell lines. This is similar to the amount of zyxin in epithelial cell lines, but zyxin is fivefold more abundant in the fibroblastic cell line.
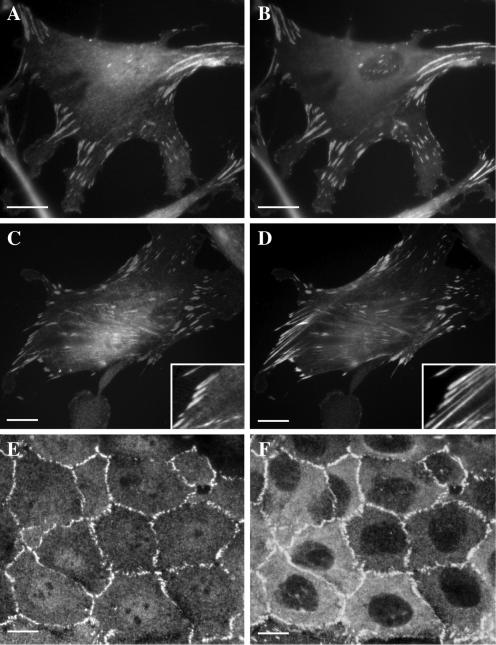
LPP Has a Similar but Not Identical Localization Compared with Zyxin
To analyze the intracellular localization of LPP, two different cell lines were double labeled by indirect immunofluorescence and examined by epifluorescence microscopy or by confocal laser scanning microscopy (Figure 2). LPP2 antibodies stained small structures at the ventral face of human foreskin fibroblasts (A and C) that were also stained by anti-vinculin mAb used to identify focal adhesions (B) and by anti-zyxin antibodies (D). In contrast to LPP, zyxin presents punctuate staining along structures previously identified as actin stress fibers (compare C and D and the insets). In human skin keratinocytes, in addition to focal adhesions, LPP2 antibodies stained the cell-to-cell contacts (E), as shown by codistribution with vinculin used as cell-to-cell contact marker (F). A faint staining obtained with LPP2 antibodies was also detectable in the nucleus of these cell types.
Figure 2.
Intracellular distribution of LPP in cultured cells. Cells were fixed, detergent permeabilized, and doubly stained by indirect immunofluorescence. (A–D) HFF cells doubly stained with affinity-purified LPP2 antibody (A and C) and with either monoclonal anti-vinculin (B) or anti-zyxin (D) antibody. Views were obtained by epifluorescence microscopy. The insets show 2.2-fold magnified details of images C and D. Ventral faces of the cells are shown. (E and F) Keratinocytes doubly stained with affinity-purified LPP2 antibody (E) and with monoclonal anti-vinculin antibody (F). Views were obtained by confocal laser scanning microscopy. Three apical sections were compiled. LPP codistributes with vinculin, which was used as a marker for focal adhesions (B) and cell-to-cell contacts (F). Note that in contrast to LPP, zyxin staining is not restricted to focal adhesions but is also found along actin stress fibers (D). LPP2 staining is also visible in the nucleus. Bars, 20 μm.
LPP Is Able to Interact with VASP In Vitro and Colocalizes In Vivo
The N-terminal domain of LPP contains FPPPP sequences at amino acids 67–74 and 88–93 (Figure 1A), which are critical for the interaction with proteins of the Ena/VASP family (Pistor et al., 1995; Niebuhr et al., 1997; Fedorov et al., 1999; Prehoda et al., 1999). To determine if LPP binds to members of this family, we tested for a direct interaction between VASP, a representative member of this family, and a recombinant proline-rich domain of LPP with the use of an overlay assay. To do so, VASP was first enriched by affinity chromatography with profilin beads. Total Vero cell extracts (Figure 3A, lane 1) and the VASP-enriched fraction (lane 2) were analyzed by SDS-PAGE and Coomassie blue staining (Figure 3A, left panel) or by immunoblotting with anti-VASP antibody (right panel). Although VASP 46 kDa and phosphorylated VASP (50 kDa) (Haffner et al., 1995) were enriched in the profilin-bound protein fraction, they was still undetected by Coomassie blue staining, whereas other profilin-binding proteins, especially actin, were easily detected. After separation by SDS-PAGE and transfer to nitrocellulose (as in A), the VASP-enriched fraction was incubated with myc-tagged GST-LPP3–414 and reprobed with anti-myc antibody to detect LPP binding (Figure 3B, lane 1). Two major bands at 46 and 90 kDa were detected (lane 1). In the presence of 100-fold excess of pure GST protein over GST-LPP3–414, the two proteins were still easily detected (lane 2). In the absence of GST-LPP3–414, the anti-myc antibody gave no signal (lane 3), whereas detection with the anti-VASP antibody showed that the 46-kDa band detected by overlay migrated exactly with VASP (lane 4). In a separate experiment, we incubated the VASP-enriched fraction with ActA recombinant protein (lane 5) in which VASP 46 kDa and phosphorylated VASP (50 kDa) were detected along with a 90-kDa protein that has previously been shown to be Mena (Reinhard et al., 1995b, 1996; Gertler et al., 1996; Golsteyn et al., 1997a). These results demonstrate a direct interaction of LPP and VASP, and likely Mena, in vitro.
