Abstract
Plants, like all eukaryotes and most prokaryotes, have evolved sophisticated mechanisms for anticipating predictable environmental changes that arise due to the rotation of the Earth on its axis. These mechanisms are collectively termed the circadian clock. Many aspects of plant physiology, metabolism and development are under circadian control and a large proportion of the transcriptome exhibits circadian regulation. In the present review, we describe the advances in determining the molecular nature of the circadian oscillator and propose an architecture of several interlocking negative-feedback loops. The adaptive advantages of circadian control, with particular reference to the regulation of metabolism, are also considered. We review the evidence for the presence of multiple circadian oscillator types located in within individual cells and in different tissues.
Keywords: biological rhythm, circadian clock, photoperiodism, plant, temperature regulation, timekeeping
Abbreviations: aRNA, antisense RNA; CAB, chlorophyll A/B-binding protein; CAT3, catalase 3; CBS, CCA1-binding site; CCA1, CIRCADIAN CLOCK ASSOCIATED 1; CHS, CHALCONE SYNTHASE; COP1, CONSTITUTIVELY PHOTOMORPHOGENIC 1; CO, CONSTANS; CRY, cryptochrome; [Ca2+]cyt, cytosolic free Ca2+ concentration; DET1, DE-ETIOLATED 1; ELF, EARLY FLOWERING; FT, FLOWERING LOCUS T; FRQ, FREQUENCY; GRP, GLYCINE-RICH PROTEIN; GI, GIGANTEA; LHY, LATE ELONGATED HYPOCOTYL; LKP2, LIGHT OXYGEN OR VOLTAGE/KELCH PROTEIN 2; LOV, LIGHT OXYGEN OR VOLTAGE; LUC, LUCIFERASE; LUX, LUX ARRHYTHMO; NR, nitrate reductase; PER, PERIOD; PHOT, phototropin; PHY, phytochrome; PRR, PSEUDO RESPONSE REGULATOR; Skp1, S-phase kinase-associated protein 1; SCF, Skp1/cullin/F-box; SCN, suprachiasmatic nucleus; SPY, SPINDLY; TOC1, TIMING OF CAB EXPRESSION 1; ZTL, ZEITLUPE
INTRODUCTION
Many aspects of the behaviour and physiology of plants and animals fluctuate over the course of each day. Some of these fluctuations occur solely in response to environmental factors, such as light/dark cycles, but a subset persist under constant conditions. Persistent rhythms that have periodicities matching that of the Earth's rotation on its axis (approx. 24 h) are termed circadian. Circadian rhythms were first measured in 1729 by the French biologist de Mairan, who noted that a heliotrope plant (probably Mimosa) sustained rhythms in leaf movement in continuous darkness [1–4]. Circadian rhythms have since been observed in most eukaryotes and in many prokaryotes [4,5]. Circadian outputs range from the relatively subtle, such as rhythms in photosynthesis, to the comparatively overt, such as rhythms in animal activity, plant leaf movement and floral opening [2,3,5].
Circadian rhythms are governed by an internal timekeeper referred to as the circadian clock. The clocks in different kingdoms are composed of a network of transcription factors arranged in interlocking negative-feedback loops [2,5–7]. The clock maintains an internal estimate of the passage of time and schedules physiological processes to occur at appropriate points in the day. In order to remain synchronized with the environment, circadian clocks are reset or entrained by specific cues that relay information about the external time. The ‘time-givers’ (or zeitgebers) are generally the light/dark and temperature cycles, although rhythms in nutrient availability may also act as a resetting signal in some organisms [5,8]. A critical property of circadian oscillators is temperature compensation, or the stability of the period of the clock over a wide range of temperatures in the physiological range. This allows clocks to maintain an accurate phase relationship between physiology and the environment in circumstances of unpredictable change in environmental temperature [2,4,5,9]. Recently, the molecular nature of the plant circadian clock, the processes regulated by circadian signals and some of the advantages conferred by temporal control of physiology have been identified. In this review, we examine the advances in identifying the genetic components of the oscillator and mechanisms of rhythmic gene control. We speculate also as to the processes by which rhythmic gene control may contribute to the enhanced fitness of plants that is conferred by the circadian clock and consider areas for future research. This emerging picture of the circadian clock describes how plants, even without a central nervous system, have sophisticated systems to tell the time.
WHY DO PLANTS HAVE CIRCADIAN CLOCKS?
