Abstract
Extended-spectrum β-lactamase (ESBL)-mediated resistance is of considerable importance in human medicine. Recently, such enzymes have been reported in bacteria from animals. We describe a longitudinal study of a dairy farm suffering calf scour with high mortality rates. In November 2004, two Escherichia coli isolates with resistance to a wide range of β-lactams (including amoxicillin-clavulanate and cefotaxime) were isolated from scouring calves. Testing by PCR and sequence analysis confirmed the isolates as being both blaCTX-M14/17 and blaTEM-35 (IRT-4) positive. They had indistinguishable plasmid and pulsed-field gel electrophoresis (PFGE) profiles. Transferability studies demonstrated that blaCTX-M was located on a conjugative 65-MDa IncK plasmid. Following a farm visit in December 2004, 31/48 calves and 2/60 cows were positive for E. coli with blaCTX-M. Also, 5/48 calf and 28/60 cow samples yielded blaCTX- and blaTEM-negative E. coli isolates that were resistant to cefotaxime, and sequence analysis confirmed that these presented mutations in the promoter region of the chromosomal ampC gene. Fingerprinting showed 11 different PFGE types (seven in blaCTX-M-positive isolates). Six different PFGE clones conjugated the same blaCTX-M-positive IncK plasmid. One clone carried a different-sized, blaCTX-M-positive, transformable plasmid. This is the first report of blaCTX-M from livestock in the United Kingdom, and this report demonstrates the complexity of ESBL epidemiology. Results indicate that horizontal plasmid transfer between strains as well as horizontal gene transfer between plasmids have contributed to the spread of resistance. We have also shown that some clones can persist for months, suggesting that clonal spread also contributes to the perpetuation of resistance.
Cefotaximases (CTX-M) are class A β-lactamases produced by a wide range of bacteria of clinical significance and in general present higher levels of hydrolytic activity against cefotaxime than against ceftazidime. CTX-M enzymes comprise a genetically diverse (five groups designated 1, 2, 8, 9, and 25 have been described) and rapidly growing family. These are the predominant extended-spectrum β-lactamases (ESBL) in human infections in parts of Eastern Europe, South America, and Japan. In the United Kingdom, the first report of a CTX-M enzyme was in a human clinical isolate of Klebsiella oxytoca with blaCTX-M9 in 2000 (1). Since then, CTX-M-producing Escherichia coli has emerged as a significant and developing problem, occurring in patients in the community as well as in those with recent hospital contact (16, 19).
In spite of this human situation, worldwide, there have been only a few recent reports of CTX-M-producing bacteria in animals (3, 4, 10, 11, 25, 29). It is unclear whether it is primarily human or veterinary use of antibiotics that has provided the selective pressures necessary for this situation. In a recent letter, we described the first reported isolation of CTX-M-producing E. coli from cattle in the United Kingdom. These resistant E. coli isolates were unlikely to be associated with the scouring problem at the farm, and other known enteric pathogens were identified during investigations (27). In the present study, the farm involved was investigated further for the occurrence of CTX-M-producing E. coli, and a longitudinal study was conducted with the aim of clarifying the molecular epidemiology of this resistance at the farm level.
MATERIALS AND METHODS
Description of farm sampling.
In November 2004, we obtained four E. coli isolates from scour samples originating from calves at a dairy farm (27), and these isolates were subjected to detailed phenotypic and genotypic characterization. A longitudinal study of the farm was designed, and the establishment was sampled on three occasions: December 2004, February 2005, and July 2005. The farm had an average population of more than 125 milking cows and more than 60 calves at any given sampling time. On each sampling visit, individual fecal samples (100 g of fresh fecal mass) were taken from randomly selected adult cows, and rectal swabs were taken from all young calves present at the farm. The sample size was arranged to be greater than that required to give 95% certainty of obtaining at least one positive sample if the prevalence is 10%. On the second and third visits, a small number of environmental samples (floor swabs, slurry, feed, and water) were also collected.
Bacteriology.