Evidence for an in vivo interaction of LPP with VASP was obtained by two complementary approaches. First, we performed coimmunoprecipitation experiments (Figure 3C). LPP was immunoprecipitated with MP2 serum from total Vero cell extracts (lane 2). Nonimmune serum was used as a negative control (lane 1). The immunoprecipitates were separated by SDS-PAGE, and the presence of LPP was confirmed by Western blotting with LPP2 antibody. No zyxin was detected by immunostudies (our unpublished results). Probing the blot with an anti-VASP antibody revealed two bands at 46 and 50 kDa (lane 2) that comigrated with VASP and phosphorylated VASP (Haffner et al., 1995) in total cell extracts (lane 3) and that were not detected when preimmune serum was used for immunoprecipitation (lane 1). In our second approach, we tested if LPP was sufficient to recruit VASP to an ectopic localization in vivo. The membrane anchor of the ActA sequence has been shown previously to be sufficient to target proteins expressed in mammalian cells to the surface of mitochondria (Pistor et al., 1994; Bubeck et al., 1997). This ectopic localization allows us to test ligand recruitment in vivo. For this purpose, we generated a chimera named GFP-LPP3–414-M (Figure 3D) made up by the Green Fluorescent Protein (GFP) fused to the N-terminal part of LPP and linked in frame to the membrane anchor of the Listeria monocytogenes protein ActA (M). Expression of this construct was confirmed by Western blotting with the use of LPP2 antibody (Figure 3E). Nontransfected (inset) or transfected HeLa cells producing this chimera were labeled with anti-VASP antibody and examined by epifluorescence microscopy. The chimera localized to mitochondria, as confirmed by staining with Rhodamine 123, a vital dye that is sequestered in functioning mitochondria (our unpublished results). In addition to its localization to focal adhesions, as in nontransfected cells, VASP staining codistributed with that of GFP-LPP3–414-M on mitochondria (Figure 3F). This staining is not seen in nontransfected cells. Altogether, these experiments show that the N-terminal portion of LPP binds VASP and can recruit this ligand to specific intracellular compartments.
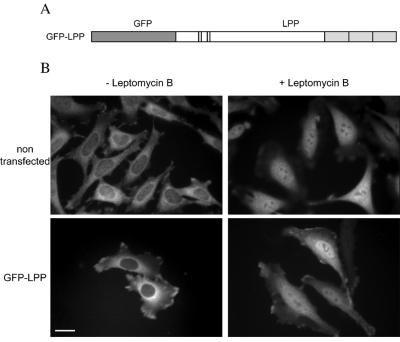
Leptomycin B Induces an Accumulation of LPP in the Nucleus
Although our cell-staining experiments showed that LPP was predominantly located at actin-rich structures, we tested if LPP might also be located at the nucleus, because a related protein, chicken zyxin, shuttles from cytoplasmic focal adhesions to the nucleus in a NES-dependent manner (Nix and Beckerle, 1997). We treated HeLa cells with leptomycin B, a drug that blocks nuclear export by preventing the formation of the NES/CRM1/Ran-GTP complex (Fornerod et al., 1997; Fukuda et al., 1997; Ossareh-Nazari et al., 1997; Wolff et al., 1997). All cells showed a nuclear accumulation of endogenous LPP after 90 min of treatment with leptomycin B, whereas only 6% of the untreated cells presented a similar staining (Figure 4B, upper panels). To rule out that nuclear staining was caused by nonspecific labeling, we used GFP as a molecular marker for full-length LPP (Figure 4A) and repeated the experiment with transfected cells producing the GFP-LPP chimera (Figure 4B, lower panels). This chimera was well expressed in transfected cells, as monitored by Western blotting (our unpublished results). Only 1% of the cells producing GFP-LPP presented nuclear staining before treatment with leptomycin B, whereas 93% of the cells accumulated the protein in the nucleus after 90 min of treatment. Although cell-staining experiments show that, at steady state, most of the LPP is in the cytoplasmic compartment, the accumulation of this protein in the nucleus in the presence of leptomycin B suggests that it shuttles between both compartments.
Figure 4.
Leptomycin B induces nuclear accumulation of LPP. (A) Scheme of GFP-LPP. GFP was fused to the N terminus of full-length LPP. (B) HeLa cells were incubated in the absence (− Leptomycin B) or the presence (+ Leptomycin B) of 20 nM leptomycin B for 90 min and then fixed. (Upper panels) Nontransfected cells that were permeabilized and stained with affinity-purified LPP2 antibodies. (Lower panels) Cells transiently transfected with a cDNA encoding GFP-LPP. Immunofluorescence and GFP were visualized by epifluorescence microscopy. A focal plane corresponding to the cell body is shown. After treatment with leptomycin B, endogenous LPP or transfected GFP-LPP signals were detected in the nucleus. Three hundred cells were counted for each treatment. Bar, 20 μm.
Characterization of the NES of LPP
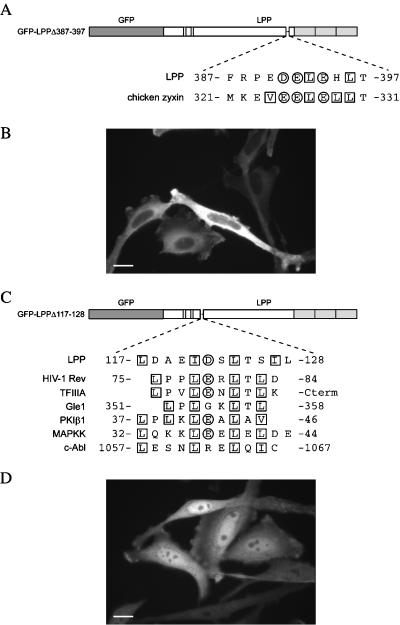
The effect of leptomycin B on the subcellular distribution of LPP suggests that LPP contains a NES that uses the CRM1-mediated nuclear export pathway. These sequences are rich in leucine residues, although there is not a strict consensus. Chicken zyxin contains a leucine-rich NES sequence situated at the hinge between the proline-rich domain and the LIM domains (Nix and Beckerle, 1997). Because this sequence is partially conserved in LPP (Figure 5A), we wanted to investigate whether it has an equivalent role in this protein. For this purpose, we generated a GFP-LPP chimera with a deletion of this putative NES sequence (amino acids 387–397) (Figure 5A). HeLa cells were transiently transfected with this GFP-LPPΔ387–397 construct (Figure 5B). The protein was found in the cytoplasm, and no difference could be observed with the distribution of full-length GFP-LPP (compare Figure 5B with Figure 4B), suggesting that this sequence was not responsible for the nuclear export of LPP. This result suggested that a different sequence must be functioning as a NES in this protein.
