Abstract
Rad52-dependent homologous recombination (HR) is regulated by the antirecombinase activities of Srs2 and Rqh1/Sgs1 DNA helicases in fission yeast and budding yeast. Functional analysis of Srs2 in Schizosaccharomyces pombe led us to the discovery of Sws1, a novel HR protein with a SWIM-type Zn finger. Inactivation of Sws1 suppresses the genotoxic sensitivity of srs2Δ and rqh1Δ mutants and rescues the inviability of srs2Δ rqh1Δ cells. Sws1 functions at an early step of recombination in a pro-recombinogenic complex with Rlp1 and Rdl1, two RecA-like proteins that are most closely related to the human Rad51 paralogs XRCC2 and RAD51D, respectively. This finding indicates that the XRCC2–RAD51D complex is conserved in lower eukaryotes. A SWS1 homolog exists in human cells. It associates with RAD51D and ablating its expression reduces the number of RAD51 foci. These studies unveil a conserved pathway for the initiation and control of HR in eukaryotic cells.
Keywords: DNA repair, genome stability, homologous recombination
Introduction
Homologous recombination (HR) is a universal process of error-free DNA repair (Paques and Haber, 1999; Sung et al, 2003; West, 2003; Krogh and Symington, 2004). Its defining feature is the use of homologous sequences as a template for repairing damaged DNA. HR is most commonly studied in the context of double-strand breaks (DSBs), but it can also be initiated by single-strand gaps that occur during DNA replication or in certain types of DNA repair, such as base excision repair or trans-lesion synthesis (Swanson et al, 1999). In the budding yeast Saccharomyces cerevisiae and fission yeast Schizosaccharomyces pombe, HR is under the control of the RAD52 epistasis group of proteins, whose members are conserved in eukaryotes (Sung et al, 2003; Krogh and Symington, 2004). Formation of a Rad51 nucleoprotein filament, a process dependent on Rad52 and mediated by the Rad51 paralogs, is a key step in HR (Sung, 1997a; Sugawara et al, 2003; Lisby et al, 2004). The Rad51 paralogs act primarily as heterodimers or larger multimeric complexes (Masson et al, 2001). For example, budding yeast Rad55 and Rad57 form a complex (Sung, 1997b), as do the equivalent proteins in fission yeast, Rhp55 and Rhp57 (Tsutsui et al, 2001). Multicellular eukaryotes have an additional Rad51 paralog subcomplex, consisting of XRCC2 and RAD51D (Braybrooke et al, 2000; Masson et al, 2001), that has not been found in yeast species.
Although HR is essential for the preservation of genome integrity, there are pathological situations in which excessive HR can destabilize the genome and cause cell death. One example involves an S. cerevisiae mutant that lacks two DNA helicases: the Sgs1 RecQ-type DNA helicase and Srs2. The inviability of this mutant can be suppressed by inactivating HR genes (Gangloff et al, 2000). Conservation of an antirecombinase activity of Sgs1 homologs is indicated by the hyper-recombination phenotype of fission yeast rqh1Δ cells and the effects of mutations of three of the five human RecQ homologs, BLM, WRN and RECQL4, which cause the cancer-prone syndromes Bloom, Werner and Rothmund–Thomson, respectively. All these syndromes are associated with genomic instability that arises from inappropriate recombination (Hickson, 2003).
The RecQ-like DNA helicases and Srs2 homologs are thought to act as antirecombinases by different mechanisms. In the case of a RecQ-like helicase complexed with a topoisomerase, in vitro studies have shown that this complex can dissolve a DNA substrate containing two Holliday junctions (HJs) into a single noncrossover product (Wu and Hickson, 2003). RecQ-like helicases might also unwind dsDNA structures that are HR intermediates, such as D-loops (van Brabant et al, 2000). In the case of Srs2, in vitro studies have shown it can disassemble the Rad51 nucleoprotein filament (Krejci et al, 2003; Veaute et al, 2003). Interestingly, inclusion of the single-strand DNA binding protein RPA in these assays has a synergistic effect that can be partially overcome by addition of Rad52. These data suggest that Srs2 and Rad52 have antagonistic effects in controlling HR.
The C-terminal noncatalytic domain of S. cerevisiae Srs2 mediates its interaction with Rad51 (Krejci et al, 2003). Curiously, this domain is apparently not conserved in S. pombe Srs2, even though the antirecombinogenic activities of the Srs2 homologs are conserved (Gangloff et al, 2000; Klein, 2001; Doe and Whitby, 2004). To better understand the interplay of pro and antirecombinogenic activities, we undertook a screen for proteins that interact with Srs2 in fission yeast. As described here, this screen led to identification of Sws1, a protein that forms a prorecombinogenic complex with XRCC2 and RAD51D-like proteins. This complex is conserved in humans, and it may be related to the Shu group of proteins recently discovered in budding yeast (Shor et al, 2005). These studies establish the existence of a conserved pathway for initiation and control of HR in eukaryotic cells.
Results
Sws1 interacts with Srs2
We performed a yeast two-hybrid screen with the C-terminal 187 amino acids of S. pombe Srs2, which is the region responsible for its interaction with Rhp51 (our unpublished results), the Rad51 homolog in fission yeast. One of the hits was SPBC11B10.06, a gene listed as a sequence orphan with a SWIM domain (Figure 1A). A SWIM domain is an uncharacterized Zn-finger-like motif found in several classes of proteins, including some members of the SWI2/SNF2 family of ATPases and the MuDR plant transposases (Makarova et al, 2002). SPBC11B10.06 was given the name Sws1 (SWIM domain-containing and Srs2-interacting protein 1).
Figure 1.
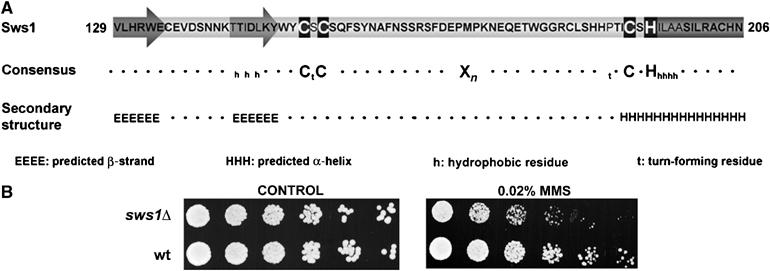
SWIM domain of Sws1 and MMS sensitivity of sws1Δ mutants. (A) SWIM domain of Sws1. The potential metal-chelating residues defined by Makarova et al (2002) are shown. The predicted α-helix and β-strands are highlighted. The secondary structure prediction was obtained using the Jpred program. (B) DNA damage survival assay. Four-fold serial dilutions of wt (PR109) and sws1Δ (VM3723) cells were plated on YES media and exposed to 0.02% MMS. Photographs were taken after 4 days at 32°C.
Typical of DNA repair proteins, GFP-tagged Sws1 localized to the nucleoplasm and was excluded from the nucleolus (Supplementary Figure 1A). This pattern was unchanged throughout the cell cycle. Sws1-GFP did not form discrete nuclear foci when cells were exposed to genotoxic agents (data not shown). The nuclear signal of Sws1-GFP was eliminated by in situ detergent extraction (Supplementary Figure 1B), indicating that Sws1 does not associate tightly with chromatin (Noguchi et al, 2003).
Analysis of sws1Δ cells revealed no obvious growth or morphological defects. They were not generally sensitive to genotoxic agents such as ionizing radiation (IR), ultraviolet (UV) light, camptothecin (CPT) or hydroxyurea (HU), but they were sensitive to 0.02% methylmethane sulphonate (MMS) (Figure 1D and Supplementary Figure 2). This finding provided the first hint that Sws1 was involved in DNA repair.