In order to understand why organisms have circadian clocks, it is necessary to understand the selective pressures that shaped their evolution [1,10]. Pittendrigh [1] hypothesized that the primary force behind the evolution of circadian clocks is the inherent advantage of phasing reactions that are adversely affected by sunlight so that they occur during the night. There also could be several other selective advantages to temporal programming. The anticipation of regular changes in the environment shortens the delay between a change in the environment and the appropriate alteration in physiology [5,10–12]. In plants, the induction of light-input pathways before dawn may allow full use to be made of the light period, while the induction of stress-response mechanisms may anticipate water-deficit stress in the late afternoon [13–15]. Scheduling of biological programmes also may be advantageous by allowing incompatible reactions to be segregated temporally. Although advantages of temporal control by the clock appear to be intuitive, there are very few examples in the plant or other kingdoms where they have been conclusively been demonstrated to occur.
Direct experimental evidence for a fitness benefit associated with the possession of a biological clock has, until recently, remained elusive. The first rigorous demonstration used cyanobacterial strains with free running circadian periods of 22, 25 or 30 h [11]. When the strains were cultured together in 22 h cycles (11 h light/11 h dark), the mutant with a 22 h circadian clock period outgrew the strains with longer periods. However, in a 30 h cycle (15 h light/15 h dark), the 30 h period mutant dominated. Similarly, under a normal 24 h cycle (12 h light/12 h dark), the wild-type (25 h period) strain outgrew the mutants [11]. These results demonstrated that cyanobacteria that had clock periods most closely matching that of their environment were able to outcompete those with mismatched clock periods [10,11]. The benefits of this ‘circadian resonance’ [16] also occur in plants. When wild-type and long- and short-period circadian clock mutants of Arabidopsis were grown in a range of environmental period lengths, plants with clock periods similar to the period of the total light/dark cycle grew more successfully than those with dissimilar periods [12]. Similarly, plants overexpressing CCA1 (CIRCADIAN CLOCK ASSOCIATED 1), which are circadian arrhythmic, performed worse for all parameters measured than wild-type plants in 12 h light/12 h dark cycles [12]. This study also identified some of the metabolic pathways that are enhanced by matching the period of the endogenous circadian oscillator with that of the environmental cycles [12]. Plants with resonant circadian periods accumulated more chlorophyll, fixed more carbon, attained approx. 45% greater biomass and grew faster [12]. In competition studies, the plants with resonant clocks out-competed those plants with clock periods that did not match the period of the light/dark cycle [12]. At least under some conditions, wild-type plants produce more viable seeds than circadian mutants, suggesting that the clock can genuinely increase fitness, i.e. fecundity [17]. However, because the clock in Arabidopsis is intimately connected with the photoperiodic response apparatus, interpreting the effects of clock mutants on fitness by measuring seed production is difficult. It is not just photosynthetic organisms that benefit from circadian clocks, as mice with mutations in the key circadian transcription factor CLK (CLOCK) have severe metabolic disorders, including hyperglycaemia, hypercholesterolaemia and hyperlipidaemia [18].
PRINCIPLES OF CLOCK ARCHITECTURE
The components of the circadian clock are not conserved between kingdoms, suggesting that clocks must have evolved independently on several occasions [15,19]. To date, the only conserved elements of the clock in plants and animals are the blue-light-sensing CRYs (cryptochromes) and protein kinase CK2α [5,20,21]. Nevertheless, there is a remarkable convergence of architectures and mechanisms among clocks from different organisms.
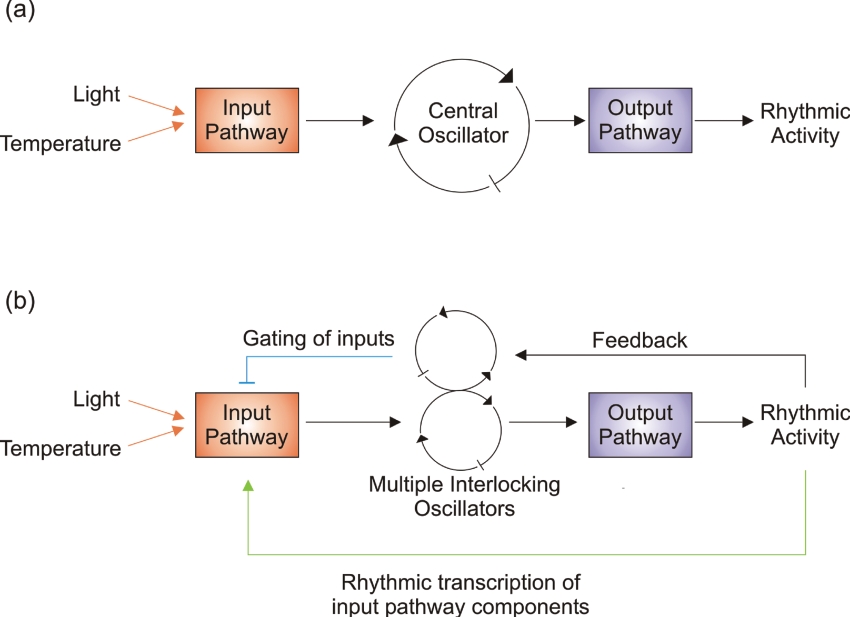
The simplest models describe the circadian clock as consisting of input pathways entraining a core oscillator which generates rhythmic outputs (Figure 1a). However, this linear progression is an oversimplification, because many components of the input pathways are themselves outputs of the clock [22,23] and rhythmic outputs from the clock may feed back to affect the functioning of the core oscillator (Figure 1b) [5]. Additionally, in Neuropsora crassa, Drosophila melanogaster, Arabidopsis and mammals, the core oscillator is composed of multiple interlocking loops (Figure 1b) [5,24,25]. It is believed that, owing to the decentralized organization of the plant body plan, a core circadian oscillator is located in each cell of the plant [26]. This contrasts with mammals, for example, where core oscillators are located in specialized cells in the SCN (suprachiasmatic nucleus) of the brain. The SCN forms a central pacemaker, whereas semi-autonomous ‘slave’ oscillators occur in the peripheral tissues [5,27,28].