Figure 5.
LPP contains a nuclear export signal. (A) Scheme of GFP-LPPΔ387–397 and amino acid sequence of the deleted region (hatched lines) aligned with the sequence of the NES of chicken zyxin (Nix and Beckerle, 1997). (B) Amino acids 387–397 of LPP are not essential for its nuclear export. HeLa cells were transiently transfected with a construct encoding GFP-LPPΔ387–397, fixed, and analyzed by epifluorescence. A focal plane corresponding to the cell body is shown. The protein localized to the cytoplasmic compartment. (C) Scheme of GFP-LPPΔ117–128 and amino acid sequence of the deleted region (hatched lines) aligned with the sequences of the NES of HIV-1 Rev (Fischer et al., 1995; Wen et al., 1995), TFIIIA (Fridell et al., 1996), Gle1 (Murphy and Wente, 1996), PKIβ1 (Wen et al., 1995), MAPKK (Fukuda et al., 1996), and c-Abl (Taagepera et al., 1998). (D) A leucine-rich stretch covering amino acids 117–128 of LPP is essential for the nuclear export of the protein. HeLa cells were transiently transfected with a construct encoding GFP-LPPΔ117–128 and processed as described in B. The protein localized to the nuclear compartment. Bar, 20 μm.
Analysis of the LPP sequence revealed another leucine-rich stretch of residues (117-LDAEIDSLTSIL-128) located in the N-terminal portion of the proline-rich domain that shows sequence similarity to well-characterized NES sequences (see Figure 5C). To test whether residues 117–128 are essential for nuclear export of LPP, we made a GFP-LPP chimera in which this sequence was deleted (GFP-LPPΔ117–128) (Figure 5C). HeLa cells were transiently transfected with this construct and analyzed by epifluorescence microscopy. The chimeric protein accumulated in the nucleus of these cells (Figure 5D), although some GFP-LPPΔ117–128 molecules were still observed in adhesion plaques. This result suggests that amino acids 117–128 of LPP are an essential part of a functional NES sequence and that this subdomain is distinct from that previously reported for a related protein, chicken zyxin (Nix and Beckerle, 1997).
LPP Harbors Transcriptional Activation Activity
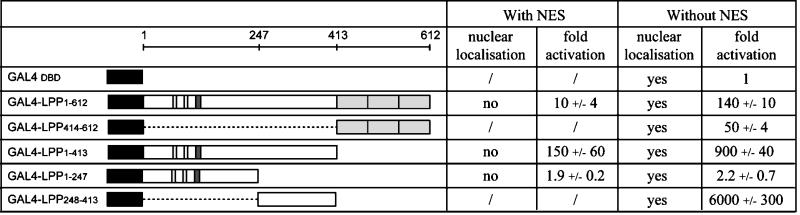
Our results show that LPP is able to localize in the nucleus, suggesting that this protein might have a role in this compartment. To investigate whether LPP may regulate transcription, we used a GAL4-based transactivation assay. An expression construct encoding the GAL4 DNA-binding domain fused to full-length LPP (GAL4-LPP1–612) was transfected into the 293 cell line together with a reporter plasmid containing the luciferase gene under the control of a GAL4-regulated promoter. We measured the luciferase enzymatic activity, which is correlated to the level of transcription of this reporter gene, in cell lysates. We found that GAL4-LPP1–612 (Figure 6, with NES) enhanced the luciferase activity 10-fold compared with the activity of the GAL4 DNA-binding domain alone. However, upon investigation of the intracellular distribution of the GAL4-LPP1–612 chimeric protein by immunocytochemistry, we found that, despite the fact that the GAL4DBD contains a strong nuclear localization signal, GAL4-LPP1–612 was predominantly localized in the cytoplasm and in adhesion plaques and that the nuclear amount was below detection level (our unpublished results). We then tested if the transcription activation capacity was related to the nuclear location of LPP. When the sequence that we identified as a NES was deleted, GAL4-LPP1–612 accumulated in the nucleus and enhanced the luciferase activity 140-fold compared with the GAL4DBD alone and 14-fold compared with the NES-containing form of GAL4-LPP1–612 (Figure 6, GAL4-LPP1–612 with and without NES).
Figure 6.
Structure and transcriptional activation capacity of GAL41–147-LPP chimeric proteins. Scheme of chimeric proteins comprising the GAL4 DNA-binding domain (amino acids 1–147) and either wild-type LPP or various N-terminal and/or C-terminal deletions of LPP. The GAL4 DNA-binding domain is represented by a black box. Domain organization of LPP is as in Figure 1. The NES of LPP is shown as a dark gray box. All chimeric constructs (with NES or without NES) were cotransfected into the 293 cell line along with a GAL4-regulated luciferase reporter and an RSV-β-galactosidase internal control, and cell lysates were assayed for luciferase activity 18–24 h after transfection. Activation (fold) is relative to the control GAL4DBD (DNA-binding domain). The results are the average of three independent experiments ± SD. Subcellular localization of the different constructs was established by immunocytochemistry with the use of a GAL4 antibody.