Sws1 inactivation suppresses srs2Δ and rqh1Δ
Genetic epistasis studies were performed to investigate the functional relationship between Sws1 and Srs2. CPT sensitivity was examined because srs2Δ mutants are particularly sensitive to this genotoxic agent (Doe and Whitby, 2004). Surprisingly, we found that sws1Δ suppressed the srs2Δ CPT sensitive phenotype (Figure 2A). This result was unanticipated because Srs2 is thought to play an active role in promoting the repair of CPT-induced DNA damage, as opposed to preventing the formation of toxic recombination intermediates (Fabre et al, 2002; Doe and Whitby, 2004).
Figure 2.
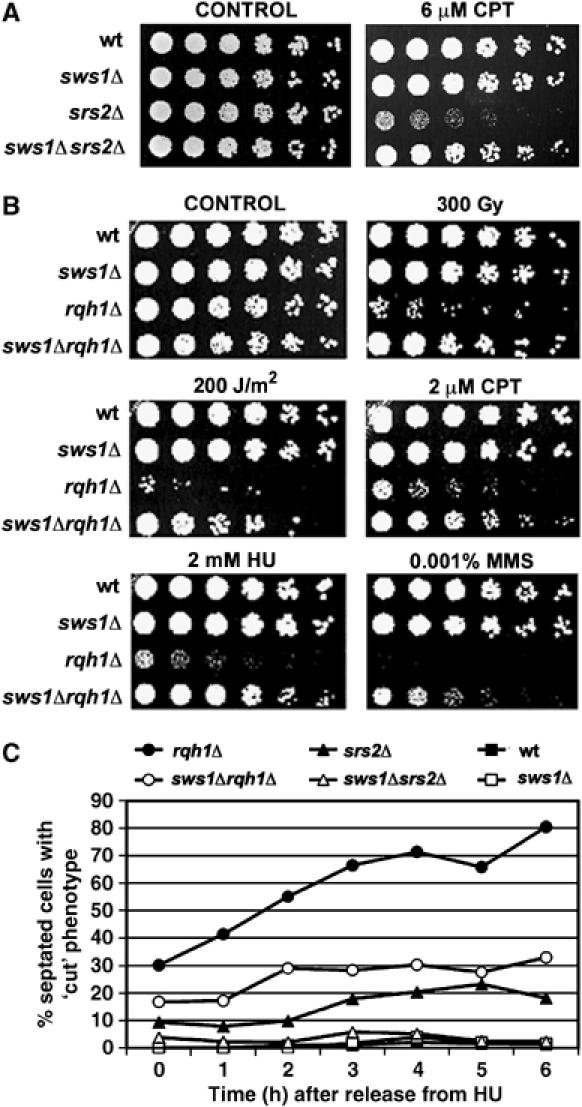
Effect of sws1+ deletion on survival of srs2Δ and rqh1Δ mutants. (A) Spot assay of wt (PR109), sws1Δ (VM3723), srs2Δ (VM3724) and sws1Δ srs2Δ (VM3721) strains. Four-fold serial dilutions of each strain were plated on YES or YES supplemented with 6 μM camptothecin. Photographs were taken after 4 days at 32°C. (B) DNA damage sensitivity of wt (PR109), sws1Δ (VM3723), rqh1Δ (SC3250), and sws1Δ rqh1Δ (VM3722) cells. Four-fold serial dilutions were plated onto YES plates and treated with the indicated DNA damaging agents. Photographs were taken after 4 days at 32°C. (C) Percentages of dividing cells with ‘cut' phenotype observed in wt (PR109), sws1Δ (VM3723), srs2Δ (VM3724), rqh1Δ (SC3250), sws1Δ srs2Δ (VM3721) and sws1Δ rqh1Δ (VM3722) strains at the indicated time points after release from 5-h-incubation in YES supplemented with 12 mM HU.
This result prompted an analysis of genetic interactions involving Sws1 and Rqh1. The HU and UV sensitivity of rqh1Δ cells is thought to arise from the formation of toxic recombination intermediates (Laursen et al, 2003; Doe and Whitby, 2004; Hope et al, 2005). Remarkably, the sws1Δ mutation not only suppressed the HU and UV sensitivity of rqh1Δ cells, it also suppressed their sensitivity to IR, CPT and low doses of MMS (Figure 2B).
Upon entering mitosis after treatment with HU, rqh1Δ cells display a characteristic ‘cut' phenotype in which unsegregated chromosomes are bisected by the cell division plate. This phenotype is thought to arise from unresolved covalent linkages of sister chromatids that have occurred through HR (Stewart et al, 1997; Doe et al, 2000). We reasoned that, if Sws1 promotes these HR events, the suppression of the HU sensitivity of rqh1Δ cells by sws1Δ should be accompanied by a reduction of the ‘cut' phenotype. Indeed, upon release from an HU arrest, the frequency of ‘cut' phenotypes was reduced ∼50% in rqh1Δ sws1Δ cells relative to rqh1Δ cells (Figure 2C). A similar effect was seen by combining the srs2Δ and sws1Δ mutations (Figure 2C). From these results we conclude that Sws1 promotes the generation of the toxic HR intermediates in rqh1Δ and srs2Δ cells.
These genetic interactions could be explained if Sws1 inhibits the antirecombinogenic activities of both Srs2 and Rqh1. To test this model, we determined whether sws1Δ rescued the extreme sickness of a rqh1Δ srs2Δ double mutant (Lee et al, 1999; Wang et al, 2001). Remarkably, sws1Δ almost completely rescued the poor growth of rqh1Δ srs2Δ cells (Figure 3A). It also rescued their poor plating efficiency, restoring it from 5 to 44% compared to wild type. The specificity of this rescue was confirmed by controlling sws1+ expression from the thiamine (vitamin B1)-repressible nmt1 promoter in sws1Δ rqh1Δ srs2Δ cells (Figure 3B). These findings showed that Sws1 can function independently of both Rqh1 and Srs2.
Figure 3.
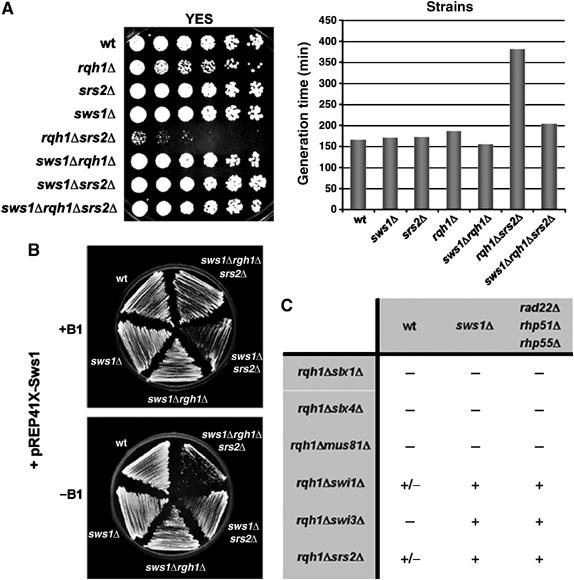
Deletion of sws1+ rescues the slow growth of srs2Δ rqh1Δ double mutants. (A) Spot assay of wt (PR109), rqh1Δ (SC3250), srs2Δ (VM3724), sws1Δ (VM3723), rqh1Δ srs2Δ (VM3756), sws1Δ rqh1Δ (VM3722) sws1Δ srs2Δ (VM3721) and sws1Δ rqh1Δ srs2Δ (VM3720) strains. Four-fold serial dilutions of each strain were plated on YES. Photographs were taken after 4 days at 32°C. The generation times of wt and some of the mutants used in the spot assay is shown. Cell growth of cultures incubated at 32°C in liquid YES was monitored by measuring optical density at 600 nm. (B) Specificity of the rescue. wt (PR109), sws1Δ (VM3723), sws1Δ rqh1Δ (VM3722), sws1Δ srs2Δ (VM3757) and sws1Δ rqh1Δ srs2Δ (VM3735) were transformed with a pREP41x-Sws1 vector and plated on EMM plates with (for overexpression of Sws1) or without thiamine (repression). When sws1+ is overproduced, sws1Δ rqh1Δ srs2Δ (VM3735) cells return to the same rates of slow growth as srs2Δ rqh1Δ (VM3756) cells. Photographs were taken after 5 days at 32°C. (C) Comparison between the subsets of double mutants rescued by sws1Δ and the already characterized recombination genes rad22Δ, rhp51Δ and rhp55Δ. Spores derived from the corresponding crosses were plated on YES and the resulting colonies screened by PCR. Crosses giving rise to triple mutants are shown as ‘+'. Crosses in which it was not possible to recover any triple mutant, or synthetic lethal combinations appeared as ‘−'. ‘+/−' indicates synthetic slow growth.