Fecal samples were directly streaked onto CHROMagar ECC (M-Tech Diagnostics) containing cefotaxime (1 μg/ml) using a cotton swab and incubated aerobically overnight at 37°C. Negative samples were enriched in buffered peptone water for 18 h at 37°C before being plated onto the same selective medium. Presumptive E. coli isolates were selected by colony morphology and color (blue colonies) and subcultured on the same selective medium for subsequent analysis. The identity of the presumptive E. coli isolates was confirmed by gadA PCR (18). The four original November 2004 E. coli isolates as well as one representative isolate from each positive sample from each of the three sampling visits were selected for further testing.
Genetic characterization of blaCTX-M genes and molecular typing.
The E. coli isolates selected were tested for the presence of blaCTX-M β-lactamase genes by PCR with primers CTX-M universal F (5′-CGA TGT GCA GTA CCA GTA A-3′) and R (5′-TTA GTG ACC AGA ATC AGC GG-3′). Identification of the specific blaCTX-M gene was carried out by sequencing of the entire CTX-M amplicon generated with specific primers designed for the CTX-M-9 group (F, 5′-ATG GTG ACA AAG AGA GTG CAA C-3′; R, 5′-TTA CAG CCC TTC GGC GAT G-3′). The PCR conditions were described previously (2). All the isolates were also typed by XbaI-pulsed-field gel electrophoresis (PFGE) as described by the Centers for Disease Control and Prevention (9) and by plasmid profiling according to methods previously described (14).
For those isolates that were cefotaxime resistant but CTX-M negative, a representative isolate from each PFGE clone from visits 1 and 2 was tested for mutations in the ampC promoter gene with primers AB1 and ampC2 (8).
The isolates obtained in November 2004 were also tested by blaTEM PCR with primers TEM-F (5′-ATT CTT GAA GAC GAA AGG GC-3′) and TEM-R (5′-ACG CTC AGT GGA ACG AAA AC-3′), and the amplicons generated were sequenced as described previously (13).
Determination of resistance phenotype.
Susceptibility to a panel of 12 β-lactams plus 13 other antibiotics (ampicillin, amoxicillin-clavulanic acid, ceftiofur, cefuroxime, ceftazidime, cefotaxime, ceftriaxone, cefoperazone, cefoxitin, cefpodixime, aztreonam, imipenem, amikacin, apramycin, chloramphenicol, colistin sulfate, furazolidone, gentamicin, nalidixic acid, ciprofloxacin, neomycin, streptomycin, sulfamethoxazole-trimethoprim, tetracycline, and sulfonamide) was determined by a disk diffusion method as described by CLSI (formerly NCCLS) guidelines (20). E. coli strain NCTC10418, which is sensitive to all the antibiotics, was used for quality control purposes.
Assessment of transferability of resistance.
Conjugations were performed with each representative PFGE clone found in the first visit and rifampin-resistant recipient Salmonella enterica serovar Typhimurium DT36 26R755 using in-broth methods (14). Conjugation mixtures were plated onto CHROMagar ECC (M-Tech Diagnostics) containing rifampin (100 mg/liter) and cefotaxime (1 mg/liter) and then incubated for 24 and 48 h at 37°C. When transfer was not achieved by conjugation, transformation experiments were conducted as follows. Plasmid DNA was prepared with a QIAGEN High Speed MIDI kit. ElectroMAX DH10B cells (Invitrogen) were transformed with 60 ng of DNA using a Bio-Rad GenePulser II electroporator under standard conditions (2 kV, 200 Ω, and 25 μF). Transformants were selected on nutrient agar containing 1 mg/liter cefotaxime after 16 h of incubation at 37°C.
Plasmid characterization.
Transferable plasmids were purified from transconjugants/transformants with a QIAGEN High Speed MIDI kit. For restriction fragment length polymorphism (RFLP), 2 μg of plasmid DNA was digested with HpaI and SmaI (Promega), and the resulting products were separated by electrophoresis on 0.8% agarose gels at 45 V for 20 h at 20°C. Determination of incompatibility groups was conducted by PCR-based replicon typing as described previously (7).
RESULTS
Characterization of the initial E. coli isolates.
The isolates from November 2004 had identical phenotypes showing resistance to ampicillin, amoxicillin-clavulanic acid, ceftiofur, cefuroxime, cefotaxime, ceftriaxone, cefoperazone, cefpodixime, chloramphenicol, streptomycin, sulfamethoxazole-trimethoprim, tetracycline, sulfonamide, nalidixic acid, and ciprofloxacin. They were all CTX-M positive, and sequence analysis confirmed the presence of a blaCTX-M17/18 gene. In addition, they carried blaTEM-35 (IRT-4). The isolates had indistinguishable PFGE and plasmid profiles.
Longitudinal study of the occurrence of cefotaxime-resistant E. coli isolates in cattle fecal samples.
Table 1 shows a summary of results from the isolation of E. coli on cefotaxime plates. The proportion of positive calf samples ranged from 62% at visit 2 to 100% at visit 3. The estimated prevalences (and exact 95% confidence intervals) were 64.6% (53.9 to 74.1%) at visit 1, 61.8% (47.1 to 74.8%) at visit 2, and 92.7% (83.1 to 97.5%) at visit 3. In the milking cow population, the proportion of positive samples was 15% at visit 2 and 50% at visit 1. The estimated prevalences (and exact 95% confidence intervals) were 3.3% (0.5 to 11.2%) at visit 1, 5.9% (1.3 to 15.9%) at visit 2, and 23.8% (14.3 to 35.8%) at visit 3. If we assume that the results on the three sampling occasions are independent, Fisher's exact test can be used to compare the proportions that are positive. This gives a P value of 0.0012 for the calves and a P value of 0.0007 for the cows; clearly, the prevalences on visit 3 are higher than those on the other visits. CTX-M-producing E. coli isolates were also isolated from floor samples from a calf house, collecting yard, and boot change area as well as from a slurry subsample of all of them taken during visit 3. At every visit, a number of cefotaxime-resistant isolates were CTX-M negative; these also showed resistance to cefoxitin, indicating possible hyperproduction of AmpC.
TABLE 1.
Occurrence of CTX-M-positive and -negative cefotaxime-resistant isolates
| Visit (date) | Sample type (no.a) | CA-positive samplesb
|
||
|---|---|---|---|---|
| No. CTX-M PCR positive | No. CTX-M PCR negative | No. of PFGE clonesc | ||
| 1 (December 2004) | Calves (48) | 31 | 5 | 9 (6) |
| Cows (60) | 2 | 28 | ||
| Environment (0) | ||||
| 2 (February 2005) | Calves (34) | 21 | 0 | 9 (6) |
| Cows (51) | 3 | 5 | ||
| Environment (7) | 0 | 0 | ||
| 3 (July 2005) | Calves (41) | 38 | 3 | 18 (13) |
| Cows (63) | 15 | 11 | ||
| Environment (16) | 4 | 1 | ||
Total number of calves, cows, or environmental samples taken at the farm.
Samples positive for E. coli on CHROMagar ECC plus 1 mg/liter cefotaxime.
Number of PFGE clonal groups identified among cefotaxime-resistant isolates. Numbers in parentheses indicate how many clones were found among CTX-M-positive isolates.
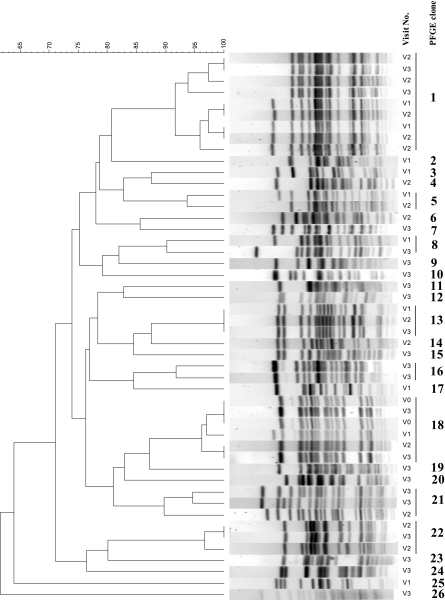
PFGE analysis showed that a diversity of clones (defined as PFGE profiles with a similarity percentage of ≥90%) was evident on each of the sampling visits to the farm, with 9, 9, and 18 different clones identified on visits 1, 2, and 3, respectively. The majority of these PFGE clones were found among CTX-M-positive isolates. Figure 1 shows a representation of the different PFGE clones identified during the three visits; using this figure, it is possible to identify the seven clonal groups (types 1, 5, 8, 13, 18, 21, and 22) that were found at more than one visit. The most persistent PFGE clone (type 18) persisted from November 2004 until July 2005. Isolates representing all PFGE types from visit 1 were also tested by conventional serotyping and belonged to types O40:K+, O33:K+, O1:K+, O20:K+, and O8:K+. Sequence analysis of the ampC promoter region of representative isolates from each of the PFGE types in visits 1 and 2 with a typical ampC resistance phenotype showed that five clones presented mutations at positions −42 (C to T), −82 (A to G), −88 (C to T), −18 (G to A), −1 (C to T), and +58 (C to T), and the remaining clone had mutations at positions +22 (C to T), +26 (T to G), +27 (A to T), +32 (G to A), and +70 (C to T).
FIG. 1.
Image generated by Bionumerics software showing the XbaI-PFGE clonal groups identified among cefoxitin-resistant isolates from visits 1 (V1), 2, and 3. A cutoff similarity value of 90% was used to define clones. The cluster analysis was performed by using the Dice coefficient and the unweighted-pair group method with arithmetic averages (optimization of 0% and band position tolerance of 2%).
Location of the CTX-M gene.
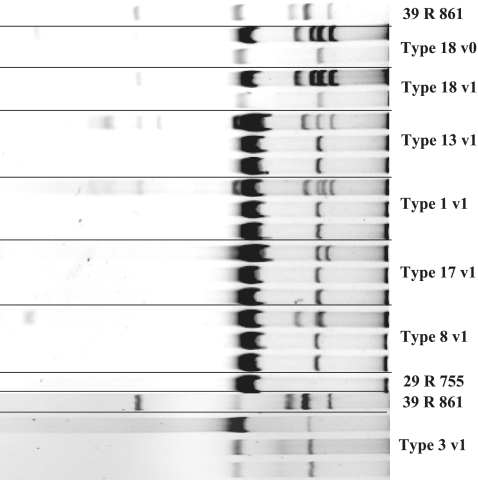
One of the original E. coli isolates as well as a representative isolate from each of the six PFGE clones within CTX-M-positive isolates from visit 1 was tested by conjugation experiments. In all but one of the PFGE types, a single plasmid of approximately 65 MDa was transferred to the Salmonella 39R755 recipient (Fig. 2). All plasmids were of an indistinguishable RFLP type and belonged to incompatibility group IncK. For one isolate (PFGE type 3), the resistance to cefotaxime could be transferred only by transformation with recipient cells acquiring a slightly smaller plasmid (Fig. 2). This plasmid was untypeable by PCR-based replicon typing. Sequence analysis is under way to identify the replicon present on this plasmid. PCR analysis of the transconjugants and transformant showed that the blaCTX-M genetic determinants were present in the specific plasmids acquired by the recipient strains. Antimicrobial susceptibility testing of the transconjugants and transformant showed that only the expected resistance to β-lactam antibiotics and to streptomycin was acquired with the transferred plasmids.
FIG. 2.
Plasmid profiles of isolates representing PFGE clones from visit 1 (v1), including type 18 (lanes 2 and 4), type 13 (lane 6), type 1 (lane 9), type 8 (lane 12), type 17 (lane 15), and type 3 (lane 20), and of transconjugants (lanes 3, 5, 7, 8, 10, 11, 13, 14, 16, and 17) and transformants (lanes 21 and 22). Reference strain E. coli 39R861 contains plasmids of 98, 42, 23.9, and 4.6 MDa. Reference strain S. enterica serovar Typhimurium 29R755 is a plasmid-free strain used as a recipient.
DISCUSSION
The first report of the CTX-M enzyme in the United Kingdom was in a clinical isolate of Klebsiella oxytoca with blaCTX-M9 (1). This enzyme was also found in Salmonella in a recent retrospective survey. Also, in the same study, and belonging to CTX-M group 9, we found blaCTX-M17-18 in several Salmonella serotypes (2). CTX-M group 9 enzymes are also the most common group found in other countries in Europe (28). However, it has recently become apparent that the predominant enzyme in the United Kingdom in humans seems to be CTX-M-15 (16). Our study represents the first report of a CTX-M enzyme from livestock in the United Kingdom.
We have shown that CTX-M enzymes are present in a farm environment and that they have spread among the E. coli intestinal flora of different animals of different age groups. We have also shown that the spread of this resistance is associated with a highly promiscuous plasmid. Plasmid analysis has indicated that horizontal transfer of these elements is a very efficient mechanism involved in the spread of CTX-M-mediated resistance at this farm. The same RFLP- and IncK-type conjugative plasmids are circulating among many different E. coli clones. Furthermore, in a single clone, we also found a smaller plasmid of an unknown replicon type with the CTX-M gene. This indicates that transfer of the genes between different plasmids is also a mechanism that contributes to dissemination of CTX-M resistance. Replicon typing of these plasmids, and of other CTX-M-harboring plasmids previously isolated from Salmonella strains of human origin in the United Kingdom (data not shown), has revealed that the plasmids found in these animal strains do not belong to the same Inc types as those found in the human Salmonella strains. The only other report of a CTX-M-14-like enzyme from animals came from poultry in Spain (5), although information on the replicon type of the plasmid is not available, and therefore, it is difficult to assess whether or not similar plasmids may be circulating in animal populations in several geographical areas.
Once one of these mobile elements is acquired, clonal spread is also a likely mechanism for the perpetuation of resistance. We have clearly shown that specific CTX-M-positive E. coli clones are able to persist for months. On the farm investigated here, various antibiotics had been used to treat clinical disease problems, including amoxicillin-clavulanic acid and marbofloxacin in scouring calves and cefquinome for mastitis treatment of milking cows. After the first visit in December 2004, all use of β-lactam antibiotics (apart from intramammary administration to lactating cattle) was stopped on the farm in an attempt to remove the selective pressure encouraging the persistence of this resistance. However, this measure has not had the desired effect, and CTX-M resistance persisted both in animals and in the farm environment for the duration of this study. Antibiograms of the transconjugants and transformant have shown that the plasmids confer resistance only to the expected β-lactams (ampicillin, ceftiofur, cefuroxime, cefotaxime, ceftriaxone, cefoperazone, and cefpodoxime) and to streptomycin. Therefore, coselection by the use of other antimicrobials may not play an important role in the maintenance of blaCTX-M-mediated resistance. At present, the mechanisms encouraging the persistence and spread of this plasmid and the CTX-M resistance it carries are unknown. We will attempt to continue to sample this farm to ascertain the evolution of the problem.
The ease with which different E. coli clones seem to be acquiring this transmissible plasmid may represent a significant risk of the spread of this resistance and eventually of an increasing incidence of resistant pathogens (21, 26). Although Salmonella enterica serovar Dublin infection was confirmed in some of the animals on the farm from which CTX-M-positive E. coli isolates were isolated, all the isolates investigated have been CTX-M negative. Nonpathogenic multidrug-resistant E. coli in the intestine is an important reservoir of resistance genes (6, 24), and these E. coli isolates of animal origin may colonize the human intestine at least temporarily (15, 17, 23). Some studies have shown that the same R plasmids can be transferred between bacterial strains from bovines and humans (22) and that transfer of R plasmids between bovine, porcine, and human E. coli strains can occur in natural microenvironments such as milk or meat (12). The close proximity among animals and their environment and humans makes it possible that these genes could be exchanged between microbial populations of different origins. It is also theoretically possible that if strict hygienic measures are not implemented, organisms carrying these genes could be entering the human food chain. The ease of transfer of this particular plasmid in the farm environment is a reminder of the importance of biosecurity measures at the farm and of proper hygiene during abattoir slaughtering and food processing.
In summary, we have reported the isolation of the first ESBL-producing animal strain in the United Kingdom. The farm had a previous history of using β-lactam antibiotics, and E. coli isolates with at least three different β-lactam enzymes were found on this farm (CTX-M, IRT, and AmpC). The resistance has persisted due to both horizontal gene transfer and clonal spread of resistant organisms. Surveys are needed to assess whether this is a unique situation on a single farm or if ESBL-mediated resistance may be emerging within animal populations in the United Kingdom.
Acknowledgments
This work was funded by the Department for Environment, Food, and Rural Affairs, United Kingdom, project VM02136.
REFERENCES
- 1.Alobwede, I., F. H. M'Zali, D. M. Livermore, J. Heritage, N. Todd, and P. M. Hawkey. 2003. CTX-M extended-spectrum beta-lactamase arrives in the UK. J. Antimicrob. Chemother. 51:470-471. [DOI] [PubMed] [Google Scholar]
- 2.Batchelor, M., K. Hopkins, E. J. Threlfall, F. A. Clifton-Hadley, A. D. Stallwood, R. H. Davies, and E. Liebana. 2005. blaCTX-M genes in clinical Salmonella isolates recovered from humans in England and Wales from 1992 to 2003. Antimicrob. Agents Chemother. 49:1319-1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Briñas, L., M. A. Moreno, T. Teshager, Y. Sáenz, M. C. Porrero, L. Domínguez, C. Torres, M. Zarazaga, and C. Porrero. 2005. Monitoring and characterization of extended-spectrum β-lactamases in Escherichia coli strains from healthy and sick animals in Spain in 2003. Antimicrob. Agents Chemother. 49:1262-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brinas, L., M. A. Moreno, T. Teshager, M. Zarazaga, Y. Saenz, C. Porrero, L. Dominguez, and C. Torres. 2003. Beta-lactamase characterization in Escherichia coli isolates with diminished susceptibility or resistance to extended-spectrum cephalosporins recovered from sick animals in Spain. Microb. Drug Resist. 9:201-209. [DOI] [PubMed] [Google Scholar]
- 5.Briñas, L., M. A. Moreno, M. Zarazaga, C. Porrero, Y. Sáenz, M. García, L. Dominguez, and C. Torres. 2003. Detection of CMY-2, CTX-M-14, and SHV-12 β-lactamases in Escherichia coli fecal-sample isolates from healthy chickens. Antimicrob. Agents Chemother. 47:2056-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calva, J. J., J. Sifuentes-Osornio, and C. Ceron. 1996. Antimicrobial resistance in fecal flora: longitudinal community-based surveillance of children from urban Mexico. Antimicrob. Agents Chemother. 40:1699-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carattoli, A., A. Bertini, L. Villa, V. Falbo, K. L. Hopkins, and E. J. Threlfall. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219-228. [DOI] [PubMed] [Google Scholar]
- 8.Caroff, N., E. Espaze, D. Gautreau, H. Richet, and A. Reynaud. 2000. Analysis of the effects of −42 and −32 ampC promoter mutations in clinical isolates of Escherichia coli hyperproducing ampC. J. Antimicrob. Chemother. 45:783-788. [DOI] [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention. 2002. Standardized molecular subtyping of foodborne bacterial pathogens by pulsed-field gel electrophoresis. Centers for Diseases Control and Prevention, Atlanta, Ga.
- 10.Costa, D., P. Poeta, L. Brinas, Y. Saenz, J. Rodrigues, and C. Torres. 2004. Detection of CTX-M-1 and TEM-52 β-lactamases in Escherichia coli strains from healthy pets in Portugal. J. Antimicrob. Chemother. 54:960-961. [DOI] [PubMed] [Google Scholar]
- 11.Hasman, H., D. Mevius, K. Veldman, I. Olesen, and F. M. Aarestrup. 2005. β-Lactamases among extended-spectrum β-lactamase (ESBL)-resistant Salmonella from poultry, poultry products and human patients in The Netherlands. J. Antimicrob. Chemother. 56:115-121. [DOI] [PubMed] [Google Scholar]
- 12.Kruse, H., and H. Sorum. 1994. Transfer of multiple drug resistance plasmids between bacteria of diverse origins in natural microenvironments. Appl. Environ. Microbiol. 60:4015-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liebana, E., M. Batchelor, C. Torres, L. Brinas, L. A. Lagos, B. Abdalhamid, N. D. Hanson, and J. Martinez-Urtaza. 2004. Pediatric infection due to multiresistant Salmonella enterica serotype Infantis in Honduras. J. Clin. Microbiol. 42:4885-4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liebana, E., M. Gibbs, C. Clouting, L. Barker, F. A. Clifton-Hadley, E. Pleydell, B. Abdalhamid, N. D. Hanson, L. Martin, C. Poppe, and R. H. Davies. 2004. Characterisation of beta-lactamases responsible for resistance to extended-spectrum cephalosporins in Escherichia coli and Salmonella enterica strains from food-producing animals in the United Kingdom. Microb. Drug Resist. 10:1-9. [DOI] [PubMed] [Google Scholar]