To characterize the portions of LPP important for this activity, we analyzed the proline-rich domain and the LIM domains separately in the same assay (Figure 6). Fusion proteins containing the three LIM domains of LPP (GAL4-LPP414–612) or the proline-rich domain (GAL4-LPP1–413) enhanced the luciferase activity by 50- or 150-fold, respectively, indicating that these domains contribute to the transcriptional activation capacity of LPP. As observed for full-length LPP, deletion of the NES in GAL4-LPP1–413 resulted in a partial nuclear accumulation of this fusion protein and in an increase of the luciferase activity by 6-fold compared with the NES-containing form (Figure 6, GAL4-LPP1–413, with and without NES). To delineate the portions of the proline-rich domain that are essential for the transcription activation capacity, we analyzed truncation variants of this domain. Successive C-terminal truncations progressively reduced the activity of the proline-rich domain (our unpublished results). We found that the activity was almost completely lost after removal of residues 248–413 (GAL4-LPP1–247). Deletion of the NES sequence of this fusion protein caused its nuclear accumulation but did not significantly increase its activity. In a complementary manner, LPP amino acids 248–413 (GAL4-LPP248–413) enhanced the luciferase activity 6000-fold compared with the GAL4DBD alone, confirming the importance of this sequence.
Characterization of HMGIC-LPP Fusions
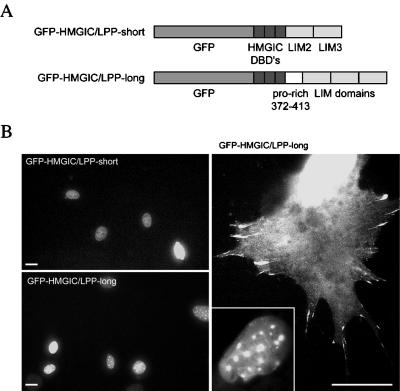
A cytogenetic subgroup of lipomas, mesenchymal tumors found in humans, is characterized by translocations involving the HMGIC and LPP genes, resulting in two different HMGIC/LPP fusion genes. These fusions encode the three DNA-binding domains of HMGIC followed by the two most C-terminal LIM domains of LPP (i.e., HMGIC/LPP-short) or a portion of the proline-rich domain (amino acids 372–413) and the three LIM domains of LPP (i.e., HMGIC/LPP-long) (Petit et al., 1996). Both fusions lack the NES of LPP. To characterize the encoded fusion proteins, we first examined their intracellular distribution. Constructs encoding GFP-tagged HMGIC/LPP fusion proteins (Figure 7A) were transiently transfected into 3T3-L1 cells. Both proteins were detected in the nucleus of these cells (Figure 7B). However, GFP-HMGIC/LPP-long could also be detected in the cytoplasm and in adhesion plaques of cells expressing very high levels of this protein, as monitored by fluorescence intensity (Figure 7B). Wild-type HMGIC, being an architectural factor, has no intrinsic transcriptional activation capacity in the GAL4-based transactivation assay (Jansen et al., 1999). Because the LIM domains of LPP have transcriptional activation activity, we determined the activity of the HMGIC/LPP fusions in this assay. However, no transcriptional activation activity for the HMGIC/LPP fusion proteins was found, suggesting that the presence of the DNA-binding domains of HMGIC impairs the transcriptional activation capacity of the LIM domains in this assay (our unpublished results).
Figure 7.
Analysis of HMGIC/LPP fusion proteins. (A) Scheme of GFP-HMGIC/LPP chimeric proteins. HMGIC-derived domains are represented by dark gray boxes. LPP-derived domains are represented as in Figure 1. DBD, DNA-binding domain. (B) 3T3-L1 cells were transiently transfected with constructs expressing GFP-HMGIC/LPP-short or GFP-HMGIC/LPP-long. Both chimeric proteins localized to the nucleus (left panels). However, in cells producing high levels of GFP-HMGIC/LPP-long, the latter can also be detected in the cytoplasm and in adhesion plaques (right panels) in addition to the nucleus (inset). Bars, 20 μm.
DISCUSSION
Analysis of chromosomal aberrations occurring in lipomas led to the identification of the LPP gene as the preferred translocation partner of HMGIC, a gene encoding an architectural factor. The deduced LPP amino acid sequence (Petit et al., 1996) revealed that LPP belongs to a family of LIM domain proteins, including transcription factors and components of cell adhesion sites (Beckerle, 1997). Our results show that despite their low sequence identity (41%), LPP and zyxin likely share overlapping functional properties, but they also have distinct features suggesting that each protein may fulfill specific roles in a cell. Moreover, we obtained evidence that LPP, a member of this new family, may participate in the control of gene transcription.
Targeting of proteins to distinct intracellular domains plays an essential role in the control of their activity. Although the localization of LPP in focal adhesions was similar to that of zyxin, LPP was not localized along stress fibers, in contrast to zyxin. Because overproduction of LPP in transfected cells did not result in its codistribution with stress fibers, it is unlikely that competition between LPP and zyxin for common binding sites would account for this difference in localization. Although zyxin and LPP share a similar domain organization, their sequences differ considerably. Therefore, it is conceivable that the ligands responsible for the targeting of LPP and zyxin, proteins that are not yet identified, are different. In the case of paxillin, an adapter protein with multiple binding motifs, it has been shown that the last LIM domain is responsible for its targeting to focal adhesions (Brown et al., 1996). Interestingly, whereas the HMGIC/LPP-short fusion localized solely to the nucleus, HMGIC/LPP-long could, in addition, localize to focal contacts when overexpressed in transfected cells. This observation opens the possibility that a small portion of the proline-rich domain (amino acids 372–413) and/or the first LIM domain of LPP may, at least partially, be responsible for directing LPP to focal adhesions.
LPP contains two FPPPP repeats, motifs that have been shown to be important for the interaction with proteins of the Ena/VASP family (Niebuhr et al., 1997; Fedorov et al., 1999; Prehoda et al., 1999). Using an overlay assay with a VASP-enriched cell fraction, we found that the N-terminal proline-rich region of LPP interacts directly with this protein in vitro. This finding suggests that at least one of the two potential VASP-binding motifs of LPP is functional, validating the consensus sequence for Ena/VASP binding determined with peptides (Niebuhr et al., 1997). The weaker binding signal obtained with LPP compared with ActA is most likely due to the fact that LPP contains only two VASP/Mena-binding sites, whereas ActA has four. Furthermore, our coimmunoprecipitation data, together with the observation that the proline-rich domain of LPP recruits VASP in vivo when targeted to an ectopic location on mitochondria, suggest that LPP and VASP are present in a complex in cultured cells.