Rqh1 is essential for viability in strains that lack the structure-specific endonucleases Mus81–Eme1 or Slx1–Slx4 (Boddy et al, 2001; Coulon et al, 2004), and in strains that lack the Swi1–Swi3 replication fork protection complex (Noguchi et al, 2003; Coulon et al, 2004). The interactions involving Swi1–Swi3 complex, but not those involving Mus81–Eme1 and Slx1–Slx4 complexes, are rescued by mutations that inactivate HR proteins (e.g. Rad22Rad52, Rhp51Rad51 and Rhp55Rad55). As summarized in Figure 3C, sws1Δ had a pattern of genetic interactions that matched those of rad22Δ, rhp51Δ and rhp55Δ mutations. These findings further supported the conclusion that Sws1 has a prorecombinogenic activity that acts in concert with the other known recombination proteins in fission yeast.
Sws1 controls an early step of HR
Rad52 has a crucial early role in HR, displacing replication protein A (RPA) and recruiting Rad51 to single-stranded DNA, leading to the formation of the Rad51 nucleoprotein filament (Sung, 1997a; Sugawara et al, 2003; Lisby et al, 2004). If Sws1 controls an early step of HR, recruitment of Rad22 (the Rad52 homolog) to DNA damage sites should be reduced in sws1Δ cells. Rad22-YFP forms bright nuclear foci at sites of DNA damage (Du et al, 2003); therefore, we monitored the effect of sws1Δ on formation of spontaneous Rad22-YFP foci in wild type, rqh1Δ and srs2Δ backgrounds. As shown in Figure 4A, the number of nuclei containing Rad22-YFP foci was substantially elevated in rqh1Δ and srs2Δ mutants relative to wild type. In all of these genetic backgrounds, sws1Δ reduced the number of spontaneous Rad22-YFP foci without affecting Rad22-YFP abundance (Figure 4A and data not shown).
Figure 4.
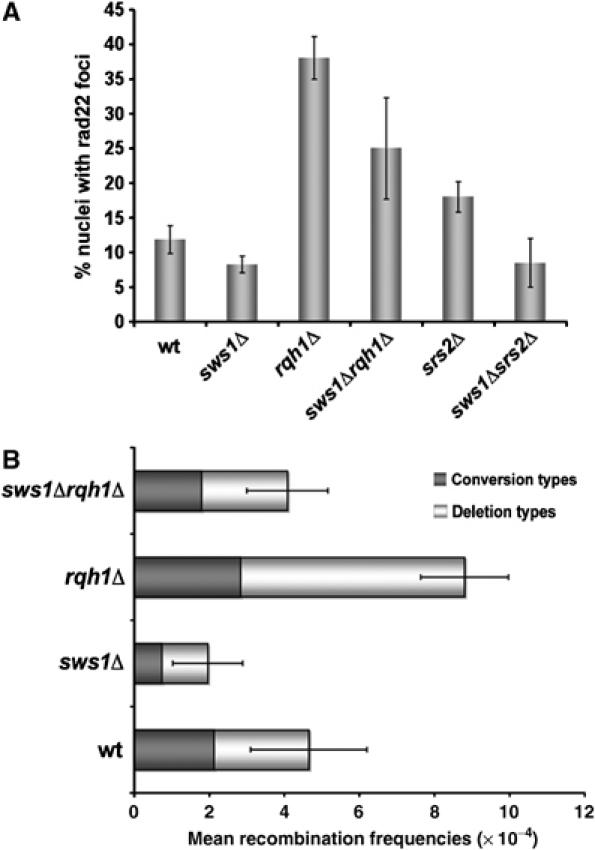
Spontaneous Rad22-YFP foci formation and recombination levels are reduced in sws1Δ background. (A) Cells containing a genomic copy of Rad22-YFP were grown in YES media at 32°C until mid-log phase and their Rad22-YFP foci were quantified. For each strain more than 300 cells were counted. The average percentage of nuclei containing at least one focus is shown. The strains used in this assay were wt (VM3725), sws1Δ (VM3729), rqh1Δ (VM3726), sws1Δrqh1Δ (VM3730), srs2Δ (VM3727), sws1Δsrs2Δ (VM3728). (B) Comparison of the spontaneous recombination frequencies of wt (PS3), sws1Δ (VM3731), rqh1Δ (VM3732) and sws1Δ rqh1Δ (VM3736). Strains containing an ura4+ marker flanked by two ade6− heteroalleles were used in this analysis. Recombination frequencies are mean values from three independent assays.
To formally address whether Sws1 promotes HR, we measured the rates of recombination between direct repeats of ade6− heteroalleles that are separated by the ura4+ gene. This assay can distinguish between two classes of recombination events: deletion types (ade6+ ura4−) and conversion types (ade6+ ura4+) (Osman et al, 1996). Rad22 is required for both types, whereas Rhp51, Rhp55 and Rhp57 are only required for conversion types. It should be noted that the overall rate of recombination is actually increased in the absence of Rhp51, Rhp55 and Rhp57, with all events being Rad22-dependent deletion types (Doe and Whitby, 2004; Doe et al, 2004). As shown in Figure 4B, sws1Δ reduced the frequency of recombination events by about half in both rqh1+ and rqh1Δ cells, with both conversion and deletion types decreased. Taken together with suppression of rqh1Δ DNA damage sensitivity by sws1Δ (Figure 2), these findings show that Sws1 controls an early step of HR.
Sws1 is related to S. cerevisiae Shu2
Before proceeding further, we decided to re-examine whether Sws1 is conserved in eukaryotes. As noted above, the only recognizable sequence motif in Sws1 is the SWIM domain (Figure 1A), which consists of a CxCxnCxH motif located downstream of two predicted β strands and followed by an α-helix (Makarova et al, 2002). To ascertain the functional significance of the SWIM domain, the first cysteine of the CxCxnCxH motif was replaced by a serine residue (C152S). This mutation is predicted to ablate metal chelation. The sws1-C152S mutation caused MMS sensitivity and suppressed the DNA damage sensitivity of rqh1Δ cells, behaving identically to sws1Δ (Figure 5A). We confirmed that Sws1-C152S-GFP localized to the nucleus with the same pattern as the wild-type protein (Supplementary Figure 1C).
Figure 5.

Sws1 is a conserved protein and the SWIM domain is required for its function. (A) Four-fold serial dilutions of wt (PR109), sws1Δ (VM3723), sws1-C152S (VM3734), rqh1Δ (SC3250), sws1Δ rqh1Δ (VM3722) and sws1-C152S rqh1Δ (VM3733) were plated on YES plates (control), YES plates supplemented with 3 mM HU or 0.02% MMS, or YES plates that afterwards were treated with 300 Gy of IR. Photographs were taken after 4 days at 32°C. (B) Model of the Zn-binding region of the SWIM domain in Sws1. The three cysteine residues and the histidine residue predicted to be involved in Zn chelation are indicated. The cysteine residue (Cys152) mutated in sws1-C152S is underlined. (C) Alignment of Sws1 and its orthologs from H. sapiens, M. musculus, A. thaliana, C. albicans and S. cerevisiae. The conserved CxC(x)nCxH motif and secondary structure elements (upstream β-strands and the downstream α-helix) are shown.
