Abstract
Legionella pneumophila replicates within alveolar macrophages, causing a severe pneumonia termed Legionnaires' disease. The bacterium resides within a vacuole that escapes immediate transport to the host lysosome. Instead, the vacuole interacts with the early secretory pathway to establish an environment suitable for rapid multiplication. A type IV secretion system is central to the pathogenicity of the bacterium, and many protein substrates that are translocated by this system to the host cell have been identified. One of these, VipD, was found to interrupt the late secretory pathway when overproduced in Saccharomyces cerevisiae. We independently identified VipD in a previous study and have further characterized this protein as well as its three paralogs. The vipD gene belongs to a family of L. pneumophila open reading frames that are predicted to contain a phospholipase A domain with sequence similarity to the type III-secreted toxin ExoU from Pseudomonas aeruginosa. Similarly to other known translocated proteins of L. pneumophila, VipD is strongly induced in early stationary phase, a time when the bacterium is most virulent. Detergent extraction studies of infected macrophages confirm that VipD is translocated into host cells via the type IV secretion system. A second assay for translocation revealed that two paralogs of VipD, VpdA and VpdB, also have translocation signals recognized by the type IV system. A strain lacking VipD and its three paralogs grew at wild-type rates in murine macrophages, although secondary mutations that cause growth defects in strains lacking VipD accumulate. The quadruple mutant displayed a growth advantage in the amoebal host Dictyostelium discoideum, indicating that the protein family may modulate intracellular growth in a complex fashion. VipD is mildly toxic when overproduced in eukaryotic cells, and the toxicity is partially dependent on the putative phospholipase active site. VipD and its paralogs therefore define a family of translocated proteins that may assist in the establishment of a vacuole suitable for bacterial replication through functioning as a phospholipase.
Legionella pneumophila is a gram-negative bacterium that causes a severe pneumonia called Legionnaires' disease in humans (26). L. pneumophila is found in freshwater amoebae and infects humans upon inhalation of contaminated aerosols, whereupon it replicates within alveolar macrophages (14). Critical to the intracellular lifestyle of this pathogen is a type IV secretion system (T4SS), encoded by the dot/icm genes (41). During engulfment of the bacteria by the macrophage and formation of the Legionella-containing vacuole (LCV), the Dot/Icm T4SS injects proteins into the host cell cytosol that enable the vacuole to avoid transport into the lysosome. The LCV interacts with the early secretory pathway soon after its closure, as seen through a close association with membranes derived from the endoplasmic reticulum (10, 21, 22). After substantial intracellular growth, the bacteria lyse the host cell and are phagocytosed by neighboring macrophages, setting the stage for subsequent rounds of intracellular growth (20).
Several translocated effectors of the Dot/Icm secretion system are known, including RalF (30), LidA (8), SidC (25), LepA/LepB (6), SdeA (1), and YlfA (5). At most, only small defects in intracellular growth are observed with mutations in any of these genes, indicating that there may be some degree of functional redundancy. RalF, LidA, SidC, YlfA, and SdeA each localize to the L. pneumophila-containing phagosome, yet the role of these proteins in pathogenesis is unknown. Recently, three additional potential Icm/Dot substrates (VipA, VipD, and VipF) that may play a role in manipulating late stages of the host secretory pathway were identified (38). Ectopic expression of each of these proteins in the yeast Saccharomyces cerevisiae causes a fraction of late transport vesicles to be missorted such that their cargo protein is inappropriately transported to an incorrect location. The molecular mechanisms behind these missorting events are unknown.
Through use of an enrichment for L. pneumophila mutants that are impaired for growth within host cells, we recently reported the identification of seven genes having insertion mutations that resulted in lowered growth in phagocytic cells (40). One of these genes is identical to the open reading frame encoding VipD. Here we report the characterization of VipD and demonstrate that this protein is a member of a family of proteins that are translocated into host cells.
MATERIALS AND METHODS
Media, plasmids, and strains.
L. pneumophila strains were grown and maintained as previously described (13, 16). Yeast strains were grown in media containing 1% Bacto yeast extract, 2% Bacto peptone, and either 2% dextrose or 2% galactose (33). Yeast transformations were performed by the method of Schiestl and Gietz (36). Axenically grown Dictyostelium discoideum was propagated in HL-5 liquid medium supplemented with penicillin and streptomycin (100 U/ml; GibcoBRL) as described previously (24).
Four plasmids (Table 1) were constructed to delete vipD and each of its paralogs. For each construct, SacI- and SalI-digested pSR47s was ligated with two PCR products, one digested with BamHI and SacI and the second digested with BamHI and SalI, which were generated using Lp02 genomic DNA as a template. For plasmid pSV27 (pΔvipD), the first PCR product was generated from primers L1-1 and L1-2 (Table 2) and the second PCR product was generated from primers L1-3 and L1-4; for plasmid pSV31 (pΔvpdA), the first PCR product was generated from primers C03-1 and C03-2 and the second PCR product was generated from primers C03-3 and C03-4; for plasmid pSV69 (pΔvpdB), the first PCR product was generated from primers A03-1 and A03-2 and the second PCR product was generated from primers A03-3 and A03-4; and for plasmid pSV58 (pΔvpdC), the first PCR product was generated with primers MF18-1 and MF18-2 and the second PCR product was generated with primers MF18-3 and MF18-4. Plasmids pSV27 and pSV31 are designed to delete the designated gene except for the first eight and last seven amino acid residues, whereas plasmids pSV69 and pSV58 allow for deletion of a large portion of the amino terminus of the designated gene and the insertion of a stop codon to block expression of downstream sequences. Plasmid pSV69 is designed to delete amino acid residues 4 through 170 of vpdB and replaces the sequence with a stop codon. Plasmid pSV58 places a stop codon after amino acid 4 and deletes the sequence from amino acids 5 to 826. Plasmid pSV88, pSV78, and pSV77, which express full-length vipD, vpdA, and vpdB, respectively, fused to the carboxy terminus of sidCΔ100 (sidC lacking its carboxy-terminal 100 amino acids), were generated by ligating BamHI- and XbaI-digested pZL204 with a similarly digested PCR product generated from Lp02 genomic DNA by using primers L1-5 and L1-6 (pSV88), C03-5 and C03-6 (pSV78), or A03-5 and A03-6 (pSV77).
TABLE 1.