By virtue of binding profilin, an actin monomer-binding protein that increases the rate of actin polymerization in vitro, Ena/VASP family members may control actin dynamics (Theriot and Mitchison, 1993; Gertler et al., 1996; Reinhard et al., 1996; Golsteyn et al., 1997a). The activity of Ena/VASP may be spatially controlled by scaffold proteins that recruit them to specific subcellular domains. Quantitation of the amount of LPP and zyxin revealed that these two proteins coexist in primary cells, as has been shown in platelets and HFF cells (Golsteyn et al., 1997b; this report) and suggested by Northern blot analysis of tissues (Macalma et al., 1996; Petit et al., 1996). In addition to zyxin and LPP, the focal adhesion protein vinculin is also able to interact with Ena/VASP family members (Reinhard et al., 1996). The biological significance of this partial functional redundancy remains unknown. Regulation by distinct signaling pathways as well as differences in the binding specificities or affinities for their ligands (Reinhard et al., 1996; Niebuhr et al., 1997) may allow each of these proteins to play a specific role in the fine tuning of actin dynamics in cells.
The accumulation of LPP in the nucleus of leptomycin B–treated cells suggests that nuclear export of LPP relies on a CRM1-dependent mechanism. Indeed, we found that an N-terminally located leucine-rich stretch of residues sharing sequence homology with well-characterized NES sequences is essential for nuclear export of LPP. Chicken zyxin contains another NES, located at the hinge between the proline-rich region and the LIM domains, that is partially conserved in LPP (Nix and Beckerle, 1997). However, the deletion of this sequence in LPP had no effect on its intracellular distribution, confirming that the NES in LPP is different from that in chicken zyxin. Differences of intramolecular location between these NES sequences may reflect a different regulation of the export of zyxin and LPP. The observation that LPP is transiently located in the nucleus raises the question of how this protein is imported into this compartment. Because LPP does not present a consensus nuclear localization signal, it may be imported via an interaction with a nuclear localization signal–containing protein.
In agreement with the observation that LPP is able to shuttle between the cytoplasm and the nucleus, we found that LPP has significant transcriptional activation capacity in a GAL4 transactivation assay in mammalian cells. While this article was under review, transcription activation capacity, measured in a similar assay, was reported for TRIP6, a protein sharing 53% sequence identity with LPP (Wang et al., 1999). This finding further emphasizes that LPP belongs to a new family, the members of which are involved in cell surface–nucleus communication. Interestingly, GAL4-LPP fusions that were undetectable in the nucleus by immunofluorescence still retained significant activity in the GAL4 assay. The activity of these fusion proteins was increased upon deletion of the NES sequence and their accumulation in the nucleus. These results suggest, first, that small amounts of endogenous LPP present in the nucleus at steady state may be sufficient for transcription activation, and second, that the spatial control of LPP and its transcription activation capacity are closely linked.
To date, the mechanism by which LPP activates transcription remains unknown. Although LIM domains have a similar structural organization as zinc-finger DNA-binding sequences, only LIM domains of hic5 have been shown to interact directly with DNA (Nishiya et al., 1998). Because there is substantial evidence that LIM domains function in protein-protein interactions, it is more likely that LPP interacts with a component (or components) of a transcriptional complex (Wadman et al., 1997). Because the activity of LPP depends on its intracellular distribution, it is possible that LPP modulates the activity of transcription factors by controlling their spatial distribution.
We found that the proline-rich domain and LIM domains contribute to the transcription activation capacity of LPP. The LPP sequence 248–413 is essential for this activity and has an ∼40-fold higher activity than the full-length GAL4-LPP fusion. Thus, it is possible that intramolecular interactions may regulate the activity of the full-length protein. Alternatively, the N-terminal portion of LPP (amino acids 1–247), which has no activity per se, may be involved in the binding of this protein to the cell surface, thereby decreasing the pool of LPP available for nuclear function.
HMGIC/LPP fusion proteins may interfere with the normal function of the HMGIC and/or LPP proteins in tumors. No transcriptional activation could be detected in the GAL4 assay for the nuclear HMGIC/LPP fusion proteins, suggesting that the transcriptional activation capacity of the LIM domains of LPP is negatively influenced by fusion to the DNA-binding domains of HMGIC in this assay. Wild-type HMGIC is thought to act as an architectural factor that binds to AT-rich sequences, thereby modulating the activity of other transcription factors (Goodwin, 1998; Mantovani et al., 1998). Thus, it cannot be excluded that if the DNA-binding domains of HMGIC bind to specific chromosomal structures in a physiological situation, the transcription activation capacity of the LIM domains is exposed, a possibility that our assay did not test.
Signaling pathways that control the organization of the actin cytoskeleton and cell proliferation are closely linked. Here we showed that LPP, a gene that is targeted by the preferential t(3;12)(q27-q28;q15) chromosomal translocation in lipomas, encodes a protein with multiple functional domains that are important for its interaction with proteins of focal adhesions and cell-to-cell contacts as well as for its transcription activation capacity. Although the physiological role of LPP is still unknown, our findings open the possibility that LPP may serve as a scaffold on which distinct protein complexes are assembled in the cytoplasm and the nucleus. Similar to the adherens junction protein β-catenin, LPP may participate in the organization of the cortical actin cytoskeleton at cell adhesion sites and in the regulation of gene transcription (Behrens et al., 1996). Knowledge obtained in the present study will constitute the basis for further investigations of the biological function of LPP and of its role as well as that of HMGIC/LPP fusions in tumorigenesis.
ACKNOWLEDGMENTS
We thank Drs. M. Arpin, G. Almouzni, and E. Jansen for carefully reading the manuscript and for their helpful comments. We also thank Sandra Meulemans and Wim Keysers for excellent technical assistance, and all our colleagues for stimulating discussions. We are indebted to Dr. J. Wehland for providing us anti-zyxin antibodies. R.M.G. and J.F. were recipients of Human Frontiers Science Program and Ministère di‘Education Nationale de la Recherche et des Technologies fellowships, respectively. M.M.R.P. is a research assistant of the Fonds voor Wetenschappelijk Onderzoek-Vlaanderen (Kom op tegen kanker). This work was supported by grants from the Association pour la Recherche Contre le Cancer (ARC 9622) and from the Centre National de la Recherche Scientifique, France, as well as by the Fonds voor Wetenschappelijk Onderzoek-Vlaanderen and the Geconcerteerde Onderzoeksacties 1997–2001, Belgium.
REFERENCES
- Aravind L, Landsman D. AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res. 1998;26:4413–4421. doi: 10.1093/nar/26.19.4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashar HR, Fejzo MS, Tkachenko A, Zhou X, Fletcher JA, Weremowicz S, Morton CC, Chada K. Disruption of the architectural factor HMGI-C: DNA-binding AT hook motifs fused in lipomas to distinct transcriptional regulatory domains. Cell. 1995;82:57–65. doi: 10.1016/0092-8674(95)90052-7. [DOI] [PubMed] [Google Scholar]
- Beckerle MC. Identification of a new protein localized at sites of cell-substratum adhesion. J Cell Biol. 1986;103:1679–1687. doi: 10.1083/jcb.103.5.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckerle MC. Zyxin: zinc fingers at sites of cell adhesion. BioEssays. 1997;19:949–957. doi: 10.1002/bies.950191104. [DOI] [PubMed] [Google Scholar]
- Behrens J, von Kries JP, Kühl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Functional interaction of β-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brindle NPJ, Holt MR, Davies JE, Price CJ, Critchley DR. The focal-adhesion vasodilator-stimulated phosphoprotein (VASP) binds to the proline-rich domain in vinculin. Biochem J. 1996;318:753–757. doi: 10.1042/bj3180753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MC, Perrotta JA, Turner CE. Identification of LIM3 as the principal determinant of paxillin focal adhesion localization and characterization of a novel motif on paxillin directing vinculin and focal adhesion kinase binding. J Cell Biol. 1996;135:1109–1123. doi: 10.1083/jcb.135.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubeck P, Pistor S, Wehland J, Jockusch BM. Ligand recruitment by vinculin domains in transfected cells. J Cell Sci. 1997;110:1361–1371. doi: 10.1242/jcs.110.12.1361. [DOI] [PubMed] [Google Scholar]
- Burnette WN. “Western blotting”: electrophoretic transfer of proteins from SDS-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981;112:195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- Dawid IB, Breen JJ, Toyama R. LIM domains: multiple roles as adapters and functional modifiers in protein interactions. Trends Genet. 1998;14:156–157. doi: 10.1016/s0168-9525(98)01424-3. [DOI] [PubMed] [Google Scholar]
- Domann E, Wehland J, Rohde M, Pistor S, Hartl M, Goebel W, Leimeister-Wächter M, Wuenscher M, Chakraborty T. A novel bacterial virulence gene in Listeria monocytogenes required for host cell microfilament interaction with homology to the proline-rich region of vinculin. EMBO J. 1992;11:1981–1990. doi: 10.1002/j.1460-2075.1992.tb05252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan GI, Lewis G, Ramsey G, Bishop JM. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov AA, Fedorov E, Gertler F, Almo SC. Structure of EVH1, a novel proline-rich ligand-binding module involved in cytoskeletal dynamics and neuronal function. Nat Struct Biol. 1999;6:661–665. doi: 10.1038/10717. [DOI] [PubMed] [Google Scholar]
- Fischer U, Huber J, Boelens WC, Mattaj IW, Luhrmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Fradelizi J, Friederich E, Beckerle MC, Golsteyn RM. Quantitative measurement of proteins by Western blotting with Cy5 coupled secondary antibodies. BioTechniques. 1999;26:484–494. [PubMed] [Google Scholar]
- Fridell RA, Fischer U, Luhrmann R, Meyer BE, Meinkoth JL, Malim MH, Cullen BR. Amphibian transcription factor IIIA proteins contain a sequence element functionally equivalent to the nuclear export signal of human immunodeficiency virus type 1 Rev. Proc Natl Acad Sci USA. 1996;93:2936–2940. doi: 10.1073/pnas.93.7.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friederich E, Gouin E, Hellio R, Kocks C, Cossart P, Louvard D. Targeting of Listeria monocytogenes ActA protein to the plasma membrane as a tool to dissect both actin-based cell morphogenesis and ActA function. EMBO J. 1995;14:2731–2744. doi: 10.1002/j.1460-2075.1995.tb07274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- Fukuda M, Gotoh I, Gotoh Y, Nishida E. Cytoplasmic localization of mitogen-activated protein kinase kinase directed by its NH2-terminal, leucine-rich short amino acid sequence, which acts as a nuclear export signal. J Biol Chem. 1996;271:20024–20028. doi: 10.1074/jbc.271.33.20024. [DOI] [PubMed] [Google Scholar]
- Gertler FB, Niebuhr K, Reinhard M, Wehland J, Soriano P. Mena, a relative of VASP and Drosophila enabled, is implicated in the control of microfilament dynamics. Cell. 1996;87:227–239. doi: 10.1016/s0092-8674(00)81341-0. [DOI] [PubMed] [Google Scholar]
- Glukhova MA, Frid MG, Kotelianski VE. Developmental changes in expression of contractile cytoskeletal proteins in human aortic smooth muscle. J Biol Chem. 1990;265:13042–13046. [PubMed] [Google Scholar]
- Golsteyn RM, Beckerle MC, Koay T, Friederich E. Structural and functional similarities between the human cytoskeletal protein, zyxin, and the ActA protein of Listeria monocytogenes. J Cell Sci. 1997a;110:1893–1906. doi: 10.1242/jcs.110.16.1893. [DOI] [PubMed] [Google Scholar]
- Golsteyn RM, Louvard D, Friederich E. The role of actin binding proteins in epithelial morphogenesis: models based upon Listeria movement. Biophys Chem. 1997b;68:73–82. doi: 10.1016/s0301-4622(97)00009-4. [DOI] [PubMed] [Google Scholar]
- Goodwin GH. The high mobility group protein, HMGI-C. Int J Biochem Cell Biol. 1998;30:761–766. doi: 10.1016/s1357-2725(98)00016-8. [DOI] [PubMed] [Google Scholar]
- Haffner C, Jarchau T, Reinhard M, Hoppe J, Lohmann SM, Walter U. Molecular cloning, structural analysis and functional expression of the proline-rich focal adhesion and microfilament-associated protein VASP. EMBO J. 1995;14:19–27. doi: 10.1002/j.1460-2075.1995.tb06971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- Jansen E, Ayoubi TAY, Meulemans SM, Van de Ven WJM. Cell type-specific protein-DNA interactions at the cAMP response elements of the prohormone convertase 1 promoter: evidence for additional transactivators distinct from CREB/ATF family members. J Biol Chem. 1997;272:2500–2508. doi: 10.1074/jbc.272.4.2500. [DOI] [PubMed] [Google Scholar]
- Jansen E, Petit MMR, Schoenmakers EFPM, Ayoubi TAY, Van de Ven WJM. High mobility group protein HMGIC: a molecular target in solid tumor formation. Gene Ther Mol Biol. 1999;3:387–395. [Google Scholar]
- Kocks C, Gouin E, Tabouret M, Berche P, Ohayon H, Cossart P. L. monocytogenes-induced actin assembly requires the actA gene product, a surface protein. Cell. 1992;68:521–531. doi: 10.1016/0092-8674(92)90188-i. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laurent V, Loisel TP, Harbeck B, Wehman A, Grobe L, Jockusch BM, Wehland J, Gertler FB, Carlier MF. Role of proteins of the Ena/VASP family in actin-based motility of Listeria monocytogenes. J Cell Biol. 1999;144:1245–1258. doi: 10.1083/jcb.144.6.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Choi HS, Gyuris J, Brent R, Moore DD. Two classes of proteins dependent on either the presence or absence of thyroid hormone for interaction with the thyroid hormone receptor. Mol Endocrinol. 1995;9:243–254. doi: 10.1210/mend.9.2.7776974. [DOI] [PubMed] [Google Scholar]
- Macalma T, Otte J, Hensler ME, Bockholt SM, Louis HA, Kalff-Suske M, Grzeschik K-H, von der Ahe D, Beckerle MC. Molecular characterization of human zyxin. J Biol Chem. 1996;271:31470–31478. doi: 10.1074/jbc.271.49.31470. [DOI] [PubMed] [Google Scholar]
- Mantovani F, Covaceuszach S, Rustighi A, Sgarra R, Heath C, Goodwin GH, Manfioletti G. NF-κB mediated transcriptional activation is enhanced by the architectural factor HMGI-C. Nucleic Acids Res. 1998;26:1433–1439. doi: 10.1093/nar/26.6.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthias PD, Bernard HU, Scott A, Brady G, Hashimoto-Gotoh T, Schütz G. A bovine papilloma virus vector with a dominant resistant marker replicates extrachromosomally in mouse and E. coli cells. EMBO J. 1983;2:1487–1492. doi: 10.1002/j.1460-2075.1983.tb01612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy R, Wente SR. An RNA-export mediator with an essential nuclear export signal. Nature. 1996;383:357–360. doi: 10.1038/383357a0. [DOI] [PubMed] [Google Scholar]
- Niebuhr K, Ebel F, Franck R, Reinhard M, Domann E, Carl UD, Walter U, Gertler FB, Wehland J, Chakraborty T. A novel proline-rich motif present in ActA of Listeria monocytogenes and cytoskeletal proteins is the ligand for the EVH1 domain, a protein module present in the Ena/VASP family. EMBO J. 1997;16:5433–5444. doi: 10.1093/emboj/16.17.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiya N, Sabe H, Nose K, Shibanuma M. The LIM domains of hic-5 protein recognize specific DNA fragments in a zinc-dependent manner in vitro. Nucleic Acids Res. 1998;26:4267–4273. doi: 10.1093/nar/26.18.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nix DA, Beckerle MC. Nuclear-cytoplasmic shuttling of the focal contact protein, zyxin: a potential mechanism for communication between sites of cell adhesion and the nucleus. J Cell Biol. 1997;138:1139–1147. doi: 10.1083/jcb.138.5.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- Petit MMR, Mols R, Schoenmakers EFPM, Mandahl N, Van de Ven WJM. LPP, the preferred fusion partner gene of HMGIC in lipomas, is a novel member of the LIM protein gene family. Genomics. 1996;36:118–129. doi: 10.1006/geno.1996.0432. [DOI] [PubMed] [Google Scholar]
- Pistor S, Chakraborty T, Niebuhr K, Domann E, Wehland J. The ActA protein of Listeria monocytogenes acts as a nucleator inducing reorganization of the actin cytoskeleton. EMBO J. 1994;13:758–763. doi: 10.1002/j.1460-2075.1994.tb06318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pistor S, Chakraborty T, Walter U, Wehland J. The bacterial actin nucleator protein ActA of Listeria monocytogenes contains multiple binding sites for host microfilament proteins. Curr Biol. 1995;5:517–525. doi: 10.1016/s0960-9822(95)00104-7. [DOI] [PubMed] [Google Scholar]