Having demonstrated the importance of the SWIM domain, we performed iterative PSI-BLAST searches with a bias towards hits containing the SWIM motif CxCxnCxH. Highly divergent homologs of Sws1 were found in most eukaryotes, with an xn variable between 15 (human) and 39 (Candida albicans) (Figure 5C). The existence of an Sws1-like gene in the yeast C. albicans prompted a closer examination of the S. cerevisiae genome. Using relaxed criteria, we found a S. cerevisiae Sws1-related gene that has a SWIM motif with an unusually long insert (xn=59) (Figure 5B and C). This gene was named Shu2 for ‘Suppressor of Sgs1 HU sensitivity' (Shor et al, 2005). As the name indicates, shu2Δ suppresses the HU sensitivity of mutants defective in Sgs1, the homolog of Rqh1. Like sws1Δ cells, shu2Δ mutants are also sensitive to MMS and they have reduced Rad52 foci (Shor et al, 2005). The implication, therefore, is that Sws1 and Shu2 are highly divergent homologs.
Sws1 associates with Rad51 paralogs
Genetic and yeast two-hybrid studies have indicated that Shu2 functions in association with three other proteins: Shu1, Psy3 and Csm2 (Huang et al, 2003; Lee et al, 2005; Shor et al, 2005). These proteins have no identifiable motifs, nor do they have homologs except in closely related yeast species (Shor et al, 2005). With the aim of identifying highly divergent homologs of Shu1, Psy3 or Csm2, we decided to use multidimensional protein identification technology (MudPIT) to identify proteins that coprecipitate with TAP-tagged Sws1 (Washburn et al, 2001). Besides Srs2, this analysis identified only one other protein known to be connected to HR (Supplementary Table II). This protein was Rlp1, a RecA-like protein that is most closely related to the RAD51 paralog XRCC2 (Khasanov et al, 2004). The most notable reported phenotype of rlp1Δ cells is their sensitivity to MMS, which was consistent with our analysis of sws1Δ cells (Figure 1B). Rlp1 has a weak two-hybrid interaction with Rhp57 (Khasanov et al, 2004), but Rhp57 was not detected in the MudPIT analysis of Sws1-TAP, nor were Rhp51 or Rhp55. However, upon careful analysis of the other proteins identified by MudPIT (Supplementary Table II), we found that one of them, encoded by SPAC17H9.03c, was related to RAD51 paralogs. Remarkably, it was most similar to vertebrate RAD51D (>45% overall similarity) (Figure 6A and B), homologs of which were heretofore thought to be absent in lower eukaryotes. The discovery of XRCC2 and RAD51 paralogs associated with Sws1 was particularly striking because vertebrate XRCC2 and RAD51D form a functional heterodimeric complex (Braybrooke et al, 2000; Masson et al, 2001). SPAC17H9.03c was named Rdl1 (Rad51D-like protein 1).
Figure 6.
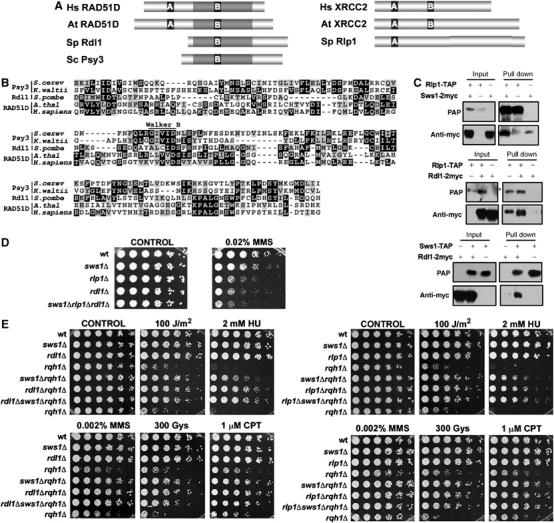
The S. pombe Rad51 paralogs, Rlp1 and Rdl1, show physical and genetic interactions with Sws1. (A) Schematic representation of the sequences of S. pombe Rdl1 and Rlp1 and their orthologs in H. sapiens (Hs), A. thaliana (At) and S. cerevisiae (Sc). A and B represent the Walker A and Walker B domains, respectively. The regions of highest similarity, centered around the Walker A and B domains are highlighted. (B) Alignment of the RAD51D family members from S. cerevisiae, K. waltii, S. pombe, A. thaliana and H. sapiens. The region of similarity between the different proteins (gray-shadowed region from the corresponding proteins in A) is shown. Conserved residues appear highlighted and the position of the Walker B motif is indicated. (C) Coimmunoprecipitation of Sws1, Rdl1 and Rlp1. Cells simultaneously transformed with the appropriate Rdl1-, Rlp1- and/or Sws1-expressing vectors were used in the assay (pREP41x for 2myc tags and pREP42x for TAP tags). Cell extracts were obtained after 21 h of incubation in the absence of thiamine. *Indicates the presence of a nonspecific band. (D) Spot assay of wt (PR109), sws1Δ (VM3723), rlp1Δ (VM3741), rdl1Δ (VM3744) and sws1Δrlp1Δrdl1Δ (VM3755) strains. Four-fold serial dilutions of each strain were plated on YES plates (CONTROL) or YES supplemented with 0.02% MMS. Photographs were taken after 4 days at 32°C. (E) Serial dilutions (fourfold) of the indicated strains were plated in YES plates in the presence of different sources of DNA damage. Photographs were taken after 4 days of incubation at 32°C. Strains used in this assays: wt (PR109), sws1Δ (VM3723), rdl1Δ (VM3744), rlp1Δ (VM3741), rqh1Δ (SC3250), sws1Δ rqh1Δ (VM3722), rlp1Δ rqh1Δ (VM3740), rdl1Δ rqh1Δ (VM3745), sws1Δ rlp1Δ rqh1Δ (VM3742) and sws1Δ rdl1Δ rqh1Δ (VM3746).
Rlp1 and Rdl1 shared no obvious sequence similarities to Shu1 or Csm2. However, Rdl1 and Psy3 were significantly similar (Figure 6B). Interestingly, this similarity was centered in a previously unnoticed Walker B-like motif of Psy3 (expectation value of <10−10 predicted by the MACAW program) that was also shared with human RAD51D (expectation value of <10−8) and RAD51D proteins from other species (Figure 6B). Importantly, Shu2 and Psy3 interact in yeast two-hybrid assays (Shor et al, 2005). These data suggest that Psy3 is a very diverged member of the RAD51D family.
Coprecipitation and immunoblot studies confirmed that Sws1 associates with Rlp1 and Rdl1 in vivo and further showed that Rdl1 coprecipitates with Rlp1 (Figure 6C), indicating that the three proteins form a stable complex. We also observed that Rlp1-GFP and Rdl1-GFP were low abundance proteins that colocalized with Sws1-GFP in the chromatin region of the nucleus (Supplementary Figure 1D).
As observed for sws1Δ cells, rdl1Δ and rlp1Δ mutants were viable and were not obviously sensitive to genotoxic agents, except for 0.02% MMS (Figure 6D). The somewhat enhanced MMS sensitivity of rdl1Δ and rlp1Δ mutants indicated that activities of Rdl1 and Rlp1 in promoting MMS survival were not fully dependent on Sws1, which is unlike the relationship between Shu2 and Psy3 (Shor et al, 2005). However, rdl1Δ and rlp1Δ suppressed the DNA damage sensitivities of rqh1Δ cells in a manner that was not additive with the suppressive effect of sws1Δ (Figure 6E), indicating that Sws1, Rlp1 and Rdl1 act in the same pathway to promote toxic recombination events in rqh1Δ cells.
Formation of Rad22 foci and recombination levels were monitored in rlp1Δ and rdl1Δ mutants alone and in combination with rqh1Δ (Figure 7A and B). In these assays, rlp1Δ behaved similarly to sws1Δ, reducing the frequency of Rad22-YFP foci and the recombination rates between ade6− heteroalleles (Figure 7). Although rdl1Δ and rlp1Δ had similar abilities to suppress rqh1Δ, they had different effects on the formation of Rad22-YFP foci and recombination rates. In rqh1+ cells, the rdl1Δ mutation increased the occurrence of spontaneous Rad22-YFP foci (Figure 7A). A similar but stronger effect was seen with the rhp55Δ and rhp57Δ mutations (Figure 7A). The rdl1Δ mutation also increased the rate of recombination between the ade6− heteroalleles, which was largely attributed to an enhanced rate of deletion types. Again, in this assay, the effect of rdl1Δ was similar although not as strong as those caused by rhp55Δ (Doe and Whitby, 2004) or rhp57Δ (Figure 7B). In contrast to rlp1Δ, the rdl1Δ mutation did not reduce the frequency of Rad22-YFP foci in the rqh1Δ background, nor did it reduce the total recombination frequency. However, rdl1Δ did accentuate the bias towards deletion type recombinants in the rqh1Δ background (Figure 7A and B).
Figure 7.
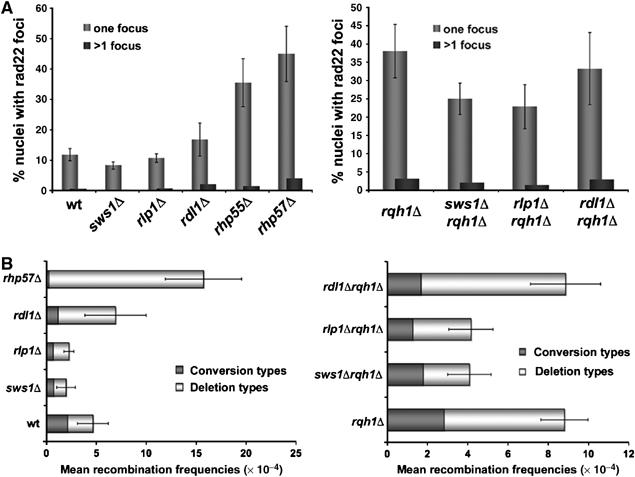
Analysis of the spontaneous rad22-YFP foci formation and recombination levels in different mutant backgrounds. (A) Cells expressing Rad22-YFP from its genomic locus were grown in YES media at 32°C until mid-log phase and their Rad22-YFP foci were quantified. For each strain, more than 300 cells were counted. The average percentage of nuclei containing at least one focus is shown (one focus). Average percentages of nuclei containing two or more foci are also indicated (>1 focus). Note that the values of wt, sws1Δ and rqh1Δ are derived from multiple experiments and are the same as those shown in Figure 4. (B) Spontaneous recombination frequencies of wt (PS3), sws1Δ (VM3731), rlp1Δ (VM3749), rdl1Δ (VM3750), rhp57Δ (VM3748), rqh1Δ (VM3732) sws1Δ rqh1Δ (VM3736), rlp1Δ rqh1Δ (VM3753) and rdl1Δ rqh1Δ (VM3750). Strains containing a non-tandem repeat of ade6− heteroalleles flanking a functional ura4+ gene were used in this study. Recombination frequencies are mean values from three independent assays, and in each assay four independent colonies were tested.
From these results we conclude that Sws1 and Rlp1 have fully interdependent functions in controlling recombination, whereas Rdl1 has a more complex set of functions. As discussed below, these data may indicate that Rdl1 functions at two stages of recombination or has locus-specific functions that are not shared by Sws1 and Rlp1.
Sws1 function is conserved in humans
Finally, we examined whether the putative Sws1 homolog in human cells (Figure 5C) has a role in HR. We first determined whether it associates with RAD51D. HeLa cells were transiently transfected with epitope-tagged constructs of SWS1, RAD51D and XRCC2. Both SWS1 and XRCC2 were detected in immunoprecipitates of RAD51D (Figure 8A). This result suggests that, in addition to its sequence similarity to fission yeast Sws1, human SWS1 shares the ability to associate with RAD51D. (We suspect that SWS1 also associates with XRCC2, but technical problems with a crossreacting protein in the immunoblots interfered with this analysis.) SWS1 failed to coprecipitate with coexpressed XRCC3 and RAD51C (our unpublished data), indicating specificity for its interactions with RAD51 paralogs.
Figure 8.
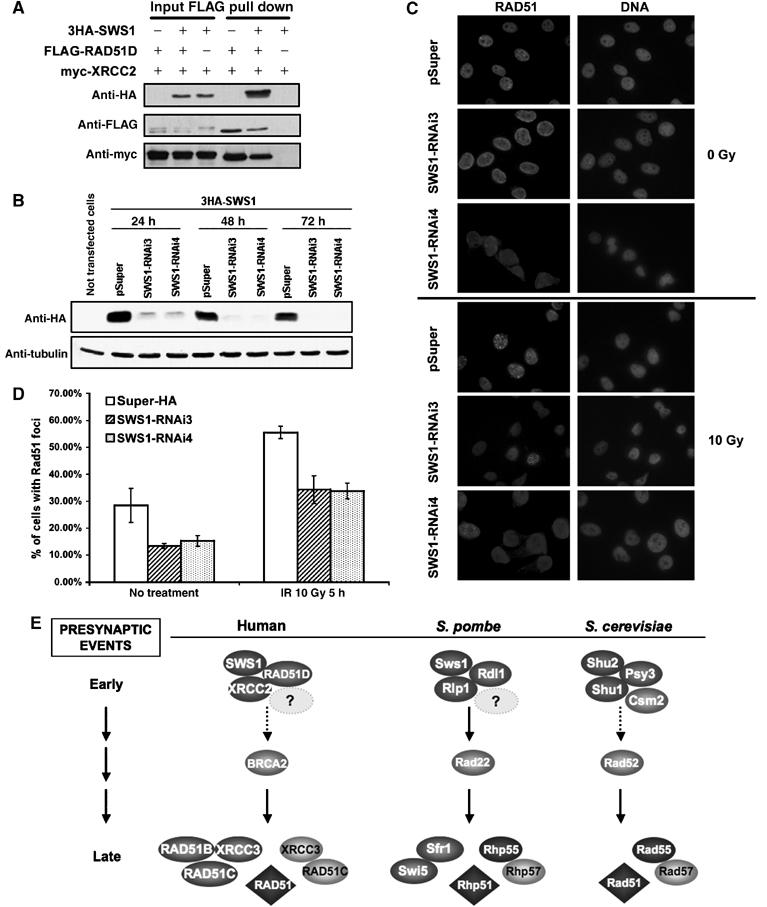
Conservation of Sws1 function in human cells. (A) Physical interaction between 3HA-SWS1, FLAG-RAD51D and myc-XRCC2. HeLa cells were transiently transfected with 3HA-SWS1 and myc-XRCC2 in the presence or absence of FLAG-RAD51D. At 48 h after transfection, lysates and FLAG-immunoprecipitates were probed for the presence of 3HA-SWS1, FLAG-RAD51D and myc-XRCC2. (B) Depletion of 3HA-SWS1 by RNAi. Four pSuper-SWS1-RNAi vectors were constructed to target different regions of the SWS1 coding sequence. Two of these vectors (containing SWS1RNAi3 and SWS1RNAi4) successfully suppressed expression of a cotransfected 3HA-SWS1 construct. Western analyses were performed using antiHA antibodies in cell lysates from cells co-transfected with 3HA-SWS1 and pSUPER vectors containing SWS1RNAi3 or SWS1RNAi4. SWS1 protein levels were substantially reduced when RNAi against SWS1 was used, while no loss of SWS1 was seen when cells were transfected with a control vector (pSuper). Tubulin was used as a loading control. (C, D) Representative Rad51 foci in cells transfected with control (pSuper-HA, which expressed HA siRNA) or pSuper-SWS1RNAi3 and 4. Untreated cells (0 Gy) and cells exposed to gamma radiation (10 Gy) were fixed 5 h after treatment for immunocytochemical analysis of Rad51. Rad51 foci were visualized by immunofluorescence staining with Abcam 13E4 anti-Rad51 monoclonal antibodies (C, left). DAPI counterstaining for cell nuclei is also shown (C, right). Quantification of Rad51 foci (D) shows a decrease in the number of cells containing at least two Rad51 foci both before and after gamma radiation. This reduction in spontaneous and gamma-induced Rad51 foci is associated with reduced SWS1 expression, as it was observed after transfection with two different constructs that cause reduced 3HA-SWS1 expression but not after transfection with the control RNAi vector (pSUPER-HA), which does not affect the number of HeLa cells showing RAD51 foci. 350 cells were counted for each sample. (E) Model for the presynaptic events in HR. Our results support a model in which S. pombe Sws1 acts together with Rlp1 and Rdl1 to promote Rad22 recruitment to the sites of DNA damage. The humans SWS1–RAD51D–XRCC2 is proposed to perform a similar function in recruiting BRCA2. We suggest that the Shu2 group of proteins performs an analogous function in S. cerevisiae. Psy3 is proposed to be the homolog of Rdl1 and RAD51D. Shu1 may be a very distantly related homolog of Rlp1 and XRCC2. Human and fission yeast proteins related to Csm2 have not yet been found. Other proteins known to be involved in HR are also shown. See text for further details. A color version of this figure is available at The EMBO Journal Online.
We reasoned that if human SWS1 was functionally similar to its homolog in fission yeast, its elimination would reduce the frequency of recombination events represented by RAD51 foci. To test this hypothesis, we used RNAi to knockdown expression of human SWS1 mRNA. Four pSuper-SWS1-RNAi vectors were constructed to target different regions of the SWS1 coding sequence. Two of these vectors successfully suppressed expression of a cotransfected 3HA-SWS1 construct (Figure 8B). HeLa cells transfected with the RNAi vectors were then scored for the presence of two or more RAD51 foci. Ablation of SWS1 expression reduced the frequency of RAD51 foci positive cells from ∼28 to ∼14% in undamaged cells (Figure 8B, C and D). SWS1 ablation also had an effect in IR-treated cells, reducing the frequency of RAD51 foci positive cells from ∼55 to ∼35% (Figure 8C and D). Flow cytometry analyses showed that cell cycle distribution 24, 48 or 72 h after transfection was unaffected by suppression of SWS1 expression, as was the abundance of Rad51 detected by immunoblotting (data not shown).
From these results we conclude that human SWS1 has a prorecombinogenic activity that is likely analogous to the activity of its homologs in fission and budding yeasts.
Discussion
Srs2 DNA helicase is a canonical antirecombinase, negatively modulating recombination by disrupting Rad51 filaments (Krejci et al, 2003; Veaute et al, 2003). In vitro, however, the inclusion of the Rad55–57 complex can overcome the inhibitory effect of Srs2 on Rad51-mediated strand exchange (Krejci et al, 2003). Rad52 was also reported to have the same effect (Krejci et al, 2003). The latter observation has at least two nonmutually exclusive interpretations. One is that in vivo Srs2 mainly targets Rad51-DNA nucleofilaments that are devoid of Rad52 and Rad55–Rad57. Alternatively, Srs2 could target other recombination proteins in addition to Rad51. With this possibility in mind, we conducted a two-hybrid screen with the C-terminal region of fission yeast Srs2. This screen uncovered a previously uncharacterized protein that we have named Sws1. Consistent with our hypothesis, elimination of Sws1 reduces recombination in fission yeast. Moreover, sws1Δ suppresses the hyper-recombination phenotypes of srs2Δ and rqh1Δ mutants and rescues the nearly lethal effects of simultaneously inactivating Srs2 and Rqh1. These striking effects contrast with the absence of IR sensitivity of sws1Δ cells. Clearly, Sws1 is not essential for HR events that repair DSBs, but it is culpable for nearly all the toxic recombination structures that form in a srs2Δ rqh1Δ strain. The MMS sensitivity of sws1Δ cells suggests that Sws1 might be involved in channeling a subset of repair events involving oxidative damage to DNA into the HR pathway. From these facts we surmise that Sws1 is a catalyst of a very early step in recombination.
Sws1 shares potentially significant sequence similarity to S. cerevisiae Shu2 and mutants that lack these proteins share a number of interesting phenotypes. Most notably, a shu2Δ mutation partially suppresses the HU sensitivity of an sgs1Δ mutant and reduces the frequency of Rad52 foci (Shor et al, 2005). It is likely, therefore, that Sws1 and Shu2 are highly divergent homologs that have analogous functions in catalyzing an early step in recombination.
Potential function of the Sws1 family in HR
Through an affinity purification scheme we identified two proteins, Rlp1 and Rdl1, in a complex with Sws1. Genetic analyses confirmed that strains defective in these proteins share many phenotypes with sws1Δ cells, indicating that they function in a complex with Sws1. Interestingly, these proteins have significant homology to Rad51 paralogs. Rlp1 is most similar to XRCC2 (Khasanov et al, 2004), while Rdl1 has the closest homology to RAD51D. Mammalian XRCC2 and RAD51D form a heterodimer that has DNA-stimulated ATPase activity (Braybrooke et al, 2000). This activity is thought to require the Walker A and Walker B motifs, implicated in ATP binding and hydrolysis. However, in vivo complementation studies suggest that while the Walker A motif in RAD51D is critical for the function of the complex, the Walker A motif of XRCC2 is largely dispensable (O'Regan et al, 2001; Gruver et al, 2005). These findings may be relevant to the observations that Rlp1 has a Walker A motif but no Walker B motif, and vice versa for Rdl1 (as confirmed by RT–PCR; Supplementary Figure 3). It is conceivable, therefore, that in the fission yeast Rlp1-Rdl1 complex, a functional ATPase activity depends on the individual contributions of a Walker A and a Walker B motif by the separate subunits. Indeed, our mutational studies have shown that the Walker A and Walker B motifs provided by Rlp1 and Rdl1, respectively, are essential for the function of the complex in vivo (Supplementary Figure 4). This situation is reminiscent of the SMC/Rad50 family of ATPases, in which Walker A and Walker B motifs that are separated by huge coiled-coil repeats are juxtaposed in three-dimensional space to form an active ATPase (de Jager et al, 2004).
The Rlp1–Rdl1 potential ATPase complex requires the Zn-finger-like SWIM domain of Sws1 for its in vivo function. This is reminiscent of the bacterial RecFOR ATPase, which contains a Zn-finger motif that is important for its prorecombinogenic function (Lee et al, 2004). This analogy may provide clues about how Sws1 promotes HR-dependent error-free repair. The RecFR ATPase has been implicated in the loading of the single-strand annealing protein RecO onto DNA bound with single-strand binding (SSB) protein. The RecFOR complex then catalyzes the loading of the nucleofilament forming protein RecA, which is the bacterial homolog of Rad51 (Morimatsu and Kowalczykowski, 2003; Kidane and Graumann, 2005). Moreover, several recFOR mutants suppress the hyper-recombination toxicity associated with loss of uvrD, a bacterial Srs2-related helicase (Veaute et al, 2005). In view of the role of RecFR in catalyzing the loading of recO onto DNA, and the importance of the ATPase activity and Zn finger in this process, we suggest that the Sws1–Rlp1–Rdl1 complex in fission yeast may fulfill an analogous role in catalyzing the loading of Rad22 onto RPA coated single-stranded DNA (Figure 8E).
This model might also apply for the XRCC2-RAD51D complex in mammalian cells. The lack of RAD51 paralogs or BRCA2 decreases RAD51 foci formation (van Veelen et al, 2005). We show here that human SWS1 associates with RAD51D. Consistent with this fact, knockdown of SWS1 reduces the number of cells with RAD51 foci. BRCA2 has been proposed to interact with the RAD51 paralogs through its BRC motifs. It is conceivable that an XRCC2–RAD51D–SWS1 complex acts very early in HR and catalyzes the recruitment of BRCA2 to damage sites. Subsequently, BRCA2 catalyzes the loading of RAD51. These observations are consistent with the model that we have proposed for Sws1 in fission yeast, with the difference being that the human XRCC2–RAD51D–SWS1 complex facilitates the loading of BRCA2, instead of Rad22.