Plasmids and strains used in this work
| Strain or plasmid | Genotype or relevant markers | Reference or source |
|---|---|---|
| Strains | ||
| L. pneumophila | ||
| Lp01 | Philadelphia-1 rpsL hsdR | 2 |
| SV8 | Lp01 dotA::miniTn10 | 24 |
| SV-L1 | Lp01 vipD1::miniTn10 (original isolate) | This work |
| SV48 | Lp01 vipD1::miniTn10 | This work |
| Lp02 | Philadelphia-1 rpsL hsdR thyA | 2 |
| Lp03 | Lp02 dotA3 | 2 |
| ZL25 | Lp02 ΔsidC | 25 |
| SV192 | Lp02 ΔvipD | This work |
| SV221 | Lp02ΔvipD ΔvpdA ΔvpdB ΔvpdC | This work |
| SV222 | Lp02ΔvipD ΔvpdA ΔvpdB ΔvpdC | This work |
| E. coli XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1lac/lacIqZΔM15 Tn10 (Tetr) | Stratagene |
| D. discoideum AX3K | Gift of D. Knecht | |
| S. cerevisiae | ||
| W303 | MATaura3-1 leu2-3,112 his3-11,15 ade2-1 trp1-1 can1-1 | Gift of A. Murray |
| WDK-1 | W303 ura3-1::pGAL | This work |
| W71-1 | W303 ura3-1::pGAL-vipD | This work |
| W71-2 | W303 ura3-1::pGAL-vipD | This work |
| W72-4 | W303 ura3-1::pGAL-vipD-D282A | This work |
| W72-5 | W303 ura3-1::pGAL-vipD-D282A | This work |
| W74-6 | W303 ura3-1::pGAL-vipD-S67A | This work |
| W74-9 | W303 ura3-1::pGAL-vipD-S67A | This work |
| Plasmids | ||
| pGEX-4T-1 | E. coli GST fusion vector | Amersham |
| pJB908 | ori RSF1010 (ΔoriT) Ampr tdΔi | J. Vogel |
| pSR47s | ori R6K ori TRP4 Kanr SacB | 28 |
| pDK20 | pGAL URA3 Yip | A. Murray |
| pZL199 | psidC | This lab |
| pZL204 | psidCΔ100 | This lab |
| pSV26 | vipD1::miniTn10 from PstI-digested genomic DNA | This work |
| pSV27 | ΔvipD construct in pSR47s | This work |
| pSV31 | ΔvpdA construct in pSR47s | This work |
| pSV58 | ΔvpdC construct in pSR47s | This work |
| pSV69 | ΔvpdB construct in pSR47s | This work |
| pSV77 | psidCΔ100-vpdB | This work |
| pSV78 | psidCΔ100-vpdA | This work |
| pSV88 | psidCΔ100-vipD | This work |
| pSV71 | pGAL-vipD URA3 Yip | This work |
| pSV72 | pGAL-vipD-D282A URA3 Yip | This work |
| pSV74 | pGAL-vipD-S67A URA3 Yip | This work |
| pSV50 | pGST-VipD expressed from pGEX-4T-1 | This work |
TABLE 2.
Oligonucleotide primers used in this work
| Primer name | Sequencea |
|---|---|
| A03-1 | CGCGGATCCCGTTTTCATAATTTCACCTG |
| A03-2 | GCGGAGCTCAGTTAATTCACTTCATCGC |
| A03-3 | CGCGGATCCTAACTGTTGCTTCTAAAGGAACC |
| A03-4 | CGAGCGTCGACTTACGCCCTATATAGGGATG |
| A03-5 | CGCGGATCCAAAACGGTAAATAGTCAAAATG |
| A03-6 | GCTCTAGATTACGCCCTATATAGGGATG |
| L1-1 | CGCGGATCCTAATTTACGGCTTTTTGTC |
| L1-2 | GCGGAGCTCTCTCGTTCAAAATAAGTAG |
| L1-3 | CGCGGATCCTCAACCACATTTGGCGG |
| L1-4 | GCAGCGTCGACTGATGCGGTAGAAGTGG |
| L1-5 | CGGGATCCACAAAAAGCCGTAAATTAAAAAG |
| L1-6 | GCTCTAGATTAATGGCCGCCAAATG |
| L1-7 | CGCGGATCCATGACAAAAAGCCGTAAATTAAA |
| L1-8 | CGCGGATCCTTAATGGCCGCCAAATGT |
| L1-9 | GGTGAATACATCGCTGCTGGAGGAATTCTGGAC |
| L1-10 | GTCCAGAATTCCTCCAGCAGCGATGTATTCACC |
| L1-11 | CATGTTAGCGGAGCAGCTGCCGGAGCAATGACG |
| L1-12 | CGTCATTGCTCCGGCAGCTGCTCCGCTAACATG |
| C03-1 | CGCGGATCCAACTTCTTGCTTTGTTTTC |
| C03-2 | GCGGAGCTCAAATCATTAAATGACCTTTG |
| C03-3 | CGCGGATCCCTGCCCAATAATAAACC |
| C03-4 | GCAGCGTCGACACATCAGACAAGGAAATC |
| C03-5 | CGCGGATCCAAAACAAAGCAAGAAGTTTC |
| C03-6 | GCTCTAGATTAATTCGGTTTATTATTGG |
| MF18-1 | CGCGGATCCTCATGGGGTCATCAGTAAATC |
| MF18-2 | GCGGAGCTCCAAATGCTTTCTGAGTTAG |
| MF18-3 | CGCGGATCCCAATGTATACGGATCTTG |
| MF18-4 | GCAGCGTCGACAGATATTACAAAATTACCTC |
Restriction enzyme sites are underlined.
Plasmid pSV26 was obtained by ligation of PstI-digested genomic DNA from strain SV-L1 (vipD1::miniTn10) prior to transformation into DH5αλpir. Only recircularized DNA containing the transposon, and thus the R6K origin of replication contained within, can be recovered. The glutathione S-transferase (GST)-VipD expression plasmid pSV50 and the pGAL-vipD vector pSV71 were constructed by ligating the BamHI-digested PCR product generated from primers L1-7 and L1-8 to BamHI-digested pGEX-4T and pDK20, respectively. Plasmids pSV72 and pSV74 were constructed from pSV71 by using the QuickChange site-directed mutagenesis kit by standard procedures (Stratagene). Primers L1-9 and L1-10 were used to construct the D282A mutation (pSV72), and primers L1-11 and L1-12 were used to create the S67A mutation (pSV74).
Strains Lp01 and Lp02, derivatives of L. pneumophila Philadelphia-1, as well as their dotA mutant derivatives (SV8 and Lp03, respectively), have been previously described (2, 24). The ΔsidC strain ZL25 contains an in-frame deletion of sidC in the Lp02 strain background (25). The strain SV-L1 (vipD1::miniTn10 in Lp01) contains a transposon insertion after amino acid residue 307 (insertion sequence GGCAGGCAC) and is the original isolate obtained in a screen for growth-defective mutants (40). Strain SV48, which reconstitutes the transposon insertion found in SV-L1 in a fresh Lp01 parental strain, was obtained by introducing plasmid pSV26 into Lp01 by natural transformation (8, 39). Strain SV192 (ΔvipD) was generated in Lp02 by two-step gene replacement by standard protocols (28) using pSV27. Strains SV221 and SV222, which contain deletions of vipD, vpdA, vpdB, and vpdC, were obtained in Lp02 through four consecutive two-step gene replacements using plasmids pSV27, pSV31, pSV58, and pSV69.
L. pneumophila infection of murine macrophages and D. discoideum.
Bone marrow-derived macrophages from the A/J mouse were prepared as described previously (8). D. discoideum AX4 was plated in MB medium prior to incubation with L. pneumophila (24). For assays of L. pneumophila growth within bone marrow-derived macrophages or D. discoideum, host cells were plated on 24-well plates and infected with L. pneumophila at a multiplicity of infection (MOI) of 0.05 for 1.5 to 2 h. At each time point, monolayers were lysed with saponin, dilutions of the lysate were plated onto bacteriological media, and CFU were determined from triplicate wells of each strain and at each time point. For immunofluorescence assays, 2 × 10 5 bone marrow-derived macrophages were seeded onto glass coverslips and incubated with bacteria at an MOI of 2.0 for 1 hour.
Saponin extraction, immunofluorescence, and Western blotting.