- 15.Linton, A. H., K. Howe, P. M. Bennett, M. H. Richmond, and E. J. Whiteside. 1977. The colonization of the human gut by antibiotic resistant Escherichia coli from chickens. J. Appl. Bacteriol. 43:465-469. [DOI] [PubMed] [Google Scholar]
- 16.Livermore, D. M., and P. M. Hawkey. 2005. CTX-M: changing the face of ESBLs in the UK. J. Antimicrob. Chemother. 56:451-454. [DOI] [PubMed] [Google Scholar]
- 17.Marshall, B., D. Petrowski, and S. B. Levy. 1990. Inter- and intraspecies spread of Escherichia coli in a farm environment in the absence of antibiotic usage. Proc. Natl. Acad. Sci. USA 87:6609-6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meiland, R., F. E. J. Coenjaerts, S. E. Geerlings, E. C. Brouwer, and A. I. M. Hoepelman. 2003. Development of a quantitative real-time PCR assay for the rapid detection of Escherichia coli in urine samples. Communication presented at the 43rd Interscience Conference on Antimicrobial Agents, Chicago, Ill., 2003.
- 19.Mushtaq, S., N. Woodford, N. Potz, and D. M. Livermore. 2003. Detection of CTX-M-15 extended-spectrum beta-lactamase in the United Kingdom. J. Antimicrob. Chemother. 52:528-529. [DOI] [PubMed] [Google Scholar]
- 20.NCCLS. 1999. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals: approved standard. NCCLS document M31-A. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 21.Nikolich, M. P., G. Hong, N. B. Shoemaker, and A. A. Salyers. 1994. Evidence for natural horizontal transfer of tetQ between bacteria that normally colonize humans and bacteria that normally colonize livestock. Appl. Environ. Microbiol. 60:3255-3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oppegaard, H., T. M. Steinum, and Y. Wasteson. 2001. Horizontal transfer of a multi-drug resistance plasmid between coliform bacteria of human and bovine origin in a farm environment. Appl. Environ. Microbiol. 67:3732-3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orskov, F., and I. Orskov. 1992. Escherichia coli serotyping and disease in man and animals. Can. J. Microbiol. 38:699-704. [PubMed] [Google Scholar]
- 24.Osterblad, M., A. Hakanen, R. Manninen, T. Leistevuo, R. Peltonen, O. Meurman, P. Huovinen, and P. Kotilainen. 2000. A between-species comparison of antimicrobial resistance in enterobacteria in fecal flora. Antimicrob. Agents Chemother. 44:1479-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shiraki, Y., N. Shibata, Y. Doi, and Y. Arakawa. 2004. Escherichia coli producing CTX-M-2 beta-lactamase in cattle, Japan. Emerg. Infect. Dis. 10:69-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shoemaker, N. B., H. Vlamakis, K. Hayes, and A. A. Salyers. 2001. Evidence for extensive resistance gene transfer among Bacteroides spp. and among Bacteroides and other genera in the human colon. Appl. Environ. Microbiol. 67:561-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teale, C. J., L. Barker, A. P. Foster, E. Liebana, M. Batchelor, D. M. Livermore, and E. J. Threlfall. 2005. Extended-spectrum beta-lactamase detected in E. coli recovered from calves in Wales. Vet. Rec. 156:186-187. [PubMed] [Google Scholar]
- 28.Walther-Rasmussen, J., and N. Hoiby. 2004. Cefotaximases (CTX-M-ases), an expanding family of extended-spectrum beta-lactamases. Can. J. Microbiol. 50:137-165. [DOI] [PubMed] [Google Scholar]
- 29.Weill, F.-X., R. Lailler, K. Praud, A. Kérouanton, L. Fabre, A. Brisabois, P. A. D. Grimont, and A. Cloeckaert. 2004. Emergence of extended-spectrum-β-lactamase (CTX-M-9)-producing multiresistant strains of Salmonella enterica serotype Virchow in poultry and humans in France. J. Clin. Microbiol. 42:5767-5773. [DOI] [PMC free article] [PubMed] [Google Scholar]