Figure 1. Schematic representation of circadian clock structures.
(a) A model depicting division of the clock into an input pathway, a central oscillator and an output pathway. (b) An elaborated description of the clock, consisting of multiple core oscillators, gated input pathways and outputs which feed back into the central oscillator. Arrows are positive arms and perpendicular lines represent negative arms of the pathway.
An alternative hypothesis is that there may be no core molecular oscillator, but rather multiple independent feedback loops that contribute collectively to timekeeping [8,29,30]. The existence of multiple oscillators has been demonstrated in Lingulodinium polyedrum (formerly Gonyaulax polyedra) [31,32] and N. crassa [33]. In N. crassa, a secondary oscillator maintains rhythms in NR (nitrate reductase) activity in the absence of the integral clock component FRQ (FREQUENCY) [34], indicating that it is a self-sustaining oscillator. A similar oscillator maintaining rhythms in NR activity may operate in plants [30].
It is likely that our understanding of the complexity of clock architecture will continue to expand as new clock genes, additional levels of molecular control and layers of cell-specificity are incorporated into the basic models. However, it is pertinent to ask why circadian clocks appear to have converged on such complex architectures. Several authors have speculated that the complexity provides the clock with stability and protection against stochastic perturbations [35–37]. More recently, modelling studies have suggested that complexity is a necessary in order to impart flexibility to circadian oscillators [38]. This flexibility forms the basis of the key clock property of entrainment [38]. It is also probable that the complexity is indicative of the need for clocks to have widespread control in order to confer effective temporal separation of diverse physiological programmes.
THE CIRCADIAN CLOCK OF ARABIDOPSIS: INPUT PATHWAYS AND ENTRAINMENT
The circadian clock of Arabidopsis has an intrinsic period of between 22 and 29 h, depending on plant accession and growth conditions [39]. Consequently, the clock mechanism is reset each dawn and/or dusk [15,40] to prevent the clock becoming increasingly desynchronized with the external light/dark cycles. Resetting of the clock involves a change in phase that does not alter the internal sequence of processes, but re-aligns the sequence with the daily environmental progression [15]. This flexibility of the clock allows the organism to adjust to changing day length and time of dawn during seasonal transitions [40].
Light is probably the predominant entrainment cue in Arabidopsis thaliana [40]. Correct entrainment of the clock to light signals is dependent, in part, on different responses of the clock to light at different times of day. At some points in the diurnal cycle, light pulses will advance the clock, and, at other points, the clock will be delayed [41]. The mechanisms by which these changes in sensitivity or ‘gating’ of the responsiveness to light during entrainment occur are obscure but provide evidence for the oscillator being responsive to resetting cues only at appropriate points in the day. Additionally, the light signal must be relatively prolonged in order to reset the clock [40,42]. These properties of the clock may provide the advantage of preventing resetting by inappropriate signals; gating preventing moonlight from resetting the oscillator and the requirement for a prolonged signal preventing responses to flashes of lightning.
In diurnal organisms such as Arabidopsis, the free-running period of the clock is shortened as the intensity of light increases, a phenomenon known as Aschoff's rule [41,43]. Aschoff's rule provides evidence for continuous readjustment of the period during the photoperiod as opposed to phase-specific responsiveness of the clock to resetting cues. It is likely that accurate synchronization and maintenance of circadian rhythms relies on both phase-responsive and continuous resetting mechanisms.