For the detergent extraction studies, a modification of previous procedures was used (11, 23). Approximately 5.0 × 107 differentiated U937 cells were plated in 10-cm tissue culture dishes and allowed to adhere for 1 hour. The monolayers were then incubated at an MOI of 5.0 with L. pneumophila grown in broth culture to postexponential phase. After a 1-hour infection, the supernatants were removed and the monolayer was lysed in 3 ml of 0.2% saponin. The detergent lysate was then incubated for at least 1 hour on ice and cleared by centrifugation at 9,000 rpm in a Beckman J2-21 centrifuge for 15 min. The supernatants were removed and subjected to methanol-chloroform precipitation (44), and the pellets were resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. Equivalent amounts of supernatant and pellet fractions were then separated by SDS-PAGE and analyzed by Western blotting.
For analysis of S. cerevisiae lysates, cells were grown for 2 days on medium containing galactose and resuspended in 3 ml water, and the equivalent of 0.3 A600 unit of cells were collected by centrifugation, resuspended in SDS-PAGE sample buffer, and lysed by boiling. For analysis of L. pneumophila lysates, cells were grown in broth to the indicated A600, isolated by centrifugation, and lysed by boiling in sample buffer. Equivalent amounts of lysate were separated by SDS-PAGE and processed for Western blot analysis.
GST-VipD was expressed in XL1-Blue from pSV50, purified by standard procedures (Amersham, Carlsbad, CA), and cleaved with thrombin. The cleaved VipD was used to raise antibodies in rabbits (Pocono Rabbit Farm and Laboratory, Canadensis, PA), and the antibodies were then affinity purified against the same protein by using standard protocols (17). Serum specific for Bacillus subtilis isocitrate dehydrogenase (ICDH) was generously provided by A. L. Sonenshein, Tufts University Medical School, Boston, MA. For immunofluorescence analyses, cells were fixed and stained by standard procedures (27, 43). Affinity-purified antibody against SidC (25) was used at a dilution of 1:500, and rat anti-L. pneumophila antibody was used at a dilution of 1:3,000. Bacteria that had been internalized by macrophages were identified through a staining procedure in which monolayers were probed with anti-L. pneumophila antibody both prior to and after methanol permeabilization, using secondary antibodies conjugated to fluors of distinct color (blue before permeabilization and red after permeabilization) at each step (8). Only intact rod-shaped bacteria were scored in these assays.
RESULTS
VipD shows high similarity to Pseudomonas aeruginosa ExoU and has three paralogs.
An enrichment was performed to identify L. pneumophila mutants that were defective for growth within phagocytic cells but still retained a functional Dot/Icm system (40). Among those mutants isolated was one with a miniTn10 insertion in the vipD gene (Fig. 1A) (38), which caused small defects in intracellular growth in the original strain background in which it was isolated (40) (see Fig. 5). Simultaneously, we also identified vipD in a screen for proteins that are translocated by the Dot/Icm system to recipient bacterial cells (25). The vipD gene is predicted to encode a 69-kDa cytosolic protein showing high sequence similarity to ExoU of the gram-negative bacterial pathogen P. aeruginosa. ExoU is a potent toxin that is translocated to mammalian cells via a type III secretion apparatus (15, 18). Furthermore, both VipD and ExoU show similarity to the phospholipase patatin, the most abundant protein found in potato tubers (19). The proteins share consensus motifs thought to be essential for phospholipase activity (Fig. 1B), which include an oxyanion hole (consensus GGGXK/R [one-letter code]) and two active-site residues, a serine (consensus GXSXG) and an aspartate (consensus DXG). The phospholipase activity of ExoU, which requires an unknown cofactor from target host cells, was found to be essential for its toxicity (31, 35). Despite this high sequence similarity, we have been unable to demonstrate a phospholipase activity for VipD (see Discussion).
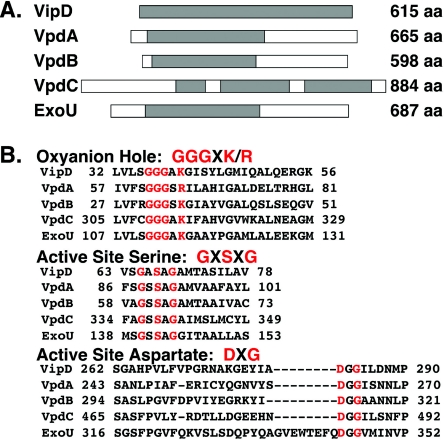
FIG. 1.
VipD and it paralogs have sequence similarity to the P. aeruginosa secreted toxin ExoU. (A) Representative diagrams of VipD, VpdA, VpdB, VpdC, and ExoU. The size of each protein (in amino acid [aa] residues) is shown on the right of each diagram. Regions of each protein that are homologous to VipD are shaded. (B) The conserved phospholipase A domains are shown, with the conserved amino acids (single-letter code) included in the oxyanion hole, the active-site serine, and the active-site aspartate indicated in red.
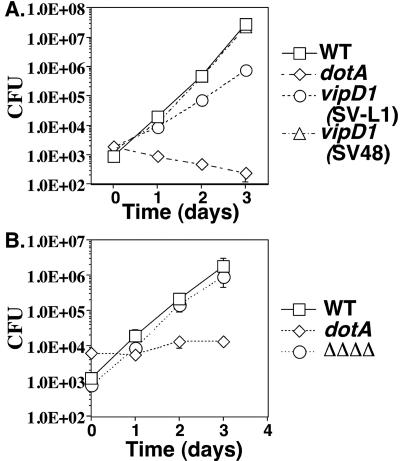
FIG. 5.
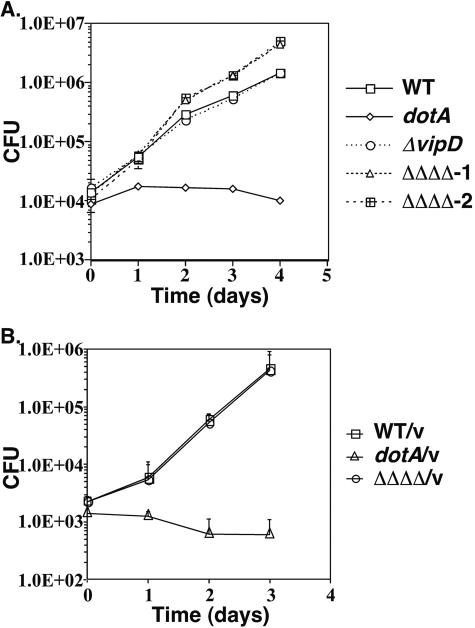
An L. pneumophila mutant lacking all vipD paralogs is proficient for intracellular growth within bone marrow-derived macrophages. (A) Defective growth phenotype of the original L. pneumophila vipD1::miniTn10 strain (strain SV-L1) is lost on moving the mutation into a fresh parental strain background (strain SV48). The wild-type (WT) strain Lp01 is the parent strain for both the vipD mutant strains (SV-L1 and SV48) and the dotA mutant strain (SV8). (B) A strain bearing deletions of vipD, vpdA, vpdB, and vpdC (strain SV221, designated ΔΔΔΔ) does not show a growth defect in murine macrophages. The wild-type strain Lp02 is the parent strain for the quadruple mutant strain and the dotA strain Lp03. (A and B) Bone marrow-derived macrophages (4 × 105 cells per well) were incubated with the indicated strains at an MOI of 0.05 for either 2 h (A) or 1.5 h (B). Viable counts were determined as described in Materials and Methods. The means and standard deviations are shown for each time point; experiments were done in triplicate.
VipD has three paralogs in the L. pneumophila Philadelphia-1 genome, which we have named VpdA, VpdB, and VpdC (Fig. 1A) (GenBank accession numbers YP_096418, YP_095258, and YP_095455, respectively). Two of these paralogs, VpdA and VpdB, are similar in size to VipD (76 and 66 kDa, respectively), whereas VpdC is significantly larger (102 kDa). Each paralog contains the consensus phospholipase domain found in VipD and ExoU (Fig. 1B).
VipD is induced in early stationary phase.
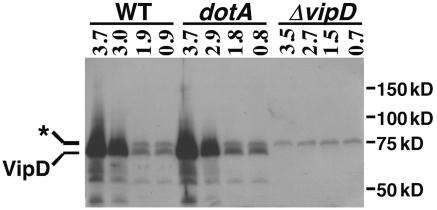
The sequence similarity of the L. pneumophila VipD family members to ExoU from P. aeruginosa, its ability to be transferred between bacteria in a recombination read-out assay (data not shown) (25), and the fact that fusion of VipD to adenylate cyclase results in a hybrid translocated by the Dot/Icm system (38) argue that these proteins are L. pneumophila translocated effectors. One characteristic of many known effectors is increased expression in early stationary phase (8, 25, 30), the growth phase at which L. pneumophila is most virulent (3). To analyze growth phase-dependent expression, an antibody against full-length VipD was raised in rabbits and used to probe bacterial lysates at distinct growth phases (see Materials and Methods). In wild-type L. pneumophila cultures, a protein with an apparent molecular mass of ∼70 kDa in was present in immunoblots of SDS-containing gels probed with affinity-purified antiserum (Fig. 2). This protein, which is close to the predicted molecular weight of VipD, was absent in a strain deleted for vipD at all growth phases (Fig. 2). L. pneumophila enters postexponential phase at an optical density of 3.0, a transition discerned by a change in morphology and induction of motility (3). At this growth phase, expression of VipD displayed a marked increase (Fig. 2). The postexponential phase induction was also observed in a dotA mutant strain and thus is not dependent on an intact dot/icm complex (Fig. 2). By this assay, the expression profile of VipD is consistent with its having a role as a translocated effector.
FIG. 2.
VipD protein is induced in postexponential phase. The indicated isogenic L. pneumophila strains were grown in broth to the indicated optical density (A600), and equivalent amounts of cells were harvested and lysed in sample buffer. Proteins in each sample were resolved by SDS-PAGE, transferred to a membrane support, and probed with an antibody specific to VipD. The band corresponding to VipD is indicated, and a nonspecific protein recognized by the antibody is marked with an asterisk. The migration of molecular mass standards is shown on the right. Wild type (WT): Lp02. dotA: Lp03. ΔvipD: SV192.
VipD is translocated into mammalian cells.
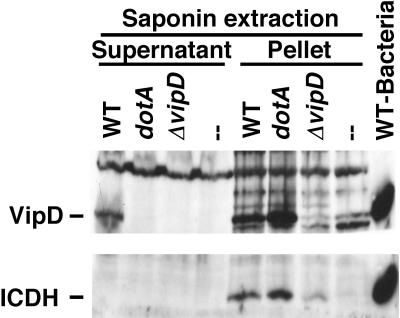
To directly demonstrate that intact VipD is a translocated substrate of the L. pneumophila Dot/Icm system, we employed a saponin extraction technique that is similar to assays utilized with Yersinia for the analysis of translocated effectors (11, 23). To this end, phorbol ester-differentiated U937 cells were incubated with L. pneumophila strains for 1 hour and then subjected to saponin extraction followed by immunoblot analysis of the detergent-soluble supernatants (see Materials and Methods). Saponin does not lyse the bacteria or release VipD from broth-grown bacteria (Fig. 3 and data not shown). As expected of a translocated protein, VipD was found in supernatants of saponin extracts when wild-type bacteria were used to infect the mammalian cells (Fig. 3). VipD was absent in this fraction when a ΔvipD strain was used and when cells were mock infected (Fig. 3). Translocation was dependent on an intact Dot/Icm T4SS, because VipD was not present in the supernatant when a dotA mutant L. pneumophila strain was used. As a control for bacterial lysis, an antibody that recognizes bacterial ICDH, a protein found in the bacterial cytoplasm, was examined, and this protein was absent from the supernatant under all conditions examined (Fig. 3). Our results thus demonstrate that VipD is specifically translocated in a Dot/Icm-dependent manner from L. pneumophila to mammalian cells.
FIG. 3.
VipD is translocated into U937 cells in a Dot/Icm-dependent manner. U937 cells were incubated with the indicated strains at an MOI of 5 for 1 h, after which host cells were harvested, extracted with 0.2% saponin, and processed as described in Materials and Methods. SDS-PAGE-fractionated samples were immunoprobed with antibodies against VipD or ICDH. Wild type (WT): U937 cells incubated with Lp02. dotA: U937 cells incubated with Lp03. ΔvipD: U937 cells incubated with strain SV192.—: U937 cells with no bacteria added. WT-Bacteria: L. pneumophila Lp02 grown to postexponential phase and lysed in SDS sample buffer to detect VipD.
VipD and its paralogs define a family of translocated proteins in L. pneumophila.
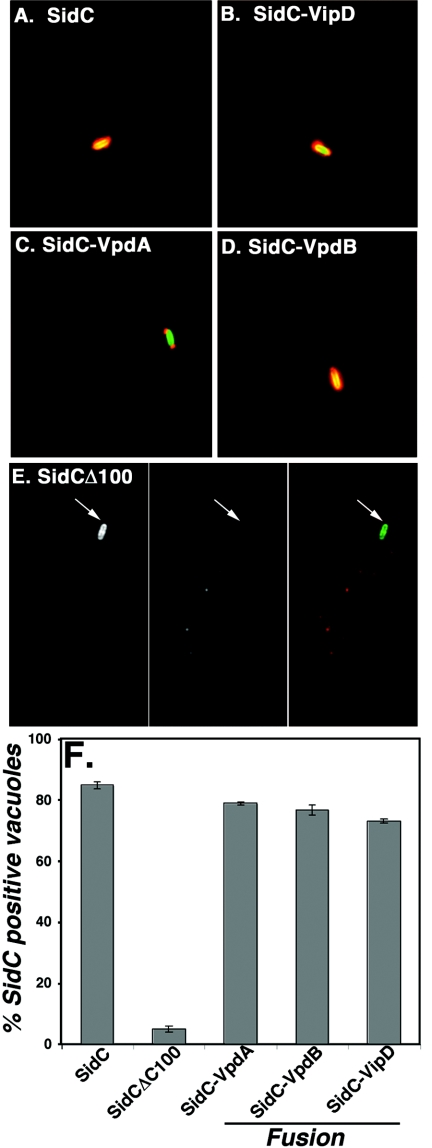
We next tested whether two paralogs of VipD, VpdA and VpdB, were also translocated into host cells. To assay translocation, we utilized the protein SidC, which is translocated into host cells where it localizes to the cytoplasmic face of the L. pneumophila vacuole (25). The translocation signal of SidC has been localized to the carboxy terminus of the protein (Z.-Q. Luo, unpublished results), similar to what has been reported for the type IV-translocated protein RalF (29). When a version of SidC that lacks its carboxy-terminal 100 amino acids (SidCΔ100) was constructed, translocation was observed for only a minor fraction of LCVs (Fig. 4F). Fusion of a translocation signal recognized by Dot/Icm to the carboxyl terminus of the SidC deletion derivative can restore deposition of the protein into host cells. After successful translocation, the fusion protein localizes to the L. pneumophila-containing vacuole as visualized with anti-SidC antibodies. To test for VpdA and VpdB translocation signals in this assay, fusion proteins in which the carboxy-terminal 100 amino acids of SidC were replaced with full-length VipD, VpdA, or VpdB were constructed. Strains deleted for sidC and harboring plasmids encoding these fusion proteins were then incubated with murine macrophages for 1 hour. The infected monolayers were then fixed, stained with anti-SidC antibody, and analyzed by immunofluorescence microscopy, scoring for the presence of SidC stain around the L. pneumophila vacuole. As described previously (25), greater than 80% of L. pneumophila-containing vacuoles stain with anti-SidC antibody (Fig. 4A and F), whereas fewer than 5% of vacuoles are SidC positive when the SidCΔ100 construct is expressed (Fig. 4E and F). As expected, fusion of SidCΔ100 to VipD yielded a high frequency of SidC-positive vacuoles, confirming the results from the detergent extraction assay (Fig. 4B and F). In addition, the SidCΔ100 fusions to both VpdA (Fig. 4C and F) and VpdB (Fig. 4D and F) displayed approximately 80% localization of the fusion protein to L. pneumophila-containing vacuoles, with intense immunofluorescence of the fusions observed. Therefore, VipD is translocated and two of its paralogs have signals that allow translocation of SidC into host cells. Interestingly, a significant portion of the phagosomes harboring the strain expressing the SidC-VpdA fusion displayed polar localization (Fig. 4C). It seems likely, therefore, that there are localization signals present in VpdA that allow localization to the termini of the bacterial vacuole or direct polar translocation of the fusion. Such polar translocation had been observed previously with LidA (8).
FIG. 4.
Evidence for the presence of translocation signals in VipD, VpdA, and VpdB. Bone marrow-derived macrophages were infected at an MOI of 2 for 1 hour with ΔsidC strains containing plasmids expressing either full-length sidC (psidC/pZL199; panel A); fusions of the 3′ end of sidCΔ100 to full-length vipD (psidCΔ100-vipD/pSV88; panel B), vpdA (psidCΔ100-vpdA/pSV78; panel C), or vpdB (psidCΔ100-vpdB/pSV77; panel D); or sidC lacking the region encoding the carboxy-terminal 100 amino acids (psidCΔ100/pZL204; panel E). Triplicate coverslips for each strain were scored for SidC staining surrounding the L. pneumophila-containing vacuole, and 100 bacteria found in singly infected macrophages were scored on each coverslip (panel F). Displayed are the means and standard deviations for each sample.
Growth of vipD mutants in murine macrophages and D. discoideum.
To explore the interaction of the vipD mutant with host cells, a growth curve in bone marrow-derived macrophages was completed. The original vipD1::Tn10 mutant (strain SV-L1) isolated in our screen (40) was used to infect murine macrophages, and CFU were determined 2, 24, 48, or 72 h postinfection. The wild-type parental strain Lp01 and a severely growth-defective dotA strain, which lacks a functional dot/icm apparatus, were also included as controls. The vipD1::miniTn10 strain SV-L1 was found to have a small growth defect at each time point analyzed compared to the wild-type strain (Fig. 5A), as was seen for other mutants isolated in this screen (40). To confirm that the growth defect was attributable to the transposon insertion in vipD, the transposon was reintroduced into the parental strain, generating strain SV48. Unexpectedly, the reconstructed vipD1 strain SV48 did not display a growth defect in murine macrophages (Fig. 5A). To further explore this phenotype, we tested the intracellular growth characteristics of many independently created ΔvipD isolates. Whereas the majority of the ΔvipD isolates displayed no growth defect in macrophages, as did the reconstructed vipD1::miniTn10 strain, a few ΔvipD strains exhibited defects in intracellular growth similar to those of the original vipD isolate SV-L1 (data not shown). We were never able to complement the growth defect of these ΔvipD strains or the original vipD1::Tn10 (SV-L1) strain in trans using a plasmid construction expressing VipD (data not shown), so we conclude that the phenotype of the growth-defective vipD mutants is due to a second mutation that arises in the strain or during its construction.
Since VipD has three paralogs, it is possible that the four proteins provide a similar function during infection of mammalian cells. To assess the requirement for the VipD paralogs for growth in mammalian cells, strains bearing deletions of individual paralogs and a strain bearing deletions of all four genes were constructed. Neither the strains bearing deletions of individual genes (data not shown) nor the strain bearing the quadruple mutation was significantly impaired for growth in macrophages during a 4-day growth curve (Fig. 5B). Therefore, neither VipD nor its paralogs are essential for bacterial replication in cultured mouse macrophages.
We next assessed the ability of an L. pneumophila strain deleted for vipD to grow within the soil amoeba D. discoideum. Using bacterial strains identical to those used in bone marrow-derived macrophages, we observed a mild enhancement of intracellular growth relative to the parental thy mutant control for a strain containing deletions of vipD and its three paralogs (Fig. 6A). A strain bearing only a deletion of vipD, in contrast, displayed growth rates nearly identical to those of the wild-type strain. As expected, the dotA mutant strain was incapable of intracellular growth (Fig. 6A). Since we had previously demonstrated that L. pneumophila thyA mutant strains have retarded growth in D. discoideum due to the inability of thymidine added to the medium to fully complement the auxotrophy (24), we monitored intracellular growth of the quadruple mutant in a thy+ background through inclusion of a plasmid expressing thyA. In the thy+ strain background, however, the growth advantage of the quadruple mutant disappeared (Fig. 6B). Therefore, the ability to detect any intracellular growth advantage that results from loss of function of multiple VipD family members appears to be limited to strain backgrounds having underlying deficiencies in intracellular growth.
FIG. 6.
Growth advantage in D. discoideum of a L. pneumophila mutant lacking vipD and each of its paralogs. (A) Lp02 (thyA mutant) strains bearing the ΔvipD ΔvpdA ΔvpdB ΔvpdC mutations (strains SV221 and SV222, designated ΔΔΔΔ-1 and ΔΔΔΔ-2) have a growth advantage in D. discoideum, whereas the ΔvipD strain (SV192) grows at a rate similar to that of the wild-type strain. (B) A Thy+ strain bearing the ΔvipD ΔvpdA ΔvpdB ΔvpdC mutations (strain SV221 containing the vector pJB908, designated ΔΔΔΔ/v) is indistinguishable from its Thy+ parent (Lp02 containing pJB908, designated WT/v) for growth in D. discoideum. (A and B) The wild-type (WT) strain Lp02 is the parent strain for the quadruple mutant strain and the dotA strain Lp03. The indicated strains were used to infect amoebae at an MOI of 0.05 for 2 hours. Viable counts were determined as described in Materials and Methods. Each time point for each strain was analyzed in triplicate, and the means and standard deviations are shown.
Toxicity of VipD to eukaryotic cells.
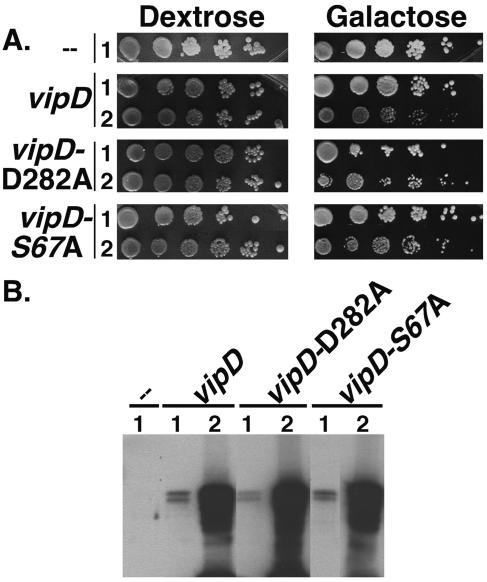
ExoU is a potent toxin for mammalian cells, and trace expression of the protein in a wide range of eukaryotic cells can lead to rapid lethality. In contrast to the behavior of ExoU, when a plasmid encoding VipD was introduced into the mammalian 293T cell line, transfectants expressing large amounts of protein could be readily identified, although viability appeared to be somewhat reduced by the presence of the protein (data not shown). To investigate toxicity of the protein in a more quantitative fashion, we expressed the protein in the yeast Saccharomyces cerevisiae. A construct in which VipD was placed under control of the galactose-regulated promoter (pGAL) was generated and integrated at the URA3 locus of a wild-type yeast strain. Tenfold serial dilutions of the resulting strain were spotted onto media containing dextrose, which represses expression from pGAL, or galactose, which induces expression. A strain in which pGAL alone was present grew well on both repressing (dextrose) and inducing (galactose) media (Fig. 7A). A strain bearing pGAL-vipD also grew well on repressing media (dextrose), whereas growth on galactose was dependent on the particular isolate analyzed. Many independent isolates grew at a rate similar to that of the control strain (pGAL), but several displayed reduced growth. Since the integration construct used is known to result in a range of expression levels (M. Dorer, personal communication), we examined VipD levels in strains bearing pGAL or pGAL-vipD. VipD was absent in yeast expressing pGAL and was present as a doublet in pGAL-vipD-expressing yeast (Fig. 7B). VipD-producing isolates that grew poorly on galactose showed far greater expression of vipD than isolates displaying wild-type growth rates. Thus, overproduction of VipD showed only mild toxicity to yeast, and based on the behavior of the protein, either VipD is likely to have functions very different from ExoU or the potency of its phospholipase activity under the conditions tested is far lower than that of ExoU.
FIG. 7.
Expression of vipD shows low levels of toxicity for yeast. (A) Either wild-type VipD or VipD containing mutations in the putative phospholipase A active site can be toxic to the yeast S. cerevisiae. Constructs containing the indicated vipD alleles under the control of the galactose-regulated promoter (pGAL) were integrated at the URA3 locus in S. cerevisiae. The resulting strains, along with a strain containing pGAL but not vipD, were grown in rich medium to stationary phase. Tenfold serial dilutions were spotted onto rich medium containing either dextrose (which represses expression of vipD) or galactose (which induces expression of vipD) and incubated for 3 days at 30°C. For each vipD-expressing construct, an isolate displaying wild-type growth on galactose medium (rows 1) and one with a slow-growth phenotype (rows 2) are shown. (B) vipD-expressing strains that are slow growing on galactose medium produce higher levels of VipD. Equivalent amounts of the strains grown on galactose medium in (A) were lysed in sample buffer, separated by SDS-PAGE, and analyzed by Western blotting using an anti-VipD antibody.
We next inquired whether the poor growth on galactose required a functional phospholipase domain. Yeast strains identical to those described above were constructed, except that the conserved active-site residues (aspartate in DXG and serine in GXSXG) in the phospholipase domain were exchanged with alanine (Fig. 1A). Each of these substitutions has been demonstrated to inactivate phospholipase A activity in the homologous proteins patatin and ExoU (19, 31, 35). Growth of yeast expressing either VipD-D282A or VipD-S67A was normal on repressing media (dextrose) (Fig. 7A). Under inducing conditions (galactose), two phenotypes were again observed, correlating with wild-type or slightly slowed growth rates. Once again, the extent of growth reduction correlated with the levels of production of VipD mutants in the strains (Fig. 7B). Interestingly, in each case in which VipD displayed high expression levels, growth on galactose of strains expressing wild-type VipD was markedly slower than that of those expressing the VipD alanine mutants. Therefore, it appears that although the phospholipase domain contributes to the toxicity exhibited by VipD, it is not the sole cause of retarded growth of yeast cells overexpressing the protein. This is reminiscent of the recent result showing that a fragment of VipD lacking the phospholipase domain interferes with yeast secretory traffic (38).
DISCUSSION
VipD was identified in our laboratory in a screen for L. pneumophila mutants having miniTn10 insertions that result in delayed completion of a single cycle of infection within bone marrow-derived macrophages (40). An assay that monitors intracellular growth over 3 days, or approximately three cycles of intracellular multiplication, verified that the original vipD strain isolated in the screen was defective for growth during each replication cycle. The growth defect disappeared, however, when the transposon mutation was recombined into a fresh strain background or when an L. pneumophila strain bearing a deletion of vipD was tested in the same assay. Through analysis of many independently isolated vipD mutants, we obtained a small number that exhibited growth defects similar to those of the original vipD isolate, indicating that there may be a mutation in the L. pneumophila background that arises and, when combined with a mutation in vipD, contributes to the intracellular defect of these strain. It is unclear what selective pressure could result in these secondary mutations, since vipD mutants do not exhibit any obvious growth defects on bacteriological medium.
A strain lacking vipD as well as its three paralogs, vpdA, vpdB, and vpdC, also grew at wild-type rates in murine macrophages, revealing that the four genes are dispensable for intracellular growth. The lack of a growth defect in L. pneumophila strains lacking the VipD family members may be due to functional redundancy between VipD and other translocated effector proteins. Alternatively, VipD may be required during stages of the infection process not represented by the assays thus far tested. For example, VipD may function in the lung prior to being engulfed by alveolar macrophages, or it may inactivate specific arms of the immune response that are involved in clearing of microorganisms from the lungs. There clearly is some selective pressure for retention of this gene family, and VipD is highly expressed in postexponential phase, arguing for a connection to other factors associated with pathogenesis. The strain background used in this study, L. pneumophila Philadelphia-1, has four members of the vipD family that possess sequences similar to those of the ExoU and patatin active site. The two other sequenced strains of L. pneumophila also have multiple vipD paralogs (including five such genes in L. pneumophila Paris-1), although an open reading frame corresponding to vipD is missing in the L. pneumophila Lens-1 strain.
A thyA mutant strain lacking vipD as well as its three paralogs displayed a growth advantage in D. discoideum compared to the parental strain (Fig. 6). We had used this auxotroph because it facilitates our ability to perform complementation experiments with plasmids that confer prototrophy. Testing of strains deleted for individual paralogs revealed that the phenotype is likely due to the absence of vpdA (data not shown). The phenotype of this quadruple mutant is clearly subtle. Under conditions in which growth in D. discoideum is accelerated by using a prototrophic strain, the growth advantage disappeared, emphasizing that the phenotype can be observed only under conditions in which bacterial growth is less than optimal.
VipD and SidCΔ100 fusions harboring two of the VipD paralogs (VpdA and VpdB) were translocated into host cells. Although at least eight translocated substrates of the Dot/Icm system have been localized to the phagosomal membrane surrounding the bacterium (1, 8, 25, 30; M. Machner and R. Isberg, unpublished data), we have been unable to localize VipD to a specific site in the host cell by immunofluorescence (data not shown). Presumably, the majority of the protein assumes a cytoplasmic locale after translocation. In contrast, a paralog of VipD, VpdA, may contain vacuolar localization signals as evidenced by the distinct polar stain of a SidCΔ100-VpdA fusion protein present on L. pneumophila-containing vacuoles. Despite the fact that VipD does not appear to localize to the phagosomal membrane, the protein behaves very similarly to other L. pneumophila translocated substrates (8, 25, 30): it displays increased expression in stationary phase, strains lacking VipD show little defect in intracellular growth, and it belongs to a family of paralogs encoded by the bacterium. Furthermore, the carboxyl termini of VipD family members, predicted to be the region harboring the translocation signal recognized by the Dot/Icm apparatus, share a feature predicted to be important for translocation of other substrates. A hydrophobic residue at position −3 of the C terminus is essential for translocation of the Dot/Icm substrate RalF, and it was proposed, based on examination of known translocated effectors, that either a hydrophobic residue or a proline at the C-3 or C-4 position may be crucial for recognition by the Dot/Icm system (29). VipD and its paralogs follow this pattern and contain similarly placed residues (VipD, Phe at −4; VpdB, Leu at −4; and VpdC, Leu at −3), although in VpdA the Pro residue is at position −2.
The VipD-related proteins patatin and ExoU have demonstrable phospholipase activities in vitro (19, 31, 34, 35), but we were unable to detect phospholipase activity from VipD by using various phospholipid substrates (data not shown; H. Sato and D. Frank, personal communication). ExoU as well as many other effector proteins from bacterial pathogens require host proteins as cofactors, but even the addition of mammalian or yeast cytosol in these assays had no effect on VipD activity. In fact, the expression of VipD in eukaryotic cells has consequences that are very different from those observed for ExoU (35). Expression of VipD in yeast (Fig. 7) and in cultured mammalian cell lines (data not shown) was relatively well tolerated. VipD caused little discernible effect on the growth rate of yeast until it was overproduced, at which point cellular growth was slowed but not halted. In contrast, expression of ExoU in S. cerevisiae causes immediate toxicity and is associated with fragmentation of the vacuole (35). In addition, the growth defect that was observed for S. cerevisiae overexpressing VipD did not appear to be totally dependent on the putative phospholipase domain, whereas similar mutations in the ExoU phospholipase active site abolished its toxicity. Shohdy and colleagues found that a version of VipD lacking the phospholipase domain perturbed the late secretory pathway to a greater extent than full-length VipD (38). If one of the functions of VipD is to interfere with the late secretory pathway, the putative phospholipase domain does not appear to be essential for this process.
Since the phospholipase active site is conserved in all four paralogs, it seems likely that VipD functions as a phospholipase at some stage in the infection process. Phospholipases have been shown to be involved in the virulence of several other pathogens, including Mycobacterium tuberculosis, Yersiniaenterocolitica, Listeria monocytogenes, and Rickettsia prowazekii (4, 32, 37, 42). Several studies indicate that proteins with phospholipase A activity play a role in membrane traffic through the mammalian secretory pathway (7, 9, 12), and VipD could therefore allow L. pneumophila to manipulate membrane flow and establish or maintain its unique vacuolar niche. Further characterization of this protein as well as its critical phospholipid substrates should allow insight into the role of this family of proteins during L. pneumophila infection.
Acknowledgments
We thank Hiromi Sato and Dara Frank for testing VipD in their in vitro phospholipase assay. We are grateful to Matt Heitman and Matthias Machner for critical reading of the manuscript.
This work was supported by the Jane Coffin Childs Memorial Fund for Cancer Research (S. M. VanRheenen), the Life Sciences Research Foundation (Z.-Q. Luo), and Program Project Award grant P30DK34928 from the National Institute of Diabetes and Kidney Diseases and the Howard Hughes Medical Institute (R. R. Isberg).
Editor: V. J. DiRita
REFERENCES
- 1.Bardill, J. P., J. L. Miller, and J. P. Vogel. 2005. IcmS-dependent translocation of SdeA into macrophages by the Legionella pneumophila type IV secretion system. Mol. Microbiol. 56:90-103. [DOI] [PubMed] [Google Scholar]
- 2.Berger, K. H., and R. R. Isberg. 1993. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol. Microbiol. 7:7-19. [DOI] [PubMed] [Google Scholar]
- 3.Byrne, B., and M. S. Swanson. 1998. Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect. Immun. 66:3029-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Camilli, A., H. Goldfine, and D. A. Portnoy. 1991. Listeria monocytogenes mutants lacking phosphatidylinositol-specific phospholipase C are avirulent. J. Exp. Med. 173:751-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campodonico, E. M., L. Chesnel, and C. R. Roy. 2005. A yeast genetic system for the identification and characterization of substrate proteins transferred into host cells by the Legionella pneumophila Dot/Icm system. Mol. Microbiol. 56:918-933. [DOI] [PubMed] [Google Scholar]
- 6.Chen, J., K. S. de Felipe, M. Clarke, H. Lu, O. R. Anderson, G. Segal, and H. A. Shuman. 2004. Legionella effectors that promote nonlytic release from protozoa. Science 303:1358-1361. [DOI] [PubMed] [Google Scholar]
- 7.Choukroun, G. J., V. Marshansky, C. E. Gustafson, M. McKee, R. J. Hajjar, A. Rosenzweig, D. Brown, and J. V. Bonventre. 2000. Cytosolic phospholipase A(2) regulates Golgi structure and modulates intracellular trafficking of membrane proteins. J. Clin. Investig. 106:983-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conover, G. M., I. Derre, J. P. Vogel, and R. R. Isberg. 2003. The Legionella pneumophila LidA protein: a translocated substrate of the Dot/Icm system associated with maintenance of bacterial integrity. Mol. Microbiol. 48:305-321. [DOI] [PubMed] [Google Scholar]
- 9.de Figueiredo, P., D. Drecktrah, R. S. Polizotto, N. B. Cole, J. Lippincott-Schwartz, and W. J. Brown. 2000. Phospholipase A2 antagonists inhibit constitutive retrograde membrane traffic to the endoplasmic reticulum. Traffic. 1:504-511. [DOI] [PubMed] [Google Scholar]
- 10.Derre, I., and R. R. Isberg. 2004. Legionella pneumophila replication vacuole formation involves rapid recruitment of proteins of the early secretory system. Infect. Immun. 72:3048-3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Derre, I., and R. R. Isberg. 2005. LidA, a translocated substrate of the Legionella pneumophila type IV secretion system, interferes with the early secretory pathway. Infect. Immun. 73:4370-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drecktrah, D., and W. J. Brown. 1999. Phospholipase A(2) antagonists inhibit nocodazole-induced Golgi ministack formation: evidence of an ER intermediate and constitutive cycling. Mol. Biol. Cell 10:4021-4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feeley, J. C., R. J. Gibson, G. W. Gorman, N. C. Langford, J. K. Rasheed, D. C. Mackel, and W. B. Baine. 1979. Charcoal-yeast extract agar: primary isolation medium for Legionella pneumophila. J. Clin. Microbiol. 10:437-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fields, B. S. 1996. The molecular ecology of legionellae. Trends Microbiol. 4:286-290. [DOI] [PubMed] [Google Scholar]
- 15.Finck-Barbancon, V., J. Goranson, L. Zhu, T. Sawa, J. P. Wiener-Kronish, S. M. Fleiszig, C. Wu, L. Mende-Mueller, and D. W. Frank. 1997. ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol. Microbiol. 25:547-557. [DOI] [PubMed] [Google Scholar]
- 16.Gabay, J. E., M. Blake, W. D. Niles, and M. A. Horwitz. 1985. Purification of Legionella pneumophila major outer membrane protein and demonstration that it is a porin. J. Bacteriol. 162:85-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harlow, E., and D. Lane. 1999. Using antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 18.Hauser, A. R., P. J. Kang, and J. N. Engel. 1998. PepA, a secreted protein of Pseudomonas aeruginosa, is necessary for cytotoxicity and virulence. Mol. Microbiol. 27:807-818. [DOI] [PubMed] [Google Scholar]
- 19.Hirschberg, H. J., J. W. Simons, N. Dekker, and M. R. Egmond. 2001. Cloning, expression, purification and characterization of patatin, a novel phospholipase A. Eur. J. Biochem. 268:5037-5044. [DOI] [PubMed] [Google Scholar]
- 20.Horwitz, M. A., and S. C. Silverstein. 1980. Legionnaires' disease bacterium (Legionella pneumophila) multiples intracellularly in human monocytes. J. Clin. Investig. 66:441-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kagan, J. C., and C. R. Roy. 2002. Legionella phagosomes intercept vesicular traffic from endoplasmic reticulum exit sites. Nat. Cell Biol. 4:945-954. [DOI] [PubMed] [Google Scholar]
- 22.Kagan, J. C., M. P. Stein, M. Pypaert, and C. R. Roy. 2004. Legionella subvert the functions of Rab1 and Sec22b to create a replicative organelle. J Exp. Med. 199:1201-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, V. T., D. M. Anderson, and O. Schneewind. 1998. Targeting of Yersinia Yop proteins into the cytosol of HeLa cells: one-step translocation of YopE across bacterial and eukaryotic membranes is dependent on SycE chaperone. Mol. Microbiol. 28:593-601. [DOI] [PubMed] [Google Scholar]
- 24.Li, Z., J. M. Solomon, and R. R. Isberg. 2005. Dictyostelium discoideum strains lacking the RtoA protein are defective for maturation of the Legionella pneumophila replication vacuole. Cell Microbiol. 7:431-442. [DOI] [PubMed] [Google Scholar]
- 25.Luo, Z. Q., and R. R. Isberg. 2004. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc. Natl. Acad. Sci. USA 101:841-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDade, J. E., C. C. Shepard, D. W. Fraser, T. R. Tsai, M. A. Redus, and W. R. Dowdle. 1977. Legionnaires' disease: isolation of a bacterium and demonstration of its role in other respiratory disease. N. Engl. J. Med. 297:1197-1203. [DOI] [PubMed] [Google Scholar]
- 27.McLean, I. W., and P. K. Nakane. 1974. Periodate-lysine-paraformaldehyde fixative. A new fixation for immunoelectron microscopy. J. Histochem. Cytochem. 22:1077-1083. [DOI] [PubMed] [Google Scholar]
- 28.Merriam, J. J., R. Mathur, R. Maxfield-Boumil, and R. R. Isberg. 1997. Analysis of the Legionella pneumophila fliI gene: intracellular growth of a defined mutant defective for flagellum biosynthesis. Infect. Immun. 65:2497-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagai, H., E. D. Cambronne, J. C. Kagan, J. C. Amor, R. A. Kahn, and C. R. Roy. 2005. A C-terminal translocation signal required for Dot/Icm-dependent delivery of the Legionella RalF protein to host cells. Proc. Natl. Acad. Sci. USA 102:826-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagai, H., J. C. Kagan, X. Zhu, R. A. Kahn, and C. R. Roy. 2002. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science 295:679-682. [DOI] [PubMed] [Google Scholar]
- 31.Phillips, R. M., D. A. Six, E. A. Dennis, and P. Ghosh. 2003. In vivo phospholipase activity of the Pseudomonas aeruginosa cytotoxin ExoU and protection of mammalian cells with phospholipase A2 inhibitors. J. Biol. Chem. 278:41326-41332. [DOI] [PubMed] [Google Scholar]
- 32.Raynaud, C., C. Guilhot, J. Rauzier, Y. Bordat, V. Pelicic, R. Manganelli, I. Smith, B. Gicquel, and M. Jackson. 2002. Phospholipases C are involved in the virulence of Mycobacterium tuberculosis. Mol. Microbiol. 45:203-217. [DOI] [PubMed] [Google Scholar]
- 33.Rose, M., F. Winston, and P. Hieter. 1990. Methods in yeast genetics. A laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Sato, H., J. B. Feix, C. J. Hillard, and D. W. Frank. 2005. Characterization of phospholipase activity of the Pseudomonas aeruginosa type III cytotoxin, ExoU. J. Bacteriol. 187:1192-1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sato, H., D. W. Frank, C. J. Hillard, J. B. Feix, R. R. Pankhaniya, K. Moriyama, V. Finck-Barbancon, A. Buchaklian, M. Lei, R. M. Long, J. Wiener-Kronish, and T. Sawa. 2003. The mechanism of action of the Pseudomonas aeruginosa-encoded type III cytotoxin, ExoU. EMBO J. 22:2959-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schiestl, R. H., and R. D. Gietz. 1989. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 16:339-346. [DOI] [PubMed] [Google Scholar]
- 37.Schmiel, D. H., E. Wagar, L. Karamanou, D. Weeks, and V. L. Miller. 1998. Phospholipase A of Yersinia enterocolitica contributes to pathogenesis in a mouse model. Infect. Immun. 66:3941-3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shohdy, N., J. A. Efe, S. D. Emr, and H. A. Shuman. 2005. Pathogen effector protein screening in yeast identifies Legionella factors that interfere with membrane trafficking. Proc. Natl. Acad. Sci. USA 102:4866-4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stone, B. J., and Y. Abu Kwaik. 1999. Natural competence for DNA transformation by Legionella pneumophila and its association with expression of type IV pili. J. Bacteriol. 181:1395-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.VanRheenen, S. M., G. Dumenil, and R. R. Isberg. 2004. IcmF and DotU are required for optimal effector translocation and trafficking of the Legionella pneumophila vacuole. Infect. Immun. 72:5972-5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vogel, J. P., and R. R. Isberg. 1999. Cell biology of Legionella pneumophila. Curr. Opin. Microbiol. 2:30-34. [DOI] [PubMed] [Google Scholar]
- 42.Walker, D. H., H. M. Feng, and V. L. Popov. 2001. Rickettsial phospholipase A2 as a pathogenic mechanism in a model of cell injury by typhus and spotted fever group rickettsiae. Am. J. Trop. Med. Hyg. 65:936-942. [DOI] [PubMed] [Google Scholar]
- 43.Watarai, M., H. L. Andrews, and R. R. Isberg. 2001. Formation of a fibrous structure on the surface of Legionella pneumophila associated with exposure of DotH and DotO proteins after intracellular growth. Mol. Microbiol. 39:313-329. [DOI] [PubMed] [Google Scholar]
- 44.Wessel, D., and U. I. Flugge. 1984. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138:141-143. [DOI] [PubMed] [Google Scholar]