Additionally, we show that Sws1 and Rdl1 are related to S. cerevisiae Shu2 and Psy3, respectively. On the basis of these similarities, we propose that Psy3 is in fact a highly divergent homolog of RAD51D and suggest that they function in a complex that possesses ATPase activity. Although not shown here, we have noticed that Shu1 exhibits potentially relevant similarities to the N-terminus of multiple Rad51 paralogs (our unpublished data), thus it is conceivable that Shu1 is an XRCC2/Rlp1-like protein. Mutational and structural studies will be needed to address this question. We cannot speculate on the Csm2 function, as we could not identify any potential homologs in Sws1-associated complexes. In S. cerevisiae, two-hybrid studies indicate that Csm2 associates most tightly with Psy3 (Shor et al, 2005), therefore, MudPIT analysis of Rdl1-associated complexes is expected to provide more insight.
Potential functions of SWIM domains
The SWIM domain, a proposed module for protein–protein or protein–DNA interactions, is most commonly found in multimodular proteins that have ATP binding domains such as translocases and protein kinases (Makarova et al, 2002). We have shown that the SWIM domain is essential for the prorecombinogenic function of Sws1. Sws1 proteins are unique in being small proteins that have no recognizable motifs other than the SWIM domain. However, as we have shown here, Sws1 proteins form a complex with Rad51 paralogs that have ATPase activity. It is therefore tempting to speculate that SWIM domains are involved in regulating some aspect of ATP binding or hydrolysis.
Additional functions of Rdl1
Rdl1 appears to have additional functions not shared with Sws1 and Rlp1. Human RAD51D might also have XRCC2-independent functions. RAD51D is the only RAD51 paralog known to be present at telomeres or in association with the Bloom helicase (Braybrooke et al, 2003; Tarsounas et al, 2004). Moreover, RAD51D knockout embryos only survive 10.5 days postconception while XRCC2 knockout mice die perinatally (Deans et al, 2000; Smiraldo et al, 2005). Biochemical fractionation and yeast two-hybrid studies have indicated the presence of multiple RAD51 paralog complexes: RAD51B–RAD51C, RAD51D–XRCC2, RAD51C–XRCC3, and a larger cocomplex of BCDX2 (Schild et al, 2000; Masson et al, 2001). Similar to RAD51D, Rdl1 might function within two Rad51 paralog complexes: an Rlp1–Sws1 containing complex that acts at an early stage of recombination, and an Rhp55–Rhp57 containing complex that acts at a later stage. The finding that potential homologs of RAD51D and XRCC2 exist in fission yeast and possibly budding yeast paves the way for further structural and functional analysis of Rad51 paralogs and their novel interacting partners.
Materials and methods
Strains, plasmids and media
Details of strains, plasmids, media and growth conditions are provided in Supplementary data. A strain list is provided in Supplementary Table I.
In situ chromatin binding assay and microscopy techniques
Log phase cells were harvested and subjected to Triton X-100 extraction to analyze chromatin association of Sws1 as previously described (Noguchi et al, 2003). DAPI (4′,6′-diamidino-2-phenylindole) was used at 0.5 μg/ml. For green fluorescent protein (GFP) and yellow fluorescent protein (YFP) visualization, exponentially growing cells were photographed using a Nikon Eclipse E800 microscope equipped with a Photometrix Quantix charge-coupled device camera.
Mass-spectrometry
Sws1-TAP protein was purified from fission yeast cells using a previously described method (Saitoh et al, 2002). The resulting peptide mixture was analyzed by multidimensional protein identification technology (MudPIT) as previously described (Boddy et al, 2001).
Recombination assays
Mitotic recombination was assayed using strains containing a non-tandem repeat of ade6 heteroalleles flanking a functional ura4+ gene (Osman et al, 1996). Further details can be found in the Supplementary data.
SWS1 RNA interference (RNAi)
3HASWS1 was cloned into pCDNA3 (Invitrogen). Two SWS1 19-nucleotide-long regions were selected and cloned into pSuper (nucleotide sequences and further information can be found in the Supplementary data).
Western blot and immunofluorescence analyses
Protocols used for whole-cell extract preparation from S. pombe and HeLa cells, as well as for preparation of HeLa cells for immunofluorescence studies are available in the Supplementary data.
Supplementary Material
Supplementary Information
Supplementary Table I
Supplementary Table II
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Acknowledgments
We thank members of the Russell laboratory and the Cell Cycle Groups at TSRI, particularly Li-Lin Du, Pierre H Gaillard and M Nick Boddy for helpful comments and discussions. This research was supported by Grants NIH (EY1328801) and MERK-MGRI-241 awarded to JRY and NIH Grants CA77325 and GM59447 awarded to PR.
References
- Boddy MN, Gaillard PH, McDonald WH, Shanahan P, Yates JR III, Russell P (2001) Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107: 537–548 [DOI] [PubMed] [Google Scholar]
- Braybrooke JP, Li JL, Wu L, Caple F, Benson FE, Hickson ID (2003) Functional interaction between the Bloom's syndrome helicase and the RAD51 paralog, RAD51L3 (RAD51D). J Biol Chem 278: 48357–48366 [DOI] [PubMed] [Google Scholar]
- Braybrooke JP, Spink KG, Thacker J, Hickson ID (2000) The RAD51 family member, RAD51L3, is a DNA-stimulated ATPase that forms a complex with XRCC2. J Biol Chem 275: 29100–29106 [DOI] [PubMed] [Google Scholar]
- Coulon S, Gaillard PH, Chahwan C, McDonald WH, Yates JR III, Russell P (2004) Slx1–Slx4 are subunits of a structure-specific endonuclease that maintains ribosomal DNA in fission yeast. Mol Biol Cell 15: 71–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deans B, Griffin CS, Maconochie M, Thacker J (2000) Xrcc2 is required for genetic stability, embryonic neurogenesis and viability in mice. EMBO J 19: 6675–6685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe CL, Dixon J, Osman F, Whitby MC (2000) Partial suppression of the fission yeast rqh1− phenotype by expression of a bacterial Holliday junction resolvase. EMBO J 19: 2751–2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe CL, Osman F, Dixon J, Whitby MC (2004) DNA repair by a Rad22–Mus81-dependent pathway that is independent of Rhp51. Nucleic Acids Res 32: 5570–5581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe CL, Whitby MC (2004) The involvement of Srs2 in post-replication repair and homologous recombination in fission yeast. Nucleic Acids Res 32: 1480–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du LL, Nakamura TM, Moser BA, Russell P (2003) Retention but not recruitment of Crb2 at double-strand breaks requires Rad1 and Rad3 complexes. Mol Cell Biol 23: 6150–6158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre F, Chan A, Heyer WD, Gangloff S (2002) Alternate pathways involving Sgs1/Top3, Mus81/Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc Natl Acad Sci USA 99: 16887–16892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff S, Soustelle C, Fabre F (2000) Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat Genet 25: 192–194 [DOI] [PubMed] [Google Scholar]
- Gruver AM, Miller KA, Rajesh C, Smiraldo PG, Kaliyaperumal S, Balder R, Stiles KM, Albala JS, Pittman DL (2005) The ATPase motif in RAD51D is required for resistance to DNA interstrand crosslinking agents and interaction with RAD51C. Mutagenesis 20: 433–440 [DOI] [PubMed] [Google Scholar]
- Hickson ID (2003) RecQ helicases: caretakers of the genome. Nat Rev Cancer 3: 169–178 [DOI] [PubMed] [Google Scholar]