Both the red-light-sensing PHYs (phytochromes) and the blue-light-sensing CRYs contribute to light input to the circadian clock in Arabidopsis [41,44]. This was established by analysing the response of PHY- and CRY-null mutants carrying the CAB2 (CHLOROPHYLL A/B-BINDING PROTEIN 2)::LUC (LUCIFERASE) reporter to increasing fluence rates of red and blue light [44]. The results indicated that PHYA acts as a receptor of low-fluence-rate red and blue light for the clock, PHYB as a high-fluence-rate red light receptor [40,44]. phyD and phyE mutants also have clock phenotypes in red light, but only in the absence of PHYB. This suggests that they share an overlapping functionality with PHYB and are partly redundant [40,45]. Intriguingly, not all red light input into the clock is accounted for by the action of PHYA, PHYB, PHYD and PHYE. It is likely that this residual input is due to the action of PHYC, but this remains to be verified experimentally [40,45]. Both CRY1 and CRY2 probably act as blue light receptors to entrain the clock with a degree of redundancy [41]. CRY1 may also act downstream of PHYA in red light signalling [40,45].
Although the CRYs and PHYs govern light input, they are also rhythmic outputs of the clock. All exhibit circadian oscillations at the RNA level, though only PHYA, PHYB and PHYC appear to oscillate at the protein level [13,46–49]. This may contribute to the rhythmic sensitivity of the clock to light [40,50] in concert with other factors, such as ELF3 (EARLY FLOWERING 3). The elf3 mutant has an arrhythmic phenotype in constant light, but displays robust rhythms in constant dark [51–53]. elf3 mutants also fail to gate the acute induction of CAB2::LUC, and show increased sensitivity to light stimuli during the subjective night. This suggests that ELF3 acts to antagonize light input into the clock during the night and also contributes to resetting of the oscillator [52,53].
It is possible that photoreceptors other than CRYs and PHYs also participate in entrainment of the core oscillator. PHOTs (phototropins) may be candidates for transducing blue light signals to the clock, since they are involved in sensing blue light in Arabidopsis. However, at the time of writing, their role in the clock has not been thoroughly tested [13,40,41]. An alternative possibility is that additional light input is mediated by ZTL (ZEITLUPE) and LKP2 [LOV (LIGHT OXYGEN OR VOLTAGE)/KELCH PROTEIN 2]. ZTL and LKP2 are two of the three members of a family of F-box, KELCH and LOV domain proteins. The LOV domain is similar to the chromophore-binding domain of the PHOT light receptors, and undergoes light-induced conformational changes in vitro [54–56]. This suggests that the ZTL family of proteins could act as blue light receptors. However, ztl mutations also cause period lengthening in darkness, and ZTL has been shown to form part of the mechanism that targets TOC1 (TIMING OF CAB EXPRESSION 1) for degradation [41,57]. Mutations in ZTL affect the ability of ZTL to interact with TOC1, and probably other proteins. Therefore it is probable that the LOV domain may function in regulating a light-dependent activity of ZTL that is involved in targeting proteins for degradation [58,59]. The role of ZTL in targeting clock proteins for degradation is discussed further below.
The circadian clock can also be entrained by temperature cycles in which the day and night temperatures differ by 4 °C or more [15,40,60]. The molecular basis of temperature entrainment of the clock is not well understood. It has been suggested that the failure of the prr7 (pseudo response regulator 7)/prr9 double mutant to maintain circadian rhythms after temperature entrainment indicates that PRR7 and PRR9 are involved in temperature input to the clock [61]. Alternatively, the observed phenotype may be a consequence of the possible involvement of these genes in the central oscillator itself [62] or light entrainment of the oscillator [41]. Temperature is a very difficult signal to study because all biochemical systems are sensitive to temperature changes, making identification of specific components of temperature-sensing pathways a challenge. With the exception of the data described above, we know little about temperature-sensing by the clock.
THE CIRCADIAN CLOCK OF ARABIDOPSIS: THE CORE OSCILLATOR
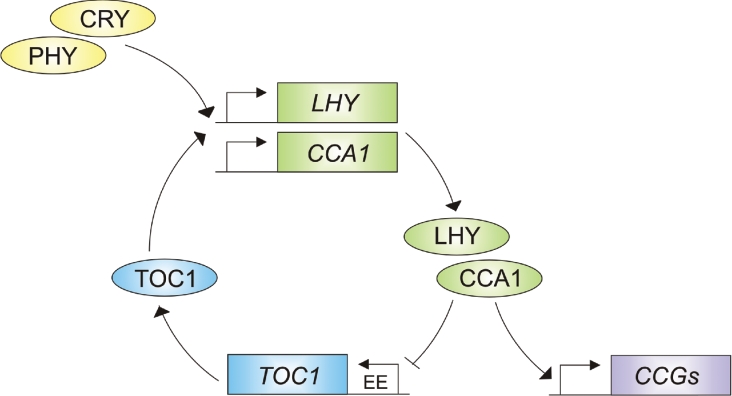
As in other organisms, the central oscillator in Arabidopsis is proposed to consist of elements arranged in interlