Abstract
The late Golgi of the yeast Saccharomyces cerevisiae receives membrane traffic from the secretory pathway as well as retrograde traffic from post-Golgi compartments, but the machinery that regulates these vesicle-docking and fusion events has not been characterized. We have identified three components of a novel protein complex that is required for protein sorting at the yeast late Golgi compartment. Mutation of VPS52, VPS53, or VPS54 results in the missorting of 70% of the vacuolar hydrolase carboxypeptidase Y as well as the mislocalization of late Golgi membrane proteins to the vacuole, whereas protein traffic through the early part of the Golgi complex is unaffected. A vps52/53/54 triple mutant strain is phenotypically indistinguishable from each of the single mutants, consistent with the model that all three are required for a common step in membrane transport. Native coimmunoprecipitation experiments indicate that Vps52p, Vps53p, and Vps54p are associated in a 1:1:1 complex that sediments as a single peak on sucrose velocity gradients. This complex, which exists both in a soluble pool and as a peripheral component of a membrane fraction, colocalizes with markers of the yeast late Golgi by immunofluorescence microscopy. Together, the phenotypic and biochemical data suggest that VPS52, VPS53, and VPS54 are required for the retrograde transport of Golgi membrane proteins from an endosomal/prevacuolar compartment. The Vps52/53/54 complex joins a growing list of distinct multisubunit complexes that regulate membrane-trafficking events.
INTRODUCTION
The last compartment of the yeast Golgi complex is the point of divergence of a number of different sorting pathways. Newly synthesized proteins transported from earlier parts of the secretory pathway receive final modifications here and are subsequently directed into a variety of different carrier vesicles for transport to the cell surface, to endosomal compartments, or to the vacuole (Harsay and Bretscher, 1995; Cowles et al., 1997; Piper et al., 1997; reviewed by Conibear and Stevens, 1998). Sorting into at least some of these different pathways is performed by coat proteins that selectively incorporate cargo into a budding vesicle, causing membrane invagination and ultimately scission. Vesicle targeting and fusion with the appropriate compartment is also regulated by a variety of integral and peripheral membrane proteins. The molecular machinery that controls these vesicle transport processes is highly conserved in eukaryotes, and related proteins form analogous complexes at different transport steps within a single organism (Ferro-Novick and Jahn, 1994; Rothman, 1994).
Crucial to the fusion process is the pairing of vesicle-associated soluble NSF attachment protein receptors (v-SNAREs) with t-SNAREs on the target membrane, which may promote membrane fusion by allowing the close apposition of the two lipid bilayers (Weber et al., 1998).
Early models suggested that the specificity of vesicle targeting lies with the SNAREs, such that interactions between specific members of the v-SNARE and t-SNARE family uniquely define a transport step (Sollner et al., 1993). However, recent work has demonstrated that v-SNAREs and t-SNAREs can pair promiscuously (Yang et al., 1999) and that a single v-SNARE can interact with multiple t-SNAREs (Fischer von Mollard et al., 1997; Lupashin et al., 1997; Fischer von Mollard and Stevens, 1999). Furthermore, a number of other proteins have been identified that are important for vesicle tethering or docking and that act before SNARE complex assembly, and it is these additional factors that may largely specify the fusion of a transport vesicle with a particular target membrane.
Rab GTPases and Sec1 family proteins have long been implicated in SNARE complex formation (Ferro-Novick and Jahn, 1994). Recently, a number of unique macromolecular protein complexes were identified that have been shown to interact physically and/or genetically with members of the Rab and Sec1 families to regulate a vesicle-docking step, which precedes SNARE complex formation. One of the best studied examples is the exocyst complex, which is proposed to link a vesicle bearing the Sec4 Rab protein with sites of polarized exocytosis (TerBush et al., 1996; Guo et al., 1999). Uso1p, which functions together with the Rab Ypt1p and Sec35p in the docking of endoplasmic reticulum (ER) vesicles at the Golgi, is required for the assembly of ER-to-Golgi SNARE complexes (Sapperstein et al., 1996; Cao et al., 1998; VanRheenen et al., 1998). The multisubunit TRAPP complex, which is found on the cis Golgi and also acts in ER-to-Golgi transport at a step preceding SNARE complex assembly, shows genetic interactions with Uso1p and Ypt1p and may also participate in vesicle docking (Sacher et al., 1998). The mammalian homologue of Uso1p, p115, acts as a docking factor to link the vesicle-associated protein giantin with a complex of GM130 and GRASP65 on Golgi membranes during intra-Golgi transport (Nakamura et al., 1997; Sonnichsen et al., 1998). In addition, the endosomal protein EEA1 binds Rab5 and is important for endosome docking in the homotypic fusion of early endosomes (Christoforidis et al., 1999). One model proposes that such docking factors are generally required to direct vesicles bearing Rab-GTP proteins to their sites of fusion, perhaps also interacting with Sec1 proteins to activate t-SNAREs and permit SNARE complex formation (Pfeffer, 1999).
Of the two parallel pathways that divert newly synthesized proteins from the secretory pathway and direct them from the yeast late Golgi to the vacuole, the carboxypeptidase Y (CPY) pathway is the best understood (Conibear and Stevens, 1998). A number of sequential budding/fusion reactions take place in the sorting of newly synthesized CPY to the vacuole. CPY bound to its receptor first enters vesicles at the late Golgi that are targeted to fuse with the prevacuolar/endosomal compartment (PVC) (Vida et al., 1993). Whereas CPY is transported to the vacuole in a second fusion step, its receptor enters retrograde vesicles that bud from the PVC and return to fuse with the late Golgi, thus allowing the receptor to carry out multiple rounds of sorting (Cereghino et al., 1995; Cooper and Stevens, 1996). Resident late Golgi membrane proteins such as Kex2p and dipeptidyl aminopeptidase (DPAP) A are also transported along early parts of the CPY pathway, and they maintain their Golgi localization by continuous retrieval from the PVC (Nothwehr et al., 1993; Bryant and Stevens, 1997). The sorting of CPY by the CPY receptor (the product of the VPS10 gene; Marcusson et al., 1994) is analogous to the sorting of newly synthesized lysosomal proteins by the mannose 6-phosphate receptor of mammalian cells (Kornfeld, 1992), and the yeast late Golgi (defined by the presence of Kex1p, Kex2p, and DPAP A) is considered to be the functional equivalent of the mammalian trans-Golgi network (TGN).
Because the CPY pathway involves a number of vesicle-budding and docking/fusion steps, it is expected to require the concerted function of a large number of accessory proteins. Genetic screens carried out in a number of laboratories have identified more than 40 genes (VPS, PEP, VAM, VAC; Jones, 1977; Bankaitis et al., 1986; Rothman and Stevens, 1986; Robinson et al., 1988; Weisman et al., 1990; Wada et al., 1992) required for the correct sorting of CPY to the vacuole. These genes were originally grouped according to their mutant vacuolar morphologies (classes A–F; Raymond et al., 1992), assuming that mutations in genes that act at the same step will produce similar phenotypes. This prediction has been borne out by more recent studies: many of the class D VPS genes encode components of a docking/fusion complex that is implicated in the fusion of late-Golgi–derived vesicles with the PVC. These include Vac1p, the yeast homologue of EEA1, which binds the Rab5 homologue Vps21p as well as the Sec1-like Vps45p and the endosomal t-SNARE Pep12p (Burd et al., 1997; Peterson et al., 1999; Tall et al., 1999). Class C Vps proteins include Vps11p, Vps18p, Vps16p, and the Sec1-like Vps33p, which form a complex that, together with the vacuolar t-SNARE Vam3p, the SNAP-25–like Vam7p, and the Rab protein Ypt7p, is required for the fusion of multiple transport intermediates with the vacuole (Haas et al., 1995; Darsow et al., 1997; Rieder and Emr, 1997; Sato et al., 1998; Ungermann and Wickner, 1998). Class E VPS gene products regulate transit through the PVC, whereas components of the retromer complex are thought to act at the PVC to sort proteins into retrograde vesicles that are targeted to fuse with the TGN (Seaman et al., 1998; Nothwehr et al., 1999).
Lacking from this catalogue of Vps proteins are candidate assembly factors that might direct the assembly of a SNARE complex during fusion with the TGN. In fact, few of the Vps proteins that have been characterized to date are even localized to the TGN. Vps1p is required for the budding of TGN vesicles, as is clathrin (Seeger and Payne, 1992; Nothwehr et al., 1995), but the adaptin subunits that are involved in lysosomal protein sorting in mammalian cells are not absolutely required for CPY sorting; in fact, entry into the CPY pathway does not appear to depend on cytoplasmic tail signals (Roberts et al., 1992; Redding et al., 1996). Therefore, it seems likely that additional proteins exist to control both budding and fusion at the TGN.
The syntaxin-like proteins Tlg1p and Tlg2p are candidate late Golgi t-SNAREs that may act at the fusion step (Abeliovich et al., 1998; Holthuis et al., 1998a,b; Nichols and Pelham, 1998; Seron et al., 1998). They are thought to reside in the yeast TGN as well as in endosomal compartments, and both colocalize in part with the late Golgi markers Kex2p and DPAP A by immunofluorescence microscopy and fractionation on sucrose gradients (Holthuis et al., 1998a). These SNAREs are required for the maintenance of normal levels of TGN proteins as well as for the correct sorting of CPY, and they have been proposed to act as targets of retrograde vesicles during the retrieval of TGN proteins from the PVC. However, other studies have suggested a requirement for these proteins in trafficking to early endosomes, and further work will be needed to clarify the role of these recently identified t-SNAREs.
We carried out a genetic screen to identify novel components of the TGN-localized vesicle transport machinery. In this paper, we report the identification of three new Vps proteins—Vps52p, Vps53p, and Vps54p—that localize to the TGN and are required for CPY sorting and for the recycling of resident TGN membrane proteins. Loss of any one of these proteins results in identical mutant phenotypes, suggesting that they form a class of Vps proteins that act together to carry out a unique step in the CPY-sorting pathway. In fact, Vps52p, Vps53p, and Vps54p were found to be physically associated in a stable complex in a 1:1:1 stoichiometry.
MATERIALS AND METHODS
Enzymes used in DNA manipulations were from New England Biolabs (Beverly, MA), Boehringer Mannheim Biochemicals (Indianapolis, IN), or GIBCO-BRL (Gaithersburg, MD). Oligonucleotides were synthesized by Keystone Laboratories (Camarillo, CA). The anti-CPY, anti-alkaline phosphatase (ALP), anti-Vps10p, anti-hemagglutinin (HA), and anti-Vph1p polyclonal antibodies have been described previously (Nothwehr et al., 1995; Cooper and Stevens, 1996). mAbs to Pep12p (mAb 24-2C3G4), ALP (mAb 1D3-A10), and CPY (mAb 10A5-B5) as well as Alexa-conjugated secondary antibodies were from Molecular Probes (Eugene, OR). Biotinylated anti-rabbit and anti-mouse secondary antibodies were from Jackson ImmunoResearch Laboratories (West Grove, PA). Anti-HA mAb HA.11 ascites fluid was purchased from BAbCO (Richmond, CA). Affinity-purified anti-myc polyclonal antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The anti-myc mAb 9E10 culture supernatant was prepared from a hybridoma obtained from the American Type Culture Collection (Rockville, MD). HRP-conjugated anti-mouse antibodies used for Western analysis were from Sigma Chemical (St. Louis, MO). 35S-Express label was purchased from DuPont New England Nuclear (Boston, MA). Fixed Staphylococcus aureus cells (IgGSorb) used to collect immune complexes were purchased from the Enzyme Center (Malden, MA). Oxalyticase was from Enzogenetics (Corvallis, OR). Zymolyase 100T was from Seikagaku (Tokyo, Japan). All other chemicals were of high-purity commercial grade.
Yeast Genetic Screen
Parent strains (LCY81 and LCY82) in which a deletion of the END4/SLA2 gene was covered by END4 on a URA3-based plasmid (pSLA2) were subjected to UV mutagenesis as described previously (Nothwehr et al., 1996) (Tables 1 and 2). Additional mutants were obtained by transforming the same strains with a NotI-digested yeast genomic library containing random insertions of a Tn3-LacZ transposon cassette (Burns et al., 1994). CPY-secreting colonies were identified by an overlay assay (Roberts et al., 1991), and those that exhibited synthetic lethality with end4Δ (as determined by the inability to grow in the presence of 5-FOA) were chosen for further study. Mutants were tested for complementation with each other and with the existing vps mutant collection. Representative mutants (containing transposon-tagged alleles) of each complementation group that did not correspond to a previously cloned gene were chosen for further study. Linkage between the transposon insertion and the CPY secretion phenotype was tested by tetrad analysis after sporulating diploids obtained by backcrossing with the parental strains SEY6210 and SEY6211. Yeast genomic DNA containing the Tn3-LacZ cassette was recovered from each backcrossed allele as described previously, except that BamHI/SacI-cut pRSQ303 was used instead of YIp5 for plasmid rescue (Burns et al., 1994; Voos and Stevens, 1998). The insertion point of the transposon was determined by sequencing the recovered genomic DNA.
Table 1.
Plasmids used in this study
| Plasmid | Description | Reference |
|---|---|---|
| pLC63 | CEN-LEU2 plasmid encoding Vps53p | This study |
| pLC64 | CEN-LEU2 plasmid encoding Vps52p | This study |
| pLC65 | CEN-LEU2 plasmid encoding Vps52p-myc | This study |
| pLC66 | CEN-URA3 plasmid encoding Vps54p | This study |
| pLC68 | CEN-LEU2 plasmid encoding Vps53p-myc | This study |
| pLC72 | CEN-LEU2 plasmid encoding Vps52p-HA | This study |
| pLC75 | CEN-LEU2 plasmid encoding Vps53p-HA | This study |
| pLC103 | CEN-URA3 plasmid encoding Vps54p-myc | This study |
| pLC104 | CEN-URA3 plasmid encoding Vps54p-HA | This study |
| pLC89 | Integrating plasmid for VPS52-myc (pRS303 based) | This study |
| pLC91 | Integrating plasmid for VPS53-myc (pRS304 based) | This study |
| pLC92 | Integrating plasmid for VPS53-HA (pRS304 based) | This study |
| pLC95 | CEN-URA3 plasmid encoding Vps52p-HA | This study |
| pSN55 | CEN-URA3 plasmid encoding A-ALP | (Nothwehr et al., 1993) |
| pSN92 | CEN-URA3 plasmid encoding ALP | (Nothwehr et al., 1993) |
| pNB6 | CEN-LEU2 plasmid encoding ALP | T.H. Stevens |
| pSN246 | CEN-LEU2 plasmid encoding A-ALP | (Nothwehr et al., 1996) |
| pFvM104 | CEN-URA3 plasmid encoding invertase | (Fischer von Mollard et al., 1997) |
| pLC115 | Integrating plasmid for Vps10p-HA | This study |
| pLC116 | Integrating plasmid for Och1p-HA | This study |
| pSLA2 | CEN-URA3 plasmid encoding End4p | D. Drubin |
| pSN273 | Plasmid containing pep4Δ∷LEU2 disruption cassette | (Nothwehr et al., 1996) |
| pLC28 | Plasmid for making vps53∷Tn3∶LacZ-URA3 disruption | This study |
| pLC121 | Plasmid for making vps52∷Tn3∶LacZ-URA3 disruption | This study |
| pRSQ306 | Derivative of pRS306 containing part of LEU2 | J. Horecka |
| pRSQ303 | Derivative of pRS303 containing part of LEU2 | J. Horecka |
Table 2.
Yeast strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| LCY81 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 end4Δ∷HIS3 | This study |
| LCY82 | MATa ura3-52 leu2-3,112 his3-Δ200 trp1-901 ade2-101 lys2-801 suc2-Δ9 end4Δ∷TRP1 | This study |
| SNY36-9B | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ∷ADE2 | (Nothwehr et al., 1993) |
| LCY222 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ∷ADE2 vps52Δ∷kanr | This study |
| LCY221 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ∷ADE2 vps53∷tn3-URA3 | This study |
| LCY200 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ∷ADE2 vps54Δ∷TRP1 | This study |
| LCY280 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ∷ADE2 vps52Δ∷kanr vps53∷tn3-URA3 vps54Δ∷TRP1 | This study |
| LCY319 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ∷ADE2 vps52Δ∷kanr pep4Δ∷LEU2 | This study |
| LCY294 | MATα ura3-52 leu2-3,112 his4-519 ade6 gal2 pep4-3 VPS10-HA | This study |
| LCY315 | MATα ura3-52 leu2-3,112 his4-519 ade6 gal2 pep4-3 VPS10-HA vps52Δ∷kanr | This study |
| LCY302 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ∷ADE2 och1∷URA3∷OCH1-HA | This study |
| LCY316 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ∷ADE2 och1∷URA3∷OCH1-HA vps52Δ∷kanr | This study |
| LCY226 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ∷ADE2 VPS52-HA | This study |
| LCY229 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ∷ADE2 vps53∷TRP1∷VPS53-HA | This study |
| LCY233 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ∷ADE2 VPS54-HA | This study |
| LCY251 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ∷ADE2 VPS52∷HA vps53∷tn3-URA3 | This study |
| LCY252 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ∷ADE2 VPS52-HA vps54Δ∷TRP1 | This study |
| LCY255 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ∷ADE2 vps53∷TRP1∷VPS53-HA vps52Δ∷LEU2 | This study |
| LCY270 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ∷ADE2 vps53∷TRP1∷VPS53-HA vps54Δ∷URA3 | This study |
| LCY253 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ∷ADE2 VPS54-HA vps52Δ∷LEU2 | This study |
| LCY254 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ∷ADE2 VPS54-HA vps53∷tn3-URA3 | This study |
| LCY228 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ∷ADE2 vps53∷TRP1∷VPS53-myc | This study |
| LCY234 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ∷ADE2 VPS54∶-myc | This study |
| LCY227 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ∷ADE2 VPS52-HA vps53∷TRP1∷VPS53-myc | This study |
| LCY256 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ∷ADE2 VPS54-myc vps52∷URA3∷VPS52-HA | This study |
| LCY237 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ∷ADE2 VPS54-HA vps53∷TRP1∷VPS53-myc | This study |
| LCY235 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ∷ADE2 VPS54-myc vps53∷TRP1∷VPS53-HA | This study |
| LCY243 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 vps27Δ∷LEU2 vps52∷URA3∷VPS52-HA | This study |
| LCY262 | MATα ura3-52 leu2-3,112 pho8Δ-X sec1-1 vps52∷tn3-URA3 | This study |
Plasmid and Strain Construction
Transposon-containing genomic sequences corresponding to the 801-base pair BglII fragment of VPS53 were recovered from a backcrossed vps53-181 strain that had been transformed with pRSQ306 (see above). The resulting plasmid (pLC28) was cut with BglII and used to disrupt VPS53 in SNY36-9B. Similarly, genomic sequences containing a transposon insertion were rescued from a backcrossed vps52 strain after transformation with pRSQ306 to create pLC121, which was digested with BglII and used to disrupt VPS52 in a sec1-containing strain (Piper et al., 1997), creating LCY262. Precise deletions of the VPS52, VPS54, and END4 ORFs were generated by PCR (Baudin et al., 1993). The appropriate selectable markers were amplified with the use of oligonucleotides that also contained 40–45 bases of identity to regions flanking the ORFs, and the resulting PCR products were used to transform yeast. Gene disruptions were verified by PCR analysis of genomic DNA and by complementation testing. The PEP4 gene was disrupted by transformation with the SacI–XhoI fragment of pSN273, and the disruption was confirmed by PCR analysis and Western blotting.
The full-length VPS52 gene was reconstructed from genomic DNA recovered from two different transposon-containing alleles of vps52. A 1.8-kilobase (kb) HindIII–NheI fragment derived from vps52-185 and a 2.2-kb NheI–SacII from vps52-54 were subcloned into pRS315 cut with HindIII and SacII to create pLC64. VPS53 and VPS54 were cloned from a yeast genomic library (American Type Culture Collection number 77162) by complementation of the growth and CPY secretion phenotypes of the respective mutant strains. Restriction analysis of the rescued library plasmids demonstrated that each contained the ORF identified previously as the site of the transposon insertion. pLC63 (CEN-VPS53) contains YJL029c subcloned as a 4.6-kb HindIII fragment into pRS315. pLC66 (CEN-VPS54) consists of a 3.6-kb BamHI–SalI fragment containing YDR027c cloned into pRS316 cut with BamHI and SalI. Plasmids pLC64, pLC63, and pLC66 were all found to fully complement the CPY secretion and growth phenotypes of the relevant mutant strains.
A two-step PCR procedure was used to add a single copy of the c-myc epitope flanked by BamHI sites to the C terminus of VPS52, VPS53, and VPS54 with the use of pLC64, pLC63, and pLC66 as templates. This introduced the sequence DPEQKLISEEDLLDP immediately before the stop codon of each ORF. PCR products containing the modified C-terminal portions of each gene were first subcloned into pKS+ (creating pLC50, pLC52, and pLC86, respectively). To create a full-length myc-tagged form of VPS52, the 1.1-kb BglII–SacI fragment from pLC50 was subcloned into pLC64 digested with BglII and SacI, resulting in plasmid pLC65. Similarly, pLC68 (CEN-VPS53-myc) was created by subcloning the 1.1-kb NcoI–ApaI fragment from pLC52 into pLC63 digested with NcoI–ApaI, and pLC103 (CEN-VPS54-myc) was created by replacing the 1-kb HpaI–SalI fragment of pLC66 with the HpaI–SalI fragment from pLC86.
An HA-tagged version of VPS52 (pLC72) was constructed by replacing the BamHI fragment of pLC65 encoding the c-myc tag with a 126-base pair BglII fragment that encodes three copies of the HA epitope tag. pLC95, a URA3-based plasmid for the expression of HA-tagged Vps52p, was made by subcloning the 4-kb SalI–SacI fragment from pLC72 into pRS316. The BglII fragment encoding the 3XHA tag was also used to replace the BamHI fragment of pLC52 and pLC86, resulting in plasmids pLC69 and pLC101, in which the 3XHA tag is fused to the C-terminal portion of VPS53 and VPS54, respectively. To make full-length HA-tagged versions of VPS53 and VPS54, the 1.1-kb NcoI–ApaI fragment from pLC69 was subcloned into pLC63 digested with NcoI–ApaI, creating pLC75 (CEN-Vps53p-HA), and the 1-kb HpaI–SalI fragment of pLC66 was replaced with the HpaI–SalI fragment from pLC101, creating pLC104 (CEN-Vps54p-HA).
To integrate the epitope-tagged alleles of VPS52, strain LCY196 (vps52Δ::LEU2) was cotransformed with pRS313 and the ClaI–SacII fragment from either pLC72 or pLC65, and the resulting His+ transformants were tested for loss of the LEU2 marker, which indicates that the epitope-tagged gene had integrated at the correct locus. To introduceVps52p-myc into other genetic backgrounds, the XbaI–SacI fragment from pLC65 was subcloned into pRS303, creating pLC89. pLC89 was linearized with HpaI and transformed into yeast to integrate VPS52-myc at the VPS52 locus. Integrating vectors for myc- or HA-tagged versions of Vps53p were made by subcloning the MunI–ApaI fragment from either pLC68 or pLC75 into pRS304, resulting in pLC91 and pLC92, which were linearized with NcoI before transformation into yeast cells. To integrate epitope-tagged versions of VPS54, pLC103 (CEN-VPS54-myc) and pLC104 (CEN-VPS54-HA) were cut with NotI and XhoI to release the fragment containing the tagged gene, which was transformed into a vps54Δ::URA3 strain. Transformants were grown on rich medium overnight and replica plated onto 5-FOA to select for loss of the URA3 marker. Each integrated, epitope-tagged allele was fully functional for growth and CPY sorting, as determined by pulse-chase immunoprecipitation.
To epitope tag Vps10p, a NotI fragment encoding three copies of the HA tag was subcloned into NotI-cut pAH101 (a gift of S. Nothwehr, University of Missouri, Columbia, MO), creating pLC115. pLC115 was linearized with SphI and transformed into yeast cells. Ura+ transformants were plated on 5-FOA, and those cells expressing Vps10p-HA were identified by Western blotting and tested for CPY sorting. To integrate the Och1p-HA–encoding allele, cells were transformed with EcoRI-cut pLC116, which contains the MfeI– HindIII fragment from pOH (Harris and Waters, 1996) subcloned into the EcoRI–HindIII sites of pRS306.
Immunoprecipitation of 35S-labeled Proteins
CPY and invertase immunoprecipitations were performed as described (Fischer von Mollard et al., 1997), except that all incubations were carried out at 30°C. Vps10p and HA-tagged proteins were immunoprecipitated under denaturing conditions from radiolabeled extracts with the same procedure that has been described previously for ALP (Nothwehr et al., 1995), with the appropriate polyclonal antibodies.
Native Immunoprecipitation of 35S-labeled Proteins
Two OD600 units of each strain were labeled with 20 μL of 35S-Express label for 30 min, chased for 30 min with unlabeled cysteine and methionine, and spheroplasted as described (Nothwehr et al., 1995). The labeled spheroplasts were resuspended together with 20 OD600 units of unlabeled spheroplasts prepared from the parental, untagged strain in 0.5 ml of lysis buffer (50 mM Tris, pH 8, 1% NP-40, 150 mM NaCl, 0.2 M sorbitol, 2 mM EDTA, and protease inhibitor cocktail) and incubated for 10 min at 4°C. Unlysed cells were removed by centrifugation at 13,000 × g for 10 min, and the lysates were precleared for 15 min with 50 μL of IgGSorb that was removed by centrifugation. The resulting supernatant was incubated with 5 μL of anti-HA antiserum for 1 h at 4°C before adding 50 μL of IgGSorb for another 1-h incubation. The pellet was washed twice with lysis buffer, once with lysis buffer containing 500 mM NaCl and 0.1% NP-40, and finally with lysis buffer lacking NaCl and NP-40. Samples were run on an 8% SDS-PAGE gel and visualized by autoradiography/fluorography.
Coprecipitation Experiments and Western Blotting
Cells were grown and spheroplasted as described previously (Graham et al., 1998). Frozen spheroplasts were resuspended in lysis buffer (50 mM Tris, pH 8.0, 0.5% Tween-20, 150 mM NaCl, 2 mM EDTA, 1 mM DTT, and protease inhibitor cocktail), and the protein concentration was adjusted to 1.3 mg/mL for each sample. A total of 0.5 ml of lysate was precleared with 50 μL of IgGSorb and incubated with 5 μL of rabbit anti-myc antiserum for 1 h at 4°C, after which 50 μL of IgGSorb was added and incubated for another 1 h at 4°C. The pellets were washed twice in lysis buffer and resuspended in 30 μL of sample buffer, and one-third of the sample was run on each of two SDS-PAGE gels. Immunoprecipitated proteins were detected by Western blotting with either anti-HA or anti-myc mAbs followed by HRP-labeled anti-mouse secondary antibody. Blots were developed with ECLplus (Amersham, Arlington Heights, IL), visualized by chemiluminescence, and quantified by chemifluorescence on a Storm phosphorimager (Molecular Dynamics, Sunnyvale, CA) with a wavelength of 450 nm.
Subcellular Fractionation and Sucrose Velocity Gradients
Fractionation of organelles by differential sedimentation was performed as described previously (Graham et al., 1998). For the separation of proteins on sucrose velocity gradients, 30 OD600 units of a strain containing integrated Vps52p-HA and Vps53p-myc were spheroplasted and frozen on dry ice. The frozen spheroplasts were lysed in 0.7 ml of lysis buffer (50 mM Tris, pH 7.5, 1% Triton X-100, 0.2 M sorbitol, 1 mM EDTA, 1 mM DTT, and protease inhibitors) for 10 min at 4°C and centrifuged for 10 min at 13,000 × g, and 0.6 ml of the supernatant was layered on top of a 5-ml 10–30% continuous sucrose gradient. A mix of molecular weight markers (BSA, aldolase, catalase, and ferritin in PBS) was loaded on top of a second gradient, and both were centrifuged for 4.5 h in a SW55 rotor at 280,000 × g. Thirteen fractions were collected from the top of each gradient. Fractions were analyzed by SDS-PAGE, followed by Western blotting to detect HA and myc epitopes, or stained with Coomassie blue to detect molecular weight markers.
Immunofluorescence Microscopy
Indirect immunofluorescence microscopy was performed essentially as described (Roberts et al., 1991). Cells were grown in YPD at 30°C to 1 OD600/ml before fixation. Strains containing plasmids were grown first in selective medium, resuspended in YPD, and grown for 3–4 h. Cells were fixed by adding 3% formaldehyde to the culture medium for 1 h, resuspended in 4% paraformaldehyde in 50 mM KPO4, pH 6.5, and incubated for 18 h at room temperature. Cells were spheroplasted and permeabilized with 5% SDS for 5 min for the immunolocalization of ALP or with 1% SDS for 2 min for all other antigens. Antibody incubations were carried out for 1 h at 22°C, with the exception of those involving anti-ALP mAb 1D3-A10, which were carried out at 4°C for 14–16 h.
RESULTS
Identification of Novel VPS Genes
To identify new genes that are involved in transport between the Golgi and the PVC, we screened for new vps mutants with the use of a transposon-based mutagenic procedure that facilitates cloning. To focus our efforts on a subset of mutants that are potentially involved in sorting at the Golgi, mutants were also tested for synthetic lethality with end4, which is a property of a subset of vps mutants that includes vps1, one of the few VPS genes known to act at the TGN (Nothwehr et al., 1995). Of 112,600 colonies screened, 960 were found to secrete CPY, and of those, 190 failed to grow on minimal plates containing 5-FOA. Mutants were divided into complementation groups and tested against representatives of the existing vps mutant collection (Table 3). For those mutants that did not correspond to previously identified VPS genes, the genomic sequence adjacent to the transposon insertion site was isolated from representative alleles and sequenced (see MATERIALS AND METHODS)
Table 3.
Complementation groups found in the end4Δ synthetic lethal screen
| Group | No. of alleles
|
Class | ||
|---|---|---|---|---|
| UV | Tn3 | Total | ||
| vps35 | 3 | 1 | 4 | A |
| vps38 | 0 | 5 | 5 | A |
| pmr1 | 1 | 1 | 2 | A |
| vps51a | 0 | 1 | 1 | B |
| vps52a | 3 | 3 | 6 | B |
| vps53a | 2 | 1 | 3 | B |
| vps54a | 2 | 1 | 3 | B |
| vps11 | 5 | 1 | 6 | C |
| vps16 | 1 | 3 | 4 | C |
| vps18 | 3 | 1 | 4 | C |
| vps33 | 4 | 0 | 4 | C |
| vps3 | 1 | 3 | 4 | D |
| vps6 | 0 | 2 | 2 | D |
| vps15 | 3 | 2 | 5 | D |
| vps19 | 2 | 2 | 4 | D |
| vps21 | 0 | 2 | 2 | D |
| vps34 | 4 | 1 | 5 | D |
| vps45 | 5 | 0 | 5 | D |
| vps1 | 8 | 1 | 9 | F |
| vps26 | 0 | 91 | 91 | F |
a New vps complementation groups identified in this study.
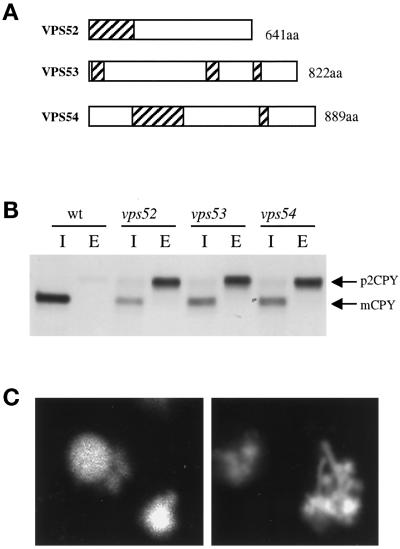
A number of complementation groups were isolated that correspond to previously identified VPS genes. The collection of vps mutants has been divided into classes according to mutant vacuolar morphology (Banta et al., 1988; Raymond et al., 1992). The synthetic lethal screen isolated a subset of these classes, including all previously identified members of classes C, D, and F as well as selected class A vps mutants. Complementation testing of a number of vps mutants that were able to grow on 5-FOA–containing medium identified known class B (vps5, vps17, vps39, vps41) and class E (vps37, vps2) mutants, indicating that synthetic lethality with end4 is a property of selected groups of vps mutants. In addition, four novel vps complementation groups were identified that had not been isolated in any of the previous vps, pep, vam, or vac genetic screens (Jones, 1977; Bankaitis et al., 1986; Rothman and Stevens, 1986; Weisman et al., 1990; Wada et al., 1992). Three of these four new vps mutants, which were named vps51, vps52, vps53, and vps54, exhibited strikingly similar phenotypes that are distinct from those of previously characterized vps mutants. vps52, vps53, and vps54 mutants are indistinguishable in terms of growth, CPY secretion, and vacuolar morphology. These mutants grow more slowly than wild-type cells at 30°C and grow very slowly at 37°C, secrete significant amounts of newly synthesized CPY, and have fragmented vacuoles that appeared as clusters of interconnecting tubules when visualized with the lumenal vacuolar dye CDCFDA (Figure 1C). The distinctive phenotype of these three mutants suggests that they share a common function that is unique among known vps mutants. Therefore, VPS52, VPS53, and VPS54 were chosen for further study. The characterization of VPS51 will be presented elsewhere (our unpublished data).
Figure 1.
(A) Scheme of the VPS52, VPS53, and VPS54 predicted ORFs. Regions with p > 0.5 of having coiled-coil structures as predicted by the COILS server are shaded. (B) CPY secretion in vps52, vps53, and vps54 mutants. The indicated strains were pulse labeled with [35S]methionine for 10 min at 30°C and chased for another 60 min in the presence of 50 μg/ml each of cysteine and methionine. CPY was immunoprecipitated from intracellular (I) and extracellular (E) fractions, resolved by SDS-PAGE, and visualized by fluorography. (C) vps52, vps53, and vps54 mutants have an unusual vacuolar morphology. Log-phase cultures were incubated with CDCFDA for 45 min at 25°C. Representative cells from wild-type (left) or vps53 (right) strains are shown. The aberrant tubulovesicular vacuolar morphology was exhibited by the vast majority of vps52, vps53, and vps54 mutant cells.
Cloning of VPS52, VPS53, and VPS54
Regions of genomic DNA flanking the transposon insertion were recovered from representative alleles and sequenced. Comparison with the Saccharomyces cerevisiae genome database revealed that the transposon insertions in mutant alleles of VPS52, VPS53, and VPS54 were present in the predicted ORFs YDR484w, YJL029c, and YDR027c, respectively. Each of these genes was also cloned by complementation of the growth and CPY secretion phenotypes, and in each case, the ORFs that contained the complementing activity were found to correspond to the ORFs containing the transposon insertion.
Each ORF is predicted to encode a hydrophilic protein with no regions of homology that are suggestive of function, although each does contain predicted regions of coiled coil (Figure 1A). YDR484w corresponds to SAC2, which was isolated as a mutant suppressor of act1-1 (Novick et al., 1989; Kolling et al., 1994). However, suppression of act1-1 by sac2 is seen only in certain strain backgrounds and requires the presence of an additional mutation. Staining with rhodamine–phalloidin revealed that actin assembly and localization was unaffected in vps52 null mutants (our unpublished results), which is consistent with the reported phenotype of sac2 mutants grown at 30°C (Novick et al., 1989). YDR027c (VPS54) has been identified in a synthetic lethal screen with rbl2 and named LUV1, but in this case too, the synthetic lethality was found to require the presence of an additional mutation (Smith et al., 1998). For the sake of simplicity, the three genes will be referred to by their VPS names throughout this report.
Although none of the three genes has a homologue in S. cerevisiae, each is evolutionarily conserved. Homologues of VPS52/SAC2 exist in mouse, human, and worm (Walter and Gunther, 1998; accession numbers AF100956.1, AL031228.1, and U29378.1, respectively), whereas VPS53 has homologues in Schizosaccharomyces pombe (accession number P87129) and worm (accession number Z27079.1), and sequences from S. pombe (accession number Z99165.1) exhibit significant similarity to VPS54. In each case, similarity extends throughout the length of the proteins, indicating that homologous proteins in higher eukaryotes may perform similar functions.
The transposon insertion in the vps53-181 allele, which truncates the protein after only seven residues, is likely to create a null mutation. However, for alleles of both vps52 and vps54, transposon insertions occur midway through the genes; therefore, null alleles of vps52 and vps54 were created in which the ORFs were precisely deleted. The resultant strains were phenotypically indistinguishable from the transposon mutants, and the null strains were used in the subsequent phenotypic analysis.
Late Golgi Membrane Proteins Are Destabilized in vps52, vps53, and vps54 Mutants
To obtain a more quantitative estimate of the CPY-sorting defect, the amount of newly synthesized CPY secreted into the medium was compared with that remaining inside the cells after radiolabeling and immunoprecipitation from both the intracellular and extracellular fractions (Figure 1B). vps52, vps53, and vps54 strains each secreted ∼70% of CPY into the medium as the p2, Golgi-modified form, whereas the wild-type strain secreted <5%. The remainder was retained intracellularly and cleaved to its mature form, indicating that it had reached a compartment containing active vacuolar proteases. In more detailed pulse-chase experiments, the fraction of CPY that was retained within the cell became processed to its mature form at a rate that was indistinguishable from that of wild-type cells (our unpublished results).
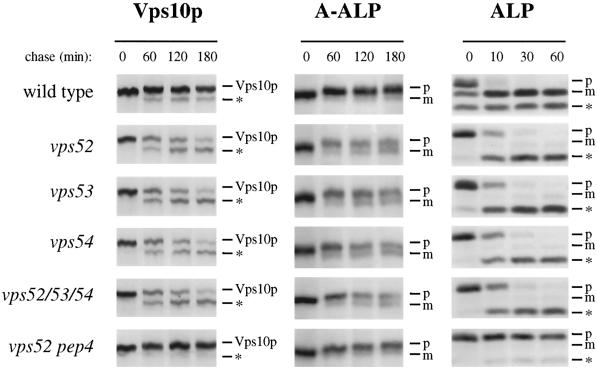
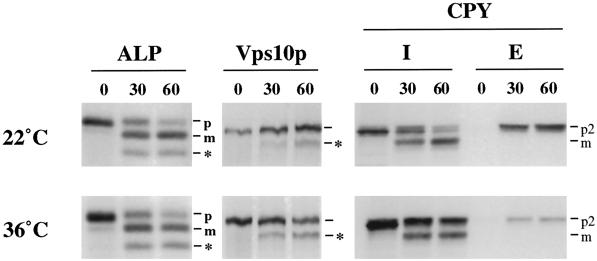
Vps10p, the receptor for CPY, is a membrane protein that cycles between the late Golgi and the endosome to perform multiple rounds of CPY sorting. Defects in the recycling of Vps10p lead to its degradation in the vacuole and result in the secretion of the Golgi-modified precursor form of CPY (p2CPY) from the cell. To determine if the CPY-sorting defects were due to loss of the sorting receptor, the stability of Vps10p was assessed by pulse-chase radiolabeling followed by immunoprecipitation (Figure 2, left panels). In vps52, vps53, and vps54 mutants, Vps10p was noticeably destabilized, with a half-life of ∼120 min. The breakdown product that appeared is characteristic of cleavage in the vacuole by PEP4-dependent proteases, and indeed, Vps10p was stabilized in vps52 mutants that also contained a pep4 mutation, suggesting that this degradation occurred in the vacuole. Furthermore, Vps10p was cleaved at the same rate in a strain that contained mutations in all three genes, suggesting that all three genes act at a common step in Vps10p recycling.
Figure 2.
Golgi membrane proteins undergo cleavage by vacuolar proteases in vps52, vps53, and vps54 mutants. (Left panels) Vacuolar cleavage of the CPY receptor Vps10p. Wild-type (SNY36-9B), vps52Δ::kanr (LCY222), vps53::Tn3URA (LCY221), vps54Δ::TRP1 (LCY200), vps52Δ::kanr vps53::Tn3URA vps54Δ::TRP1 (LCY280), and vps52Δ::kanr pep4Δ::LEU2 (LCY319) strains were radiolabeled for 10 min with [35S]methionine. After chasing for the indicated times, aliquots of cells were removed and subjected to immunoprecipitation with antibodies to Vps10p. Samples were analyzed by SDS-PAGE and fluorography. The positions of full-length Vps10p and its PEP4-dependent cleavage product (*) are indicated. (Center panels) Processing of the late Golgi marker protein A-ALP. Strains containing a centromere-based plasmid encoding A-ALP (pSN55 for SNY36-9B, LCY280, LCY200, and LCY319; pSN246 for LCY221 and LCY280) were subjected to pulse-chase immunoprecipitation and analyzed as described above except that anti-ALP antibodies were added to immunoprecipitate A-ALP from the cell extracts. ProA-ALP (p) is converted to the faster-migrating mature form (m) by PEP4-dependent processing in the vacuole. (Right panels) Maturation of the vacuolar membrane protein ALP. Strains harboring plasmids for the expression of ALP (pSN92 for SNY36-9B, LCY280, LCY200, and LCY319; pNB6 for LCY221 and LCY280) were pulse chased for the indicated times, immunoprecipitated with anti-ALP antibodies, and analyzed as described above. PEP4-dependent cleavage of proALP (p) results in the formation of the mature form (m) as well as an additional commonly observed degradation product (*).
The localization of a number of resident late Golgi proteins also depends on recycling from the prevacuolar compartment, and a failure to recycle results in transport to the vacuole, where they are degraded (Wilcox et al., 1992; Nothwehr et al., 1993; Cooper and Stevens, 1996). A-ALP is a model late Golgi protein that consists of the membrane and lumenal domains of ALP fused to the cytoplasmic domain of DPAP A, which contains the sorting information required for localization of the chimera to the late Golgi (Nothwehr et al., 1993). The fate of newly synthesized A-ALP was followed by pulse-chase immunoprecipitation. In wild-type cells, A-ALP was very stable, and very little of the A-ALP underwent the PEP4-dependent cleavage to the mature form during the 180-min chase (Figure 2, center panels). In contrast, in each of the mutant strains, more than half of the A-ALP was processed to the mature form at the end of the chase period. The PEP4-dependent cleavage of A-ALP suggests that Golgi membrane proteins were mislocalized to the vacuole in vps52, vps53, and vps54 cells. Indeed, the late Golgi protein Kex2p, an endopeptidase that cleaves proalpha factor to its mature form, was also destabilized in each of the mutant strains; thus, these strains secreted proalpha factor (our unpublished results).
There are multiple vesicular transport pathways that lead from the Golgi to the vacuole (Conibear and Stevens, 1998). Vps10p, A-ALP, and Kex2p all follow the CPY pathway. In contrast, the vacuolar membrane protein ALP is sorted into a distinct class of vesicles at the Golgi and follows an alternative route to the vacuole that bypasses the PVC. In wild-type cells, newly synthesized ALP is delivered to the vacuole and proteolytically processed to its mature form with a half-time of 5 min. In each of the mutant strains, ALP processing was delayed only slightly (Figure 2, right panels). The alternative pathway to the vacuole does not seem to be significantly perturbed in the mutants, because >90% of ALP was processed after the 60-min chase. A similar delay was seen for vps45 mutants (our unpublished results), which specifically block transport along the CPY pathway; this may be an indirect effect of blocking traffic along the CPY pathway, or alternatively, it may be due to altered levels of proteases in the vacuole.
The degradation of Golgi membrane proteins by vacuolar proteases is characteristic of mutants that are defective in some part of the cycling pathway between the yeast late Golgi and the PVC (Nothwehr and Hindes, 1997; Seaman et al., 1997). Because the phenotypes of each of the single-mutant strains are remarkably similar, it is possible that Vps52p, Vps53p, and Vps54p perform a similar function. The rates at which Vps10p, A-ALP, and ALP are transported to the vacuole are no different in a strain containing all three mutations (vps52, vps53, and vps54), strongly suggesting that VPS52, VPS53, and VPS54 act together at a common transport step (Figure 2).
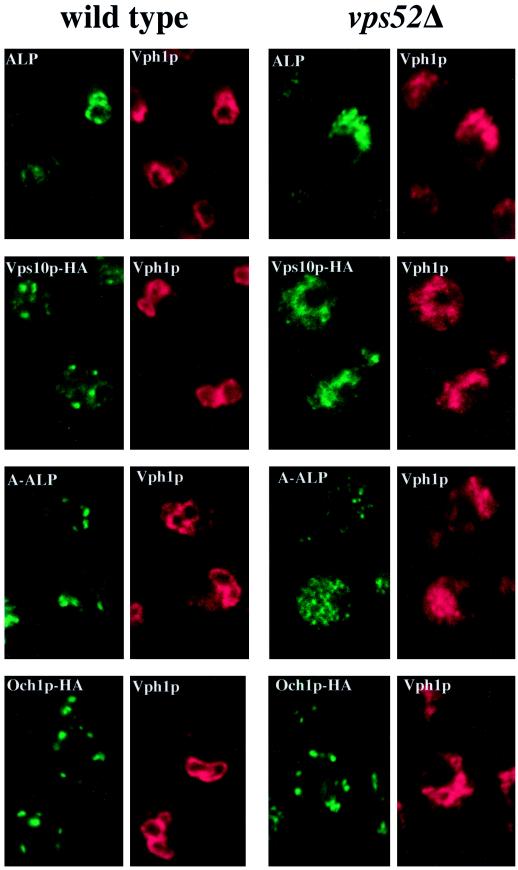
Membrane Protein Localization in vps52 Mutants
To confirm the results of the pulse-chase studies, the steady-state localization of a variety of marker proteins was examined by indirect immunofluorescence microscopy. Because vps52, vps53, and vps54 mutants were phenotypically indistinguishable, we limited our analysis to vps52 and vps54 mutants, which gave identical results. Only the data for vps52 mutants are shown in Figure 3. Vph1p is an integral membrane subunit of the vacuolar H+-ATPase, which is transported to the vacuole along the CPY pathway, where it is responsible for vacuolar acidification (Stevens and Forgac, 1997). In wild-type cells, Vph1p colocalized with the vacuolar membrane protein ALP in ring-like structures characteristic of vacuolar membranes (Figure 3A). In vps52Δ cells, ALP and Vph1p also colocalized to the same intracellular structures, which did not form rings but instead were seen as irregularly shaped clusters of vesicles and tubules similar in appearance to the intracellular structures that stain with CDCFDA (Figure 1C) and that correspond to the fragmented vacuole. This suggests that there are no obstacles to the transport of membrane proteins from the Golgi to the vacuole by way of either the CPY pathway or the alternative ALP pathway.
Figure 3.
Localization of membrane proteins by double-label immunofluorescence in wild-type and vps52Δ cells. (A) The vacuolar membrane protein ALP (green) colocalizes with the vacuolar ATPase subunit Vph1p (red) in wild-type (SNY36-9B) and vps52Δ (LCY222) cells containing centromere plasmid pSN92 (ALP). (B) Vps10p-HA (green) is localized to dots that are distinct from the vacuoles that label with Vph1p (red) in wild-type cells (LCY294), whereas both markers label the fragmented vacuoles of vps52Δ cells (LCY315). (C) A-ALP (green) is localized to dots that are distinct from the vacuoles that label with Vph1p (red) in wild-type cells (SNY36-9B containing pSN55), whereas in vps52Δ cells, A-ALP is found in smaller punctate structures as well as in the vacuoles of vps52Δ cells (LCY222 containing pSN55). (D) The cis-Golgi marker protein Och1p-HA (green) exhibits a similar punctate distribution in both wild-type (LCY302) and vps52Δ (LCY316) strains that is distinct from the vacuolar staining of Vph1p (red). Strains were fixed, spheroplasted, and double labeled with rabbit anti-Vph1p antiserum and a mouse mAb to either ALP (A and C) or HA (B and D) followed by fluorescently labeled secondary antibodies.
Vps10p, which was found in a number of discrete punctate structures in wild-type cells that correspond to the late Golgi (Figure 3B) (Cooper and Stevens, 1996), was mislocalized in vps52 mutants to the same vacuolar structures that contained Vph1p (Figure 3B). This is consistent with the results of the kinetic analysis of Vps10p turnover (Figure 2), which demonstrated that Vps10p was cleaved by vacuolar proteases in vps52 mutant cells. Together, these data indicate that Vps10p was not efficiently recycled from the PVC back to the Golgi in these mutants but instead was mislocalized to the vacuole and exposed to vacuolar proteases.
The distribution of A-ALP was also significantly altered in vps52 mutants. In wild-type cells, A-ALP, like Vps10p, localized to a number of distinct dots that are typical of Golgi proteins (Figure 3C) (Nothwehr et al., 1993). In a vps52 strain, A-ALP showed vacuolar staining that is consistent with the observed processing by vacuolar proteases. However, in many cells, A-ALP was additionally found in small, diffuse punctate structures (Figure 3C). Immunolocalization of Kex2p showed a similar staining pattern that combined vacuolar structures with small dots (our unpublished results). Although the A-ALP– and Kex2p-labeled structures could represent fragmented Golgi, such labeling is often associated with transport vesicles that collect in a number of other vps mutants (Piper et al., 1994; Holthuis et al., 1998b). The distribution of the cis-Golgi marker Och1p-HA was found to be unchanged in vps52 mutants compared with wild-type cells (Figure 3D). Therefore, vps52 mutants did not exhibit gross alterations in the overall morphology of the Golgi complex but instead were specifically defective in the localization of a number of late Golgi membrane proteins.
Invertase Is Secreted in Its Fully Glycosylated Form from vps52 Mutants
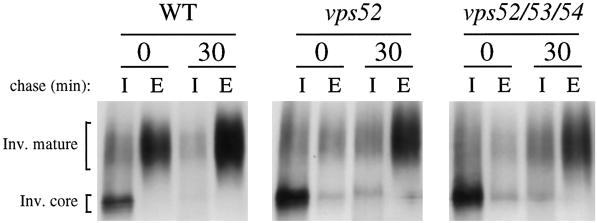
The glycosylation and transport of the secretory protein invertase was examined to assess the integrity of the Golgi compartment and early Golgi functions (Figure 4). Strains containing a plasmid-borne SUC2 gene were incubated for 45 min in low-glucose medium to derepress the synthesis of invertase before pulse labeling for 7 min and chasing for 0 and 30 min. Invertase immunoprecipitated from the intracellular fraction immediately after the pulse was present inside the cell as the core-glycosylated ER form. In wild-type cells, a significant amount of the outer-chain modified (fully glycosylated) form of invertase was secreted into the medium during the labeling period, where it appeared as a slowly migrating, diffuse band. Secretion of invertase was delayed in vps52 and vps52/53/54 cells, which showed little accumulation of invertase in the medium during the 7-min pulse. Slowed transport through the Golgi may be a consequence of the slow growth of these strains. However, most of the invertase was secreted into the medium in a fully glycosylated form after 30 min, indicating that transport through the secretory pathway was not blocked in the mutant strains. Similar results were found for vps53 and vps54 cells (our unpublished results). Therefore, despite the mislocalization of TGN membrane proteins, there is no indication of defects in the localization or function of the early and medial Golgi mannosyltransferases responsible for invertase glycosylation.
Figure 4.
Invertase is fully glycosylated and secreted from vps52Δ and triple mutant strains. Wild-type (SNY36-9B), vps52Δ::kanr (LCY222), and vps52Δ::kanr vps53::Tn3URA vps54Δ::TRP1 (LCY280) strains containing a plasmid encoding SUC2 (pFvM104) were grown in low-glucose medium for 45 min to induce invertase expression before labeling with [35S]methionine for 7 min and chasing for 30 min. Invertase was immunoprecipitated from intracellular (I) and extracellular (E) fractions and analyzed by SDS-PAGE and fluorography.
ALP and Vps10p Do Not Reach the Vacuole by Way of the Plasma Membrane in vps52 Mutants
VPS1 is required for the formation of two different types of vesicles from the TGN, which divert proteins from the secretory pathway into the ALP and CPY pathways. In vps1 mutants, membrane proteins such as ALP are missorted to the plasma membrane and rely on endocytosis to reach the vacuole (Nothwehr et al., 1995). Clathrin is required for the formation of only one class of vesicles at the TGN, and clathrin temperature-sensitive mutants missort TGN proteins (but not ALP) to the cell surface (Seeger and Payne, 1992).
To determine if membrane proteins reach the vacuole in vps52 mutants by way of the plasma membrane, vacuolar cleavage of Vps10p and ALP was examined in a sec1-ts vps52 double mutant strain (Figure 5). In cells grown at 22°C, p2CPY was secreted into the medium, whereas a portion of the CPY was retained inside the cell and processed to the mature form, as described previously for vps52 mutants. Inactivation of SEC1 after a shift to the nonpermissive temperature of 36°C resulted in the intracellular accumulation of p2CPY due to a block in the fusion of secretory vesicles with the plasma membrane. However, this late block did not affect maturation of the 30% of newly synthesized CPY that was correctly sorted in vps52 mutants. Moreover, vacuolar processing of ALP and Vps10p was not blocked at the nonpermissive temperature, indicating that protein traffic is not rerouted to the plasma membrane in vps52 mutants.
Figure 5.
Vacuolar transport of ALP and Vps10p in vps52 mutants is independent of SEC1. A vps52 s1-1 double mutant strain (LCY262 containing pNB6) was preincubated for 30 min, labeled for 10 min, and chased for the indicated times at 22°C (top panels) or 36°C (bottom panels). ALP, Vps10p, and CPY were immunoprecipitated and analyzed as described above. I, intracellular; E, extracellular.
vps52, vps53, and vps54 mutants were also tested for the missorting of ALP to the cell surface by following the kinetics of ALP maturation in end4-ts vps52 (or vps53 or vps54) double mutant cells. Because ALP was transported to the vacuole in all three mutant strains independent of END4 function (our unpublished results), VPS52, VPS53, and VPS54 are unlikely to participate in the formation of vesicles at the TGN that sort membrane proteins into the CPY and ALP pathways.
Vps52p, Vps53p, and Vps54p Depend on Each Other for Stability
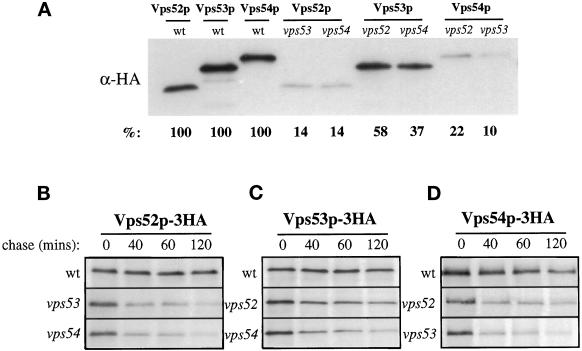
The similar phenotypes of vps52, vps53, and vps54 single and triple mutants suggest that the functions of these three genes are related. Because some of these genes may regulate the synthesis or stability of some of the others, we investigated the relative levels of each gene product in different mutant strains. Three copies of the influenza HA tag were introduced at the C terminus of the VPS52, VPS53, and VPS54 ORFs, and the resulting constructs were integrated into the genome. In each case, the tagged protein was found to be fully functional, as assessed by CPY sorting and growth. Analysis by SDS-PAGE followed by Western blotting identified, for each protein, a major band that migrated at the predicted molecular weight (Figure 6A). Interestingly, the steady-state level of each of the tagged proteins was dependent on the presence of the other two proteins. For example, in vps53 and vps54 mutant strains, Vps52p-HA was present at only 14% of the level found in wild-type cells. Decreases in the levels of Vps53p-HA and Vps54p-HA were also seen in the corresponding vps52, vps53, and vps54 mutant backgrounds.
Figure 6.
Vps52p, Vps53p, and Vps54p are stable only when coexpressed. (A) Steady-state levels of integrated, HA-tagged versions of Vps52p, Vps53p, and Vps54p in mutant strains. Detergent extracts were made from each of the indicated strains (lane 1, LCY226; lane 2, LCY229; lane 3, LCY233; lane 4, LCY251; lane 5, LCY252; lane 6, LCY255; lane 7, LCY270; lane 8, LCY253; lane 9, LCY254), adjusted to give equal protein concentrations as determined by Bradford assay, and analyzed by Western blotting with an anti-HA mAb. Blots were visualized by chemiluminescence and quantified by chemifluorescence (see MATERIALS AND METHODS). The percentage of each HA-tagged protein remaining in the delete strains, normalized to wild-type values, is shown. (B–D) Pulse-chase analysis of protein stability in delete strains. The strains described above were labeled with [35S]methionine for 10 min and chased for the times indicated before immunoprecipitating with HA antibodies. Samples were analyzed by SDS-PAGE and fluorography.
Pulse-chase analysis was performed to determine whether the decreased steady-state levels were the result of a decreased rate of synthesis or an increase in turnover rate (Figure 6, B–D). Mutant strains were labeled for 10 min and chased for varying lengths of time. Vps52p-HA, Vps53p-HA, and Vps54p-HA were all quite stable in wild-type cells during the 2-h chase but were destabilized in each of the mutant strains. The extent of the destabilization seen by pulse-chase immunoprecipitation was found to correlate with the steady-state levels seen in Western blotting. Therefore, it seems that each of these three proteins was unstable and rapidly turned over in the absence of the other two.
Vps52p, Vps53p, and Vps54p Form a 1:1:1 Complex
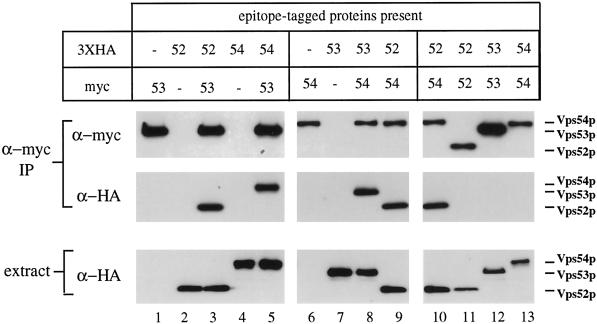
The observation that Vps52p, Vps53p, and Vps54p each was subject to rapid degradation in the absence of the other two suggested that physical associations between the three proteins may contribute to their stability in wild-type cells. Immunoprecipitation experiments were carried out under nondenaturing conditions to determine if the proteins form a complex. Triple HA- or myc-tagged forms of each protein were created and integrated in different pairwise combinations. CPY sorting and growth were unimpaired in strains containing one or more epitope-tagged proteins, indicating that the modified forms are fully functional. Strains containing one or more epitope-tagged proteins were subjected to immunoprecipitation under nondenaturing conditions with an affinity-purified anti-myc antiserum and analyzed by Western blotting for copurifying proteins (Figure 7). When anti-myc antiserum was used to immunoisolate Vps53p-myc from a strain that coexpressed Vps52p-HA, >90% of the Vps52p-HA present in the extract was detected in the precipitate, indicating that these proteins do indeed exist in a complex (Figure 7, lane 3). No detectable Vps52p-HA was present in anti-myc precipitates from a strain that did not also contain myc-tagged Vps53p. Immunoprecipitation of Vps53p-myc also led to the copurification of >90% of Vps54p-HA (Figure 7, lane 5), suggesting that the intracellular pools of all three proteins are present in a single complex. Consistent with this interpretation, Vps53p-HA and Vps52p-HA were coprecipitated with equal efficiency from cells that also expressed Vps54p-myc (Figure 7, lanes 8 and 9).
Figure 7.
Vps52p, Vps53p, and Vps54p coprecipitate as a 1:1:1 complex. Strains containing one or more epitope-tagged proteins (as indicated) were subjected to immunoprecipitation under nondenaturing conditions with affinity-purified rabbit anti-myc antibodies. Copurifying proteins were subsequently detected by Western blotting with anti-myc (top panels) or anti-HA (center panels) mouse mAbs and species-specific secondary antibodies. Equal amounts of total cell lysates, as determined by Bradford assay, were also analyzed by Western blotting to determine the relative amounts of each HA-tagged protein in the extracts (bottom panels). Blots were visualized by chemiluminescence and quantified by chemifluorescence (see MATERIALS AND METHODS). All samples were processed in parallel; lanes 9 and 10 are identical. Apparent differences in the levels of Vps52p-myc and Vps54p-myc relative to Vps53p-myc are due to context dependence of the anti-myc mAb 9E10; all three proteins are recognized equally well by the rabbit anti-myc serum (our unpublished results). The following strains were used: lane 1, LCY228; lane 2, LCY226; lane 3, LCY227; lane 4, LCY233; lane 5, LCY237; lane 6, LCY234; lane 7, LCY229; lane 8, LCY235; lane 9, LCY256; lane 10, LCY256; lane 11, LCY226 + pLC65; lane 12, LCY228 + pLC75; lane 13, LCY234 + pLC104.
Immunoisolation of a myc-tagged protein resulted in the nearly complete depletion of the coexpressed HA-tagged protein from the cell lysates (>90%). In addition, no coprecipitation was observed when Vps53p-myc and Vps52p-HA were expressed in separate strains and cell lysates were mixed immediately before immunoprecipitation, indicating that complexes do not separate and reform under the experimental conditions used (our unpublished results). Therefore, Vps52p, Vps53p, and Vps54p appear to be subunits of a stable complex.
To determine if any of these components are present in the complex in more than one copy, native immunoprecipitation experiments were performed with strains in which differently tagged forms of the same protein were coexpressed (Figure 7, lanes 10–13). If each complex contained two copies of Vps52p, then in a strain expressing both Vps52p-myc and Vps52p-HA, approximately half of the complexes should contain both Vps52p-myc and Vps52p-HA, and precipitation of Vps52p-myc should result in the coprecipitation of 50% of the Vps52p-HA. However, in cells expressing equivalent levels of both Vps52p-HA and Vps52p-myc, HA-tagged Vps52p was not detected in Vps52p-myc immunoprecipitates at levels above background, even after very long exposures. The same result was obtained for Vps53p and Vps54p (Figure 7, lanes 10–13), indicating that each of these proteins is present in the complex in a single copy.
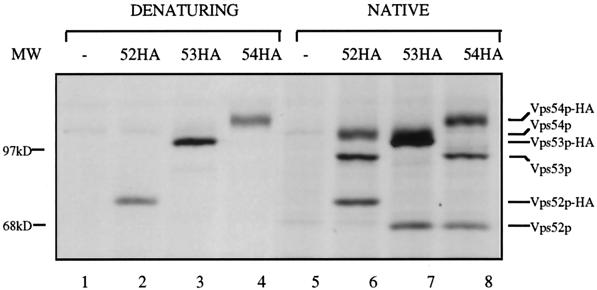
The Vps52/53/54 complex may contain additional components that were not identified in our screen or that, when mutated, result in different phenotypes. To look for additional copurifying proteins, we metabolically labeled cells expressing Vps52p-HA, Vps53p-HA, or Vps54p-HA. When anti-HA immunoprecipitations were carried out on lysates prepared from radiolabeled strains under denaturing conditions, a prominent band of the expected molecular weight was seen in each of the tagged strains that was not present in the untagged strains (Figure 8, lanes 1–4). When extracts from cells expressing Vps52p-HA were prepared under nondenaturing conditions, two other prominent bands were seen in addition to Vps52p-HA. Because the addition of the 3XHA epitope tag adds ∼5 kDa to the molecular mass of each protein, the tagged proteins would be expected to migrate more slowly than the untagged forms. Indeed, the two proteins that copurified with Vps52p-HA migrated as expected for the untagged forms of Vps53p and Vps54p. An analysis of the copurifying bands from Vps53p-HA and Vps54p-HA strains confirmed that Vps52p, Vps53p, and Vps54p could be immunoprecipitated from labeled extracts under native conditions. These data indicate that Vps52p, Vps53p, and Vps54p form a discrete complex that may contain only these three proteins.
Figure 8.
Vps52p, Vps53p, and Vps54p are major components of the immunopurified Vps52/53/54 complex. Radiolabeled strains expressing integrated, HA-tagged forms of Vps52p (LCY226; lanes 2 and 6), Vps53p (LCY229; lanes 3 and 7), Vps54p (LCY233; lanes 4 and 8), and the untagged parent strain (SNY36-9B; lanes 1 and 5) were spheroplasted and lysed under denaturing (lanes 1–4) or nondenaturing (lanes 5–8) conditions, and the extracts were subjected to immunoprecipitation with anti-HA antiserum. Four times as many cells were used for the native immunoprecipitation (see MATERIALS AND METHODS). The positions of wild-type and tagged forms of Vps52p, Vps53p, and Vps54p are indicated on the right. The presence of the triple-HA epitope tag is predicted to add an additional 5 kDa to the molecular mass of each protein.
No other copurifying bands apart from the three known subunits of the complex could be identified in these experiments. Additional components bound to the complex by low-affinity interactions may have been released during the immunoprecipitation procedure. In addition, nonspecific binding of lower-molecular-weight proteins may have obscured copurifying proteins in the low-molecular-weight range, which would be expected to contain fewer cysteine/methionine residues and therefore radiolabel less intensely.
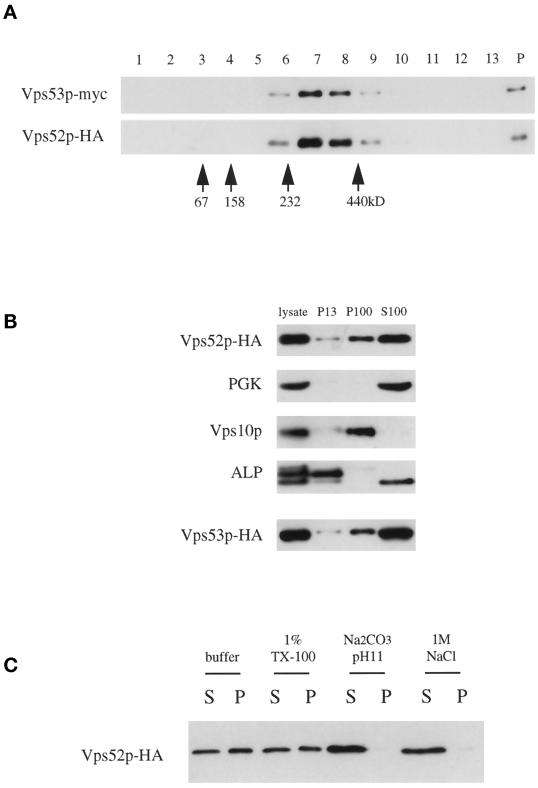
Vps53p-myc and Vps52p-HA present in detergent extracts were found to cosediment as a single peak on sucrose velocity gradients as a complex of ∼300 kDa (Figure 9A). This value is close to the sum of the predicted molecular masses of Vps52p-HA, Vps53p-myc, and Vps54p (278 kDa), which suggests that there are not a large number of additional, unidentified subunits in the complex. These findings are consistent with the data from coimmunoprecipitation (Figure 7) and stability (Figure 6) experiments, which suggest that the proteins do not exist in stable monomeric pools but instead are associated in a single complex at steady state.
Figure 9.
(A) Vps52p and Vps53p cosediment on sucrose velocity gradients. The low-speed supernatant from LCY227 cells lysed in buffer containing 1% Triton X-100 was loaded at the top of a continuous 10–30% sucrose gradient and spun for 4.5 h, and a mixture of molecular mass markers was run in parallel on a second gradient. Three percent of each fraction, collected from the top of the gradient (lanes 1–13), as well as 10% of the material that sedimented to the bottom of the tube (lane P), was analyzed by Western blotting for myc and HA epitopes. The positions of the molecular mass markers (determined by Coomassie staining of SDS-PAGE gels) are indicated. (B) Association of Vps52p-HA and Vps53p-HA with the high-speed pellet. Subcellular fractions were prepared from a vps52Δ strain containing pLC72 (Vps52p-HA) by differential sedimentation as described in MATERIALS AND METHODS, separated by SDS-PAGE, and analyzed by Western blotting for HA, phosphoglycerate kinase (cytosolic marker protein), Vps10p (Golgi marker), and ALP (vacuolar marker; the faster-migrating band present in the S100 is a soluble degradation product). Subcellular fractions were prepared in parallel from a vps53::Tn3 strain containing pLC75 (Vps53p-HA) and analyzed by blotting for the same markers; only the anti-HA blot is shown. (C) Vps52p is peripherally associated with P100 membranes. S13 supernatant fractions from LCY226 were treated with 1% Triton X-100 (TX-100), 0.1 M Na2CO3, pH 11, or 1 M NaCl for 10 min at 4°C before centrifugation at 100,000 × g for 45 min. Equal amounts of the supernatant (S) and pellet (P) were analyzed by Western blotting with anti-HA antibodies.
The Vps52/53/54 Complex Is Peripherally Associated with the Late Golgi Compartment
Subcellular fractionation by differential sedimentation was performed to determine the subcellular distribution of the complex (Figure 9B). Although the majority of the complex was in a cytoplasmic pool (S100), a somewhat variable fraction (30–50%) sedimented with the high-speed membrane pellet (P100) that contains the Golgi, endosomes, vesicles, and other membranes (Marcusson et al., 1994; Becherer et al., 1996). However, the complex is not an integral component of these membranes, because treatment with 0.1 M Na2CO3, pH 11, or 1 M NaCl released the complex into the supernatant (Figure 9C). The Vps52p-HA present in the high-speed pellet was insoluble in 1% Triton X-100, indicating that at least a fraction of the complex is involved in more extensive protein–protein interactions. These interactions may be disrupted under certain experimental conditions, which may explain the variability in the amount of P100-associated Vps52p-HA as well as the presence of a single peak in sucrose velocity gradients.
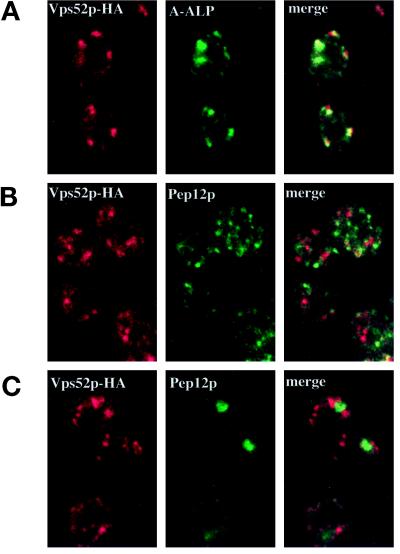
By immunofluorescence microscopy, Vps52p-HA localized to a number of distinct dots (Figure 10). Vps53p-HA and Vps54p-HA exhibited a similar staining pattern (our unpublished results), which is typical of many Golgi membrane proteins (Redding et al., 1991; Nothwehr et al., 1993; Cooper and Stevens, 1996). In S. cerevisiae, the Golgi complex is not organized into stacked cisternae, and components of early and late Golgi compartments can readily be distinguished by immunofluorescence microscopy (Harris and Waters, 1996; Holthuis et al., 1998a). Most of the structures that contained Vps52p-HA were also found to label with the late Golgi marker A-ALP, although there were some differences in the relative intensities of the structures labeled by each antibody (Figure 10A). In contrast, antibodies to the prevacuolar marker Pep12p labeled much smaller and more numerous structures distributed throughout the cytoplasm (Figure 10B). In merged images, the staining patterns of Vps52p-HA and Pep12p did not overlap, whereas Vps52p-HA showed extensive colocalization with A-ALP; therefore, the complex is likely to be associated with the TGN and not with the PVC.
Figure 10.
Vps52p-HA colocalizes with A-ALP at the late Golgi by indirect immunofluorescence microscopy. (A) Vps52p-HA–expressing cells (LCY226) containing pSN55 (A-ALP) were fixed, spheroplasted, and double labeled with an anti-HA polyclonal antiserum and an anti-ALP mAb. Pairs of images corresponding to each antigen were collected with the use of confocal microscopy, and the images were merged to show coincidence of the two staining patterns. A majority of the structures that labeled with the HA antibody also contained A-ALP, although the relative intensities of the two staining patterns varied. (B) LCY226 (Vps52p-HA) cells and (C) LCY243 (Vps52p-HA vps27Δ) cells double labeled with an anti-HA polyclonal antiserum and a mAb to Pep12p.
In class E vps mutants such as vps27, membrane traffic from the prevacuolar compartment is blocked, leading to the accumulation of an aberrant class E structure, which contains prevacuolar markers such as Pep12p as well as late Golgi proteins such as A-ALP and Vps10p, which continuously recycle through the prevacuole (Cereghino et al., 1995; Piper et al., 1995; Voos and Stevens, 1998). A number of peripheral membrane proteins involved in vacuolar protein sorting that are localized diffusely in wild-type cells (e.g., Mvp1p, Grd19p, Vps5p) are more readily visualized in vps27 mutants, where they are associated with the class E compartment (Ekena and Stevens, 1995; Nothwehr and Hindes, 1997; Voos and Stevens, 1998). However, the distribution of Vps52p-HA did not change in vps27 mutants, whereas Pep12p accumulated in one or two large, brightly staining structures typical of the class E compartment (Figure 10C). Therefore, in contrast to many of the proteins required for the recycling of TGN proteins from the PVC, the Vps52/53/54 complex does not appear to be localized to the PVC but instead is peripherally associated with the late Golgi complex.
DISCUSSION
A large number of different factors might be expected to regulate the variety of vesicle formation and fusion reactions that take place at the TGN, but to date few such factors have been identified in yeast. In this paper, we describe the identification and characterization of three components of a novel TGN-localized complex—the Vps52/53/54 complex—that is required for CPY sorting as well as for the localization of resident TGN membrane proteins.
Vps52p, Vps53p, and Vps54p Form a Stable Complex
We have shown that Vps52p, Vps53p, and Vps54p are subunits of a novel complex and that these proteins coimmunoprecipitate from cell lysates and cofractionate on sucrose velocity gradients. They do not seem to exist in monomeric pools, and the observation that loss of any one of these proteins leads to the degradation of each of the other subunits suggests that they are components of a stable multimeric complex. When this complex was purified from radiolabeled extracts, the major copurifying bands were found to correspond to Vps52p, Vps53p, and Vps54p. The immunoprecipitation data indicate that each ‘of the three proteins is present in a single copy, and although the combined predicted molecular weight of these three subunits corresponds roughly to the apparent size of the complex on sucrose velocity gradients, we cannot exclude the possibility of other as-yet-unidentified components. Vps54p/Luv1p has been reported to interact in vitro with Rbl2p, a molecular chaperone that binds β-tubulin and participates in its folding and assembly (Smith et al., 1998). β-Tubulin was not enriched in Vps54p immunoprecipitates, and microtubules are not required for CPY sorting (Vater et al., 1992). Although the in vivo interaction with Rbl2p may reflect the affinity of chaperones for unfolded proteins in the cell extract, it may also indicate a more general role for Rbl2p in the assembly of diverse macromolecular complexes.
Vps52p, Vps53p, and Vps54p each contain regions of predicted coiled-coil structure, which may contribute to the protein–protein interactions that mediate complex assembly, but none of these proteins contains regions of sequence homology to any well-characterized protein or functional domain. However, the complex is clearly conserved throughout evolution, and homologues of each subunit exist in eukaryotic organisms ranging from worms to humans. Many factors involved in vesicle transport processes are members of protein families that perform related functions at different transport steps, and these have been the focus of intensive study (Ferro-Novick and Jahn, 1994; Conibear and Stevens, 1998). Recently, a number of unique multiprotein complexes have been described that play important roles at individual trafficking steps, and such complexes may contribute to the fidelity of transport events (Pfeffer, 1999).
Function of the Vps52/53/54 Complex in Golgi Sorting
The results from our phenotypic analysis of vps52, vps53, and vps54 mutants are consistent with the idea that Vps52p, Vps53p, and Vps54p function together at a common step in protein transport between the late Golgi and the prevacuolar compartment. Null alleles of each gene have identical phenotypes, and simultaneous deletion of all three genes does not present any more serious consequences for the cell than deletion of a single subunit. Loss of the complex results in the secretion of ∼70% of the newly synthesized CPY. Despite this rather severe CPY-missorting phenotype, none of these genes was identified in any of the previous vps, pep, or vam screens. This may be due in part to the relatively poor growth of these mutant strains at all temperatures; a number of earlier screens relied on growth advantages conferred by the secretion of CPY or of CPY–invertase fusions on selective medium (Bankaitis et al., 1986; Rothman and Stevens, 1986).
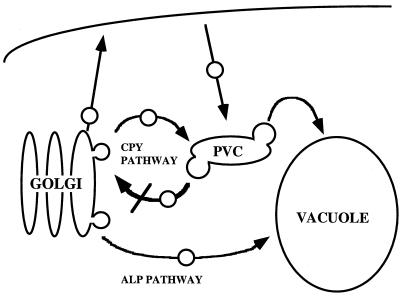
In addition to CPY missorting, all three mutant strains exhibit defects in the localization and stability of a number of late Golgi membrane proteins, which points to a defect in the recycling pathway between the Golgi and the prevacuole (Figure 11). Vps10p, A-ALP, and Kex2p are not efficiently retrieved from the PVC; instead, they are mislocalized to the vacuole, where they are cleaved by vacuolar proteases. However, early Golgi functions and endocytosis of the styryl dye FM-464 to the vacuole (our unpublished results) are relatively unaffected in the mutant strains. A number of observations indicate that transport from the TGN to the vacuole also proceeds normally in vps52, vps53, and vps54 mutants. Localization of the vacuolar H+-ATPase, which is transported from the Golgi to the vacuole by way of the PVC, is not perturbed. The vacuolar membrane protein ALP, which follows an alternative pathway to the vacuole that bypasses the PVC, also reaches the vacuole in mutant strains with normal kinetics.
Figure 11.
Model of transport pathways in vps52, vps53, and vps54 mutants. Resident late Golgi membrane proteins such as Kex2p and the marker protein A-ALP are transported along the CPY pathway and maintain their Golgi distribution by recycling from the PVC. Loss of retrograde transport from the PVC back to the late Golgi in vps52, vps53, or vps54 mutants leads to the slow delivery of TGN proteins, including the CPY receptor Vps10p, to the vacuole. This results in the vacuolar degradation of Golgi proteins and the secretion of CPY.
In vps1 mutants, which are defective in vesicle formation at the TGN, newly synthesized vacuolar membrane proteins such as ALP are transported to the cell surface and rely on endocytosis to reach the vacuole (Nothwehr et al., 1995). The rerouting of membrane proteins to the plasma membrane has been postulated to explain the synthetic lethality of vps1 and end4Δ. However, defects in a number of other cellular functions are also lethal in combination with end4 mutations, including defects in endocytosis, actin assembly, and vacuolar acidification (Holtzman et al., 1993; Munn and Riezman, 1994), and vps mutants such as vps45 do not missort ALP to the plasma membrane (Piper et al., 1997) yet were also isolated in the end4Δ synthetic lethal screen. vps52 mutants do not missort ALP or Vps10p into the secretory pathway, because these membrane proteins reach the vacuole independent of SEC1. Therefore, it is unlikely that the Vps52/53/54 complex is involved in vesicle formation at the TGN. Instead, the transport defects in vps52/53/54 mutant strains appear to be limited to the retrograde pathway that recycles Golgi membrane proteins from the PVC back to the TGN.
A number of Vps proteins have been characterized that are also required for the retrieval of Golgi proteins from the PVC, including Vps35p, Vps29p, Vps26p, Vps5p, and Vps17p, which make up the PVC-localized retromer complex (Nothwehr and Hindes, 1997; Seaman et al., 1998; Nothwehr et al., 1999). In these mutants, Vps10p is not recycled from the PVC back to the Golgi but instead is transported to the vacuole with a half time of ∼30 min, which is comparable to the rate at which mutant forms of Vps10p lacking a cytosolic domain are delivered to the vacuole (Cooper and Stevens, 1996). The retromer complex is thought to act at the PVC to select cargo for retrograde transport. However, the Vps52/53/54 complex is found at the TGN, where it colocalizes with the late Golgi marker A-ALP. Although A-ALP, like other resident TGN proteins, cycles continuously through the PVC, its steady-state distribution is distinct from that of the PVC marker protein Pep12p. Blocking or slowing membrane traffic from the PVC by mutation of VPS27 results in the accumulation of A-ALP and other TGN proteins together with Pep12p in the class E compartment, which is an exaggerated form of the PVC (Piper et al., 1995). Retromer proteins as well as other peripheral membrane proteins implicated in retrieval from the PVC are also associated with this class E compartment (Nothwehr and Hindes, 1997). However, the distribution of the Vps52/53/54 complex is unchanged in vps27 mutants and is clearly distinct from that of Pep12p. Therefore, the Vps52/53/54 complex does not appear to be associated with the PVC or to follow a recycling pathway through the PVC; instead, the complex is targeted to the TGN by a mechanism that is independent of efficient retrograde transport from the PVC.
The TGN localization of the Vps52/53/54 complex suggests that if it does indeed mediate retrograde transport, its localization to the target membrane is more indicative of a role in docking or fusion of retrograde vesicles. One might expect transport vesicles containing recycling TGN proteins to accumulate in such mutants. However, disruption of retrograde transport at a fusion step may prevent the recycling of membrane proteins that are required for the formation of new vesicles, thus slowing the rate of exit from the TGN and inhibiting the formation of new vesicles from the PVC. Therefore, a defect in the fusion of retrograde vesicles with the TGN could indirectly lead to the slow delivery of TGN membrane proteins to the vacuole rather than to a dramatic accumulation of vesicular intermediates. In fact, although a portion of A-ALP and Kex2p can be detected in the vacuoles of vps52 mutants, much of the A-ALP and Kex2p is found in a large number of dispersed small punctate structures that are similar to transport vesicles and distinct from the fewer, larger dots that characterize the late Golgi in wild-type cells. An accumulation of vesicles has also been reported in sac2 mutants (allelic to vps52) grown at 30°C and examined by electron microscopy (Novick et al., 1989).
Vps10p is found primarily in the vacuole in vps52/53/54 mutants rather than in vesicles. Differences in the trafficking of Vps10p and TGN proteins such as Kex2p have been demonstrated previously (Voos and Stevens, 1998; Nothwehr et al., 1999). One model to account for these differences is that Kex2p and Vps10p cycle through different endosomal compartments (e.g., early endosomes versus PVC; Holthuis et al., 1998a). Loss of the Vps52/53/54 complex may block the fusion of a single class of recycling vesicles with the TGN, resulting in the accumulation of vesicles from just one population of endosomes. Alternatively, cargo molecules present in the same endosomal compartment could enter recycling vesicles with different efficiencies. The differential effects of grd19 and certain vps35-ts mutations on Vps10p and Kex2p trafficking have been ascribed to differences in the machinery that recruits cargo into recycling vesicles (Voos and Stevens, 1998; Nothwehr et al., 1999).
Because no mutants have yet been described that specifically prevent the fusion of retrograde vesicles with the yeast TGN, it is difficult to predict the phenotypic consequences of blocking this transport step. Tlg1p and Tlg2p are t-SNAREs that localize to both the TGN and the endosomal compartments (Abeliovich et al., 1998; Holthuis et al., 1998a,b) and have been proposed to mediate the fusion of endosome-derived vesicles with the yeast TGN (Nichols and Pelham, 1998), although other roles for these proteins are possible (Seron et al., 1998). These t-SNAREs, like the Vps52/53/54 complex, are required to maintain normal levels of late Golgi membrane proteins and for the correct sorting of CPY. tlg1 and tlg2 mutants secrete significant amounts of newly synthesized CPY (50 and 35%, respectively; Nichols et al., 1998), although vps52, vps53, and vps54 strains secrete still more. However, the promiscuity of SNARE–SNARE interactions may allow other post-Golgi t-SNAREs to substitute at least in part for the loss of the relevant late Golgi t-SNARE. In fact, loss of the vacuolar t-SNAREs Vam3p and Vam7p results in less severe defects in vacuolar morphology and function than loss of the class C proteins Vps11p, Vps18p, Vps16p, and Vps33p, which act together to regulate the fusion of multiple transport intermediates with the vacuole, and high levels of the prevacuolar t-SNARE Pep12p suppress mutations in VAM3 (Darsow et al., 1997; Rieder and Emr, 1997). It is also difficult to extrapolate from the phenotypes resulting from defects in t-SNAREs, because these proteins are likely to regulate a number of different transport events. For example, a putative TGN t-SNARE might also be expected to mediate the fusion of recycling vesicles from the plasma membrane and early endosomal compartments as well as from the PVC, events that may involve different sets of v-SNAREs and associated docking factors.
Now that an obligatory role for unique accessory proteins has been demonstrated for the formation of SNARE complexes at many different trafficking steps (Pfeffer, 1999), it seems likely that such factors will be found to be generally required for all vesicle docking and/or fusion reactions. Further experiments will be needed to determine if the Vps52/53/54 complex acts as a docking factor in retrograde traffic to the TGN, perhaps in conjunction with Tlg1p and/or Tlg2p. Alternatively, the Vps52/53/54 complex could be required to maintain the integrity of the TGN. The two possibilities are not mutually exclusive: GM130 functions both as a docking factor, interacting with p115 in intra-Golgi transport in mammalian cells, and as a matrix protein, mediating the assembly and disassembly of stacked Golgi cisternae through successive cell divisions coupled to a cycle of phosphorylation/dephosphorylation (Nakamura et al., 1997; Lowe et al., 1998). The isolation of temperature-sensitive-for-function alleles of vps52, vps53, and vps54 should help in separating the direct effects of these mutations from those resulting from long-term loss of late Golgi function. Such mutations would permit an epistasis analysis to order the function of these three new VPS genes relative to those of other genes in the CPY pathway.
ACKNOWLEDGMENTS
We thank S. Nothwehr, J. Horecka, G. Waters, and D. Drubin for providing plasmids, M. Snyder for providing the yeast transposon library, and D. Jackson for preparing affinity-purified anti-HA antiserum. We are grateful to Steve Nothwehr for critical evaluation of the manuscript and to members of the Stevens laboratory past and present for helpful discussions. Mike Marusich and the University of Oregon Monoclonal Antibody Facility is acknowledged for the preparation of mouse mAbs, and DNA sequencing was performed by Yanling Wang. This work was supported by a postdoctoral fellowship from the American Heart Association, Oregon Affiliate (to E.C.), and by grant GM32448 from the National Institutes of Health (to T.H.S.)
Abbreviations:
- ALP
alkaline phosphatase
- CPY
carboxypeptidase Y
- DPAP
dipeptidyl aminopeptidase
- ER
endoplasmic reticulum
- HA
hemagglutinin
- PVC
prevacuolar/endosomal compartment
- SNARE
soluble NSF attachment protein receptor
- TGN
trans-Golgi network
REFERENCES
- Abeliovich H, Grote E, Novick P, Ferro-Novick S. Tlg2p, a yeast syntaxin homolog that resides on the Golgi and endocytic structures. J Biol Chem. 1998;273:11719–11727. doi: 10.1074/jbc.273.19.11719. [DOI] [PubMed] [Google Scholar]
- Bankaitis VA, Johnson LM, Emr SD. Isolation of yeast mutants defective in protein targeting to the vacuole. Proc Natl Acad Sci USA. 1986;83:9075–9079. doi: 10.1073/pnas.83.23.9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banta LM, Robinson JS, Klionsky DJ, Emr SD. Organelle assembly in yeast: characterization of yeast mutants defective in vacuolar biogenesis and protein sorting. J Cell Biol. 1988;107:1369–1383. doi: 10.1083/jcb.107.4.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becherer KA, Rieder SE, Emr SD, Jones EW. Novel syntaxin homologue, Pep12p, required for the sorting of lumenal hydrolases to the lysosome-like vacuole in yeast. Mol Biol Cell. 1996;7:579–594. doi: 10.1091/mbc.7.4.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant NJ, Stevens TH. Two separate signals act independently to localize a yeast late Golgi membrane protein through a combination of retrieval and retention. J Cell Biol. 1997;136:287–297. doi: 10.1083/jcb.136.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd CG, Peterson M, Cowles CR, Emr SD. A novel Sec18p/NSF-dependent complex required for Golgi-to-endosome transport in yeast. Mol Biol Cell. 1997;8:1089–1104. doi: 10.1091/mbc.8.6.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns N, Grimwade B, Ross-Macdonald PB, Choi EY, Finberg K, Roeder GS, Snyder M. Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae. Genes Dev. 1994;8:1087–1105. doi: 10.1101/gad.8.9.1087. [DOI] [PubMed] [Google Scholar]
- Cao X, Ballew N, Barlowe C. Initial docking of ER-derived vesicles requires Uso1p and Ypt1p but is independent of SNARE proteins. EMBO J. 1998;17:2156–2165. doi: 10.1093/emboj/17.8.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereghino JL, Marcusson EG, Emr SD. The cytoplasmic tail domain of the vacuolar protein sorting receptor Vps10p and a subset of VPS gene products regulate receptor stability, function, and localization. Mol Biol Cell. 1995;6:1089–1102. doi: 10.1091/mbc.6.9.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoforidis S, McBride HM, Burgoyne RD, Zerial M. The Rab5 effector EEA1 is a core component of endosome docking. Nature. 1999;397:621–625. doi: 10.1038/17618. [DOI] [PubMed] [Google Scholar]
- Conibear E, Stevens TH. Multiple sorting pathways between the late Golgi and the vacuole in yeast. Biochim Biophys Acta. 1998;1404:211–230. doi: 10.1016/s0167-4889(98)00058-5. [DOI] [PubMed] [Google Scholar]
- Cooper AA, Stevens TH. Vps10p cycles between the late-Golgi and prevacuolar compartments in its function as the sorting receptor for multiple yeast vacuolar hydrolases. J Cell Biol. 1996;133:529–541. doi: 10.1083/jcb.133.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles CR, Odorizzi G, Payne GS, Emr SD. The AP-3 adaptor complex is essential for cargo-selective transport to the yeast vacuole. Cell. 1997;91:109–118. doi: 10.1016/s0092-8674(01)80013-1. [DOI] [PubMed] [Google Scholar]
- Darsow T, Rieder SE, Emr SD. A multispecificity syntaxin homologue, Vam3p, essential for autophagic and biosynthetic protein transport to the vacuole. J Cell Biol. 1997;138:517–529. doi: 10.1083/jcb.138.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekena K, Stevens TH. The Saccharomyces cerevisiae MVP1 gene interacts with VPS1 and is required for vacuolar protein sorting. Mol Cell Biol. 1995;15:1671–1678. doi: 10.1128/mcb.15.3.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro-Novick S, Jahn R. Vesicle fusion from yeast to man. Nature. 1994;370:191–193. doi: 10.1038/370191a0. [DOI] [PubMed] [Google Scholar]
- Fischer von Mollard G, Nothwehr SF, Stevens TH. The yeast v-SNARE Vti1p mediates two vesicle transport pathways through interactions with the t-SNAREs Sed5p and Pep12p. J Cell Biol. 1997;137:1511–1524. doi: 10.1083/jcb.137.7.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer von Mollard G, Stevens TH. The Saccharomyces cerevisiae v-SNARE Vti1p is required for multiple membrane transport pathways to the vacuole. Mol Biol Cell. 1999;10:1719–1732. doi: 10.1091/mbc.10.6.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham LA, Hill KJ, Stevens TH. Assembly of the yeast vacuolar H+-ATPase occurs in the endoplasmic reticulum and requires a Vma12p/Vma22p assembly complex. J Cell Biol. 1998;142:39–49. doi: 10.1083/jcb.142.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Roth D, Walch-Solimena C, Novick P. The exocyst is an effector for Sec4p, targeting secretory vesicles to sites of exocytosis. EMBO J. 1999;18:1071–1080. doi: 10.1093/emboj/18.4.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas A, Scheglmann D, Lazar T, Gallwitz D, Wickner W. The GTPase Ypt7p of Saccharomyces cerevisiae is required on both partner vacuoles for the homotypic fusion step of vacuole inheritance. EMBO J. 1995;14:5258–5270. doi: 10.1002/j.1460-2075.1995.tb00210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris SL, Waters MG. Localization of a yeast early Golgi mannosyltransferase, Och1p, involves retrograde transport. J Cell Biol. 1996;132:985–998. doi: 10.1083/jcb.132.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harsay E, Bretscher A. Parallel secretory pathways to the cell surface in yeast. J Cell Biol. 1995;131:297–310. doi: 10.1083/jcb.131.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holthuis JC, Nichols BJ, Dhruvakumar S, Pelham HR. Two syntaxin homologues in the TGN/endosomal system of yeast. EMBO J. 1998a;17:113–126. doi: 10.1093/emboj/17.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holthuis JCM, Nichols BJ, Pelham HRB. The syntaxin Tlg1p mediates trafficking of chitin synthase III to polarized growth sites in yeast. Mol Biol Cell. 1998b;9:3383–3397. doi: 10.1091/mbc.9.12.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman DA, Yang S, Drubin DG. Synthetic-lethal interactions identify two novel genes, SLA1 and SLA2, that control membrane cytoskeleton assembly in Saccharomyces cerevisiae. J Cell Biol. 1993;122:635–644. doi: 10.1083/jcb.122.3.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones EW. Proteinase mutants of Saccharomyces cerevisiae. Genetics. 1977;85:23–33. doi: 10.1093/genetics/85.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolling R, Lee A, Chen EY, Botstein D. Nucleotide sequence of the SAC2 gene of Saccharomyces cerevisiae. Yeast. 1994;10:1211–1216. doi: 10.1002/yea.320100909. [DOI] [PubMed] [Google Scholar]
- Kornfeld S. Structure and function of the mannose 6-phosphate/insulinlike growth factor II receptors. Annu Rev Biochem. 1992;61:307–330. doi: 10.1146/annurev.bi.61.070192.001515. [DOI] [PubMed] [Google Scholar]
- Lowe M, Rabouille C, Nakamura N, Watson R, Jackman M, Jamsa E, Rahman D, Pappin DJ, Warren G. Cdc2 kinase directly phosphorylates the cis-Golgi matrix protein GM130 and is required for Golgi fragmentation in mitosis. Cell. 1998;94:783–793. doi: 10.1016/s0092-8674(00)81737-7. [DOI] [PubMed] [Google Scholar]
- Lupashin VV, Pokrovskaya ID, McNew JA, Waters MG. Characterization of a novel yeast SNARE protein implicated in Golgi retrograde traffic. Mol Biol Cell. 1997;8:2659–2676. doi: 10.1091/mbc.8.12.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcusson EG, Horazdovsky BF, Cereghino JL, Gharakhanian E, Emr SD. The sorting receptor for yeast vacuolar carboxypeptidase Y is encoded by the VPS10 gene. Cell. 1994;77:579–586. doi: 10.1016/0092-8674(94)90219-4. [DOI] [PubMed] [Google Scholar]
- Munn AL, Riezman H. Endocytosis is required for the growth of vacuolar H(+)-ATPase-defective yeast: identification of six new END genes. J Cell Biol. 1994;127:373–386. doi: 10.1083/jcb.127.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N, Lowe M, Levine TP, Rabouille C, Warren G. The vesicle docking protein p115 binds GM130, a cis-Golgi matrix protein, in a mitotically regulated manner. Cell. 1997;89:445–455. doi: 10.1016/s0092-8674(00)80225-1. [DOI] [PubMed] [Google Scholar]
- Nichols BJ, Holthuis JC, Pelham HR. The Sec1p homologue Vps45p binds to the syntaxin Tlg2p. Eur J Cell Biol. 1998;77:263–268. doi: 10.1016/s0171-9335(98)80084-8. [DOI] [PubMed] [Google Scholar]
- Nichols BJ, Pelham HR. SNAREs and membrane fusion in the Golgi apparatus. Biochim Biophys Acta. 1998;1404:9–31. doi: 10.1016/s0167-4889(98)00044-5. [DOI] [PubMed] [Google Scholar]
- Nothwehr SF, Bruinsma P, Strawn LA. Distinct domains within Vps35p mediate the retrieval of two different cargo proteins from the yeast prevacuolar/endosomal compartment. Mol Biol Cell. 1999;10:875–890. doi: 10.1091/mbc.10.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr SF, Bryant NJ, Stevens TH. The newly identified yeast GRD genes are required for retention of late-Golgi membrane proteins. Mol Cell Biol. 1996;16:2700–2707. doi: 10.1128/mcb.16.6.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr SF, Conibear E, Stevens TH. Golgi and vacuolar membrane proteins reach the vacuole in vps1 mutant yeast cells via the plasma membrane. J Cell Biol. 1995;129:35–46. doi: 10.1083/jcb.129.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr SF, Hindes AE. The yeast VPS5/GRD2 gene encodes a sorting nexin-1-like protein required for localizing membrane proteins to the late Golgi. J Cell Sci. 1997;110:1063–1072. doi: 10.1242/jcs.110.9.1063. [DOI] [PubMed] [Google Scholar]
- Nothwehr SF, Roberts CJ, Stevens TH. Membrane protein retention in the yeast Golgi apparatus: dipeptidyl aminopeptidase A is retained by a cytoplasmic signal containing aromatic residues. J Cell Biol. 1993;121:1197–1209. doi: 10.1083/jcb.121.6.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P, Osmond BC, Botstein D. Suppressors of yeast actin mutations. Genetics. 1989;121:659–674. doi: 10.1093/genetics/121.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson MR, Burd CG, Emr SD. Vac1p coordinates Rab and phosphatidylinositol 3-kinase signaling in Vps45p-dependent vesicle docking/fusion at the endosome. Curr Biol. 1999;9:159–162. doi: 10.1016/s0960-9822(99)80071-2. [DOI] [PubMed] [Google Scholar]
- Pfeffer SR. Transport-vesicle targeting: tethers before SNAREs. Nat Cell Biol. 1999;1:E17–E22. doi: 10.1038/8967. [DOI] [PubMed] [Google Scholar]
- Piper RC, Bryant NJ, Stevens TH. The membrane protein alkaline phosphatase is delivered to the vacuole by a route that is distinct from the VPS-dependent pathway. J Cell Biol. 1997;138:531–545. doi: 10.1083/jcb.138.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper RC, Cooper AA, Yang H, Stevens TH. VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. J Cell Biol. 1995;131:603–617. doi: 10.1083/jcb.131.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper RC, Whitters EA, Stevens TH. Yeast Vps45p is a Sec1p-like protein required for the consumption of vacuole-targeted, postGolgi transport vesicles. Eur J Cell Biol. 1994;65:305–318. [PubMed] [Google Scholar]
- Raymond CK, Howald-Stevenson I, Vater CA, Stevens TH. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol Biol Cell. 1992;3:1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redding K, Holcomb C, Fuller RS. Immunolocalization of Kex2 protease identifies a putative late Golgi compartment in the yeast Saccharomyces cerevisiae. J Cell Biol. 1991;113:527–538. doi: 10.1083/jcb.113.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redding K, Seeger M, Payne GS, Fuller RS. The effects of clathrin inactivation on localization of Kex2 protease are independent of the TGN localization signal in the cytosolic tail of Kex2p. Mol Biol Cell. 1996;7:1667–1677. doi: 10.1091/mbc.7.11.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder SE, Emr SD. A novel RING finger protein complex essential for a late step in protein transport to the yeast vacuole. Mol Biol Cell. 1997;8:2307–2327. doi: 10.1091/mbc.8.11.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CJ, Nothwehr SF, Stevens TH. Membrane protein sorting in the yeast secretory pathway: evidence that the vacuole may be the default compartment. J Cell Biol. 1992;119:69–83. doi: 10.1083/jcb.119.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CJ, Raymond CK, Yamashiro CT, Stevens TH. Methods for studying the yeast vacuole. Methods Enzymol. 1991;194:644–661. doi: 10.1016/0076-6879(91)94047-g. [DOI] [PubMed] [Google Scholar]
- Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman JE. Mechanisms of intracellular protein transport. Nature. 1994;372:55–63. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- Rothman JH, Stevens TH. Protein sorting in yeast: mutants defective in vacuole biogenesis mislocalize vacuolar proteins into the late secretory pathway. Cell. 1986;47:1041–1051. doi: 10.1016/0092-8674(86)90819-6. [DOI] [PubMed] [Google Scholar]
- Sacher M, Jiang Y, Barrowman J, Scarpa A, Burston J, Zhang L, Schieltz D, Yates JRR, Abeliovich H, Ferro-Novick S. TRAPP, a highly conserved novel complex on the cis-Golgi that mediates vesicle docking and fusion. EMBO J. 1998;17:2494–2503. doi: 10.1093/emboj/17.9.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapperstein SK, Lupashin VV, Schmitt HD, Waters MG. Assembly of the ER to Golgi SNARE complex requires Uso1p. J Cell Biol. 1996;132:755–767. doi: 10.1083/jcb.132.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato TK, Darsow T, Emr SD. Vam7p, a SNAP-25-like molecule, and Vam3p, a syntaxin homolog, function together in yeast vacuolar protein trafficking. Mol Cell Biol. 1998;18:5308–5319. doi: 10.1128/mcb.18.9.5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MN, Marcusson EG, Cereghino JL, Emr SD. Endosome to Golgi retrieval of the vacuolar protein sorting receptor, Vps10p, requires the function of the VPS29, VPS30, and VPS35 gene products. J Cell Biol. 1997;137:79–92. doi: 10.1083/jcb.137.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MN, McCaffery JM, Emr SD. A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J Cell Biol. 1998;142:665–681. doi: 10.1083/jcb.142.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger M, Payne GS. Selective and immediate effects of clathrin heavy chain mutations on Golgi membrane protein retention in Saccharomyces cerevisiae. J Cell Biol. 1992;118:531–540. doi: 10.1083/jcb.118.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seron K, et al. A yeast t-SNARE involved in endocytosis. Mol Biol Cell. 1998;9:2873–2889. doi: 10.1091/mbc.9.10.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Archer JE, Solomon F. Regulation of tubulin polypeptides and microtubule function: Luv1p [correction of Rki1p] interacts with the beta-tubulin binding protein Rbl2p. Chromosoma. 1998;107:471–478. doi: 10.1007/s004120050331. [DOI] [PubMed] [Google Scholar]
- Sollner T, Whiteheart SW, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman JE. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- Sonnichsen B, Lowe M, Levine T, Jamsa E, Dirac-Svejstrup B, Warren G. A role for giantin in docking COPI vesicles to Golgi membranes. J Cell Biol. 1998;140:1013–1021. doi: 10.1083/jcb.140.5.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens TH, Forgac M. Structure, function and regulation of the vacuolar (H+)-ATPase. Annu Rev Cell Dev Biol. 1997;13:779–808. doi: 10.1146/annurev.cellbio.13.1.779. [DOI] [PubMed] [Google Scholar]
- Tall GG, Hama H, DeWald DB, Horazdovsky BF. The phosphatidylinositol 3-phosphate binding protein vac1p interacts with a rab GTPase and a Sec1p homologue to facilitate vesicle-mediated vacuolar protein sorting. Mol Biol Cell. 1999;10:1873–1889. doi: 10.1091/mbc.10.6.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TerBush DR, Maurice T, Roth D, Novick P. The exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J. 1996;15:6483–6494. [PMC free article] [PubMed] [Google Scholar]
- Ungermann C, Wickner W. Vam7p, a vacuolar SNAP-25 homolog, is required for SNARE complex integrity and vacuole docking and fusion. EMBO J. 1998;17:3269–3276. doi: 10.1093/emboj/17.12.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanRheenen SM, Cao X, Lupashin VV, Barlowe C, Waters MG. Sec35p, a novel peripheral membrane protein, is required for ER to Golgi vesicle docking. J Cell Biol. 1998;141:1107–1119. doi: 10.1083/jcb.141.5.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vater CA, Raymond CK, Ekena K, Howald-Stevenson I, Stevens TH. The VPS1 protein, a homolog of dynamin required for vacuolar protein sorting in Saccharomyces cerevisiae, is a GTPase with two functionally separable domains. J Cell Biol. 1992;119:773–786. doi: 10.1083/jcb.119.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida TA, Huyer G, Emr SD. Yeast vacuolar proenzymes are sorted in the late Golgi complex and transported to the vacuole via a prevacuolar endosome-like compartment. J Cell Biol. 1993;121:1245–1256. doi: 10.1083/jcb.121.6.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voos W, Stevens TH. Retrieval of resident late-Golgi membrane proteins from the prevacuolar compartment of Saccharomyces cerevisiae is dependent on the function of Grd19p. J Cell Biol. 1998;140:577–590. doi: 10.1083/jcb.140.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada Y, Ohsumi Y, Anraku Y. Genes for directing vacuolar morphogenesis in Saccharomyces cerevisiae. I. Isolation and characterization of two classes of vam mutants. J Biol Chem. 1992;267:18665–18670. [PubMed] [Google Scholar]
- Walter L, Gunther E. Identification of a novel highly conserved gene in the centromeric part of the major histocompatibility complex. Genomics. 1998;52:298–304. doi: 10.1006/geno.1998.5463. [DOI] [PubMed] [Google Scholar]
- Weber T, Zemelman BV, McNew JA, Westermann B, Gmachl M, Parlati F, Sollner TH, Rothman JE. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- Weisman LS, Emr SD, Wickner WT. Mutants of Saccharomyces cerevisiae that block intervacuole vesicular traffic and vacuole division and segregation. Proc Natl Acad Sci USA. 1990;87:1076–1080. doi: 10.1073/pnas.87.3.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox CA, Redding K, Wright R, Fuller RS. Mutation of a tyrosine localization signal in the cytosolic tail of yeast Kex2 protease disrupts Golgi retention and results in default transport to the vacuole. Mol Biol Cell. 1992;3:1353–1371. doi: 10.1091/mbc.3.12.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Gonzalez L, Jr, Prekeris R, Steegmaier M, Advani RJ, Scheller RH. SNARE interactions are not selective: implications for membrane fusion specificity. J Biol Chem. 1999;274:5649–5653. doi: 10.1074/jbc.274.9.5649. [DOI] [PubMed] [Google Scholar]