- Pollard TD. Actin cytoskeleton: missing link for intracellular bacterial motility? Curr Biol. 1995;5:837–840. doi: 10.1016/s0960-9822(95)00167-9. [DOI] [PubMed] [Google Scholar]
- Prehoda KE, Lee DJ, Lim WA. Structure of the enabled/VASP homology 1 domain-peptide complex: a key component in the spatial control of actin assembly. Cell. 1999;97:471–480. doi: 10.1016/s0092-8674(00)80757-6. [DOI] [PubMed] [Google Scholar]
- Price CJ, Jones P, Davidson MD, Patel B, Bendori R, Geiger B, Critchley DR. Primary sequence and domain structure of chicken vinculin. Biochem J. 1989;259:453–461. doi: 10.1042/bj2590453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard M, Giehl K, Abel K, Haffner C, Jarchau T, Hoppe V, Jockusch BM, Walter U. The proline-rich focal adhesion and microfilament protein VASP is a ligand for profilins. EMBO J. 1995a;14:1583–1589. doi: 10.1002/j.1460-2075.1995.tb07146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard M, Halbrugge M, Scheer U, Wiegand C, Jockusch BM, Walter U. The 46/50 kDa phosphoprotein VASP purified from human platelets is a novel protein associated with actin filaments and focal contacts. EMBO J. 1992;11:2063–2070. doi: 10.1002/j.1460-2075.1992.tb05264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard M, Jouvenal K, Tripier D, Walter U. Identification, purification, and characterization of a zyxin-related protein that binds the focal adhesion and microfilament protein VASP (vasodilator-stimulated phosphoprotein) Proc Natl Acad Sci USA. 1995b;92:7956–7960. doi: 10.1073/pnas.92.17.7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard M, Rüdiger M, Jockusch BM, Walter U. VASP interaction with vinculin: a recurring theme of interactions with proline-rich motifs. FEBS Lett. 1996;399:103–107. doi: 10.1016/s0014-5793(96)01295-1. [DOI] [PubMed] [Google Scholar]
- Robine S, Sahuquillo-Merino C, Louvard D, Pringault E. Regulatory sequences on the human villin gene trigger the expression of a reporter gene in differentiating HT29 intestinal cell line. J Biol Chem. 1993;268:11426–11434. [PubMed] [Google Scholar]
- Rogel-Gaillard C, Breitburd F, Orth G. Human papillomavirus type 1 E4 proteins differing by their N-terminal ends have distinct cellular localizations when transiently expressed in vitro. J Virol. 1992;66:816–823. doi: 10.1128/jvi.66.2.816-823.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski I, Bell B, Broad P, Hollis M. GAL4 fusion vectors for expression in yeast or mammalian cells. Gene. 1992;118:137–141. doi: 10.1016/0378-1119(92)90261-m. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Schoenmakers EFPM, Wanschura S, Mols R, Bullerdiek J, Van den Berghe H, Van de Ven WJM. Recurrent rearrangements in the high mobility group protein gene, HMGI-C, in benign mesenchymal tumors. Nat Genet. 1995;10:436–444. doi: 10.1038/ng0895-436. [DOI] [PubMed] [Google Scholar]
- Smith DB, Johnson KS. Single step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Taagepera S, McDonald D, Loeb JE, Whitaker LL, McElroy AK, Wang JY, Hope TJ. Nuclear-cytoplasmic shuttling of C-ABL tyrosine kinase. Proc Natl Acad Sci USA. 1998;95:7457–7462. doi: 10.1073/pnas.95.13.7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theriot JA, Mitchison TJ. The three faces of profilin. Cell. 1993;75:835–838. doi: 10.1016/0092-8674(93)90527-w. [DOI] [PubMed] [Google Scholar]
- van de Loo JW, Creemers JW, Bright NA, Young BD, Roebroek AJ, Van de Ven WJ. Biosynthesis, distinct posttranslational modifications, and functional characterization of lymphoma proprotein convertase. J Biol Chem. 1997;272:27116–27123. doi: 10.1074/jbc.272.43.27116. [DOI] [PubMed] [Google Scholar]
- Wadman IA, Osada H, Grutz GG, Agulnick AD, Westphal H, Forster A, Rabbitts TH. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Dooher JE, Koedood Z, Gilmore TD. Characterization of mouse trip6: a putative intracellular signaling protein. Gene. 1999;234:403–409. doi: 10.1016/s0378-1119(99)00168-7. [DOI] [PubMed] [Google Scholar]
- Wen W, Meinkoth JL, Tsien RY, Taylor SS. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- Wolff B, Sanglier J-J, Wang Y. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem Biol. 1997;4:139–147. doi: 10.1016/s1074-5521(97)90257-x. [DOI] [PubMed] [Google Scholar]
- Wolffe AP. Architectural transcription factors. Science. 1994;264:1100–1101. doi: 10.1126/science.8178167. [DOI] [PubMed] [Google Scholar]
- Yi J, Beckerle MC. The human TRIP6 gene encodes a LIM domain protein and maps to chromosome 7q22, a region associated with tumorigenesis. Genomics. 1998a;49:314–316. doi: 10.1006/geno.1998.5248. [DOI] [PubMed] [Google Scholar]
- Yi, J., and Beckerle, M.C. (1998b). Molecular characterization of Trip6, a new member of the zyxin family. Mol. Biol. Cell (Abstract 2393), 413a.
- Zumbrunn J, Trueb B. A zyxin-related protein whose synthesis is reduced in virally transformed fibroblasts. Eur J Biochem. 1996;241:657–663. doi: 10.1111/j.1432-1033.1996.00657.x. [DOI] [PubMed] [Google Scholar]