- Hope JC, Maftahi M, Freyer GA (2005) A postsynaptic role for Rhp55/57 that is responsible for cell death in Deltarqh1 mutants following replication arrest in Schizosaccharomyces pombe. Genetics 170: 519–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ME, Rio AG, Nicolas A, Kolodner RD (2003) A genomewide screen in Saccharomyces cerevisiae for genes that suppress the accumulation of mutations. Proc Natl Acad Sci USA 100: 11529–11534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jager M, Trujillo KM, Sung P, Hopfner KP, Carney JP, Tainer JA, Connelly JC, Leach DR, Kanaar R, Wyman C (2004) Differential arrangements of conserved building blocks among homologs of the Rad50/Mre11 DNA repair protein complex. J Mol Biol 339: 937–949 [DOI] [PubMed] [Google Scholar]
- Khasanov FK, Salakhova AF, Chepurnaja OV, Korolev VG, Bashkirov VI (2004) Identification and characterization of the rlp1+, the novel Rad51 paralog in the fission yeast Schizosaccharomyces pombe. DNA Repair 3: 1363–1374 [DOI] [PubMed] [Google Scholar]
- Kidane D, Graumann PL (2005) Dynamic formation of RecA filaments at DNA double strand break repair centers in live cells. J Cell Biol 170: 357–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein HL (2001) Mutations in recombinational repair and in checkpoint control genes suppress the lethal combination of srs2Delta with other DNA repair genes in Saccharomyces cerevisiae. Genetics 157: 557–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, Klein H, Ellenberger T, Sung P (2003) DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423: 305–309 [DOI] [PubMed] [Google Scholar]
- Krogh BO, Symington LS (2004) Recombination proteins in yeast. Annu Rev Genet 38: 233–271 [DOI] [PubMed] [Google Scholar]
- Laursen LV, Ampatzidou E, Andersen AH, Murray JM (2003) Role for the fission yeast RecQ helicase in DNA repair in G2. Mol Cell Biol 23: 3692–3705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Johnson RE, Yu SL, Prakash L, Prakash S (1999) Requirement of yeast SGS1 and SRS2 genes for replication and transcription. Science 286: 2339–2342 [DOI] [PubMed] [Google Scholar]
- Lee BI, Kim KH, Park SJ, Eom SH, Song HK, Suh SW (2004) Ring-shaped architecture of RecR: implications for its role in homologous recombinational DNA repair. EMBO J 23: 2029–2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, St Onge RP, Proctor M, Flaherty P, Jordan MI, Arkin AP, Davis RW, Nislow C, Giaever G (2005) Genome-wide requirements for resistance to functionally distinct DNA-damaging agents. PLoS Genet 1: e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Barlow JH, Burgess RC, Rothstein R (2004) Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118: 699–713 [DOI] [PubMed] [Google Scholar]
- Makarova KS, Aravind L, Koonin EV (2002) SWIM, a novel Zn-chelating domain present in bacteria, archaea and eukaryotes. Trends Biochem Sci 27: 384–386 [DOI] [PubMed] [Google Scholar]
- Masson JY, Tarsounas MC, Stasiak AZ, Stasiak A, Shah R, McIlwraith MJ, Benson FE, West SC (2001) Identification and purification of two distinct complexes containing the five RAD51 paralogs. Genes Dev 15: 3296–3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimatsu K, Kowalczykowski SC (2003) RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: a universal step of recombinational repair. Mol Cell 11: 1337–1347 [DOI] [PubMed] [Google Scholar]
- Noguchi E, Noguchi C, Du LL, Russell P (2003) Swi1 prevents replication fork collapse and controls checkpoint kinase Cds1. Mol Cell Biol 23: 7861–7874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Regan P, Wilson C, Townsend S, Thacker J (2001) XRCC2 is a nuclear RAD51-like protein required for damage-dependent RAD51 focus formation without the need for ATP binding. J Biol Chem 276: 22148–22153 [DOI] [PubMed] [Google Scholar]
- Osman F, Fortunato EA, Subramani S (1996) Double-strand break-induced mitotic intrachromosomal recombination in the fission yeast Schizosaccharomyces pombe. Genetics 142: 341–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F, Haber JE (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 63: 349–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh S, Chabes A, McDonald WH, Thelander L, Yates JR, Russell P (2002) Cid13 is a cytoplasmic poly(A) polymerase that regulates ribonucleotide reductase mRNA. Cell 109: 563–573 [DOI] [PubMed] [Google Scholar]
- Schild D, Lio YC, Collins DW, Tsomondo T, Chen DJ (2000) Evidence for simultaneous protein interactions between human Rad51 paralogs. J Biol Chem 275: 16443–16449 [DOI] [PubMed] [Google Scholar]
- Shor E, Weinstein J, Rothstein R (2005) A genetic screen for top3 suppressors in Saccharomyces cerevisiae identifies SHU1, SHU2, PSY3 and CSM2: four genes involved in error-free DNA repair. Genetics 169: 1275–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smiraldo PG, Gruver AM, Osborn JC, Pittman DL (2005) Extensive chromosomal instability in Rad51d-deficient mouse cells. Cancer Res 65: 2089–2096 [DOI] [PubMed] [Google Scholar]
- Stewart E, Chapman CR, Al-Khodairy F, Carr AM, Enoch T (1997) rqh1+, a fission yeast gene related to the Bloom's and Werner's syndrome genes, is required for reversible S phase arrest. EMBO J 16: 2682–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N, Wang X, Haber JE (2003) In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol Cell 12: 209–219 [DOI] [PubMed] [Google Scholar]
- Sung P (1997a) Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J Biol Chem 272: 28194–28197 [DOI] [PubMed] [Google Scholar]
- Sung P (1997b) Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev 11: 1111–1121 [DOI] [PubMed] [Google Scholar]
- Sung P, Krejci L, Van Komen S, Sehorn MG (2003) Rad51 recombinase and recombination mediators. J Biol Chem 278: 42729–42732 [DOI] [PubMed] [Google Scholar]
- Swanson RL, Morey NJ, Doetsch PW, Jinks-Robertson S (1999) Overlapping specificities of base excision repair, nucleotide excision repair, recombination, and translesion synthesis pathways for DNA base damage in Saccharomyces cerevisiae. Mol Cell Biol 19: 2929–2935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarsounas M, Munoz P, Claas A, Smiraldo PG, Pittman DL, Blasco MA, West SC (2004) Telomere maintenance requires the RAD51D recombination/repair protein. Cell 117: 337–347 [DOI] [PubMed] [Google Scholar]
- Tsutsui Y, Khasanov FK, Shinagawa H, Iwasaki H, Bashkirov VI (2001) Multiple interactions among the components of the recombinational DNA repair system in Schizosaccharomyces pombe. Genetics 159: 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Brabant AJ, Ye T, Sanz M, German IJ, Ellis NA, Holloman WK (2000) Binding and melting of D-loops by the Bloom syndrome helicase. Biochemistry 39: 14617–14625 [DOI] [PubMed] [Google Scholar]
- van Veelen LR, Essers J, van de Rakt MW, Odijk H, Pastink A, Zdzienicka MZ, Paulusma CC, Kanaar R (2005) Ionizing radiation-induced foci formation of mammalian Rad51 and Rad54 depends on the Rad51 paralogs, but not on Rad52. Mutat Res 574: 34–49 [DOI] [PubMed] [Google Scholar]
- Veaute X, Delmas S, Selva M, Jeusset J, Le Cam E, Matic I, Fabre F, Petit MA (2005) UvrD helicase, unlike Rep helicase, dismantles RecA nucleoprotein filaments in Escherichia coli. EMBO J 24: 180–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F (2003) The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423: 309–312 [DOI] [PubMed] [Google Scholar]
- Wang SW, Goodwin A, Hickson ID, Norbury CJ (2001) Involvement of Schizosaccharomyces pombe Srs2 in cellular responses to DNA damage. Nucleic Acids Res 29: 2963–2972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn MP, Wolters D, Yates JR III (2001) Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol 19: 242–247 [DOI] [PubMed] [Google Scholar]
- West SC (2003) Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol 4: 435–445 [DOI] [PubMed] [Google Scholar]
- Wu L, Hickson ID (2003) The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature 426: 870–874 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Table I
Supplementary Table II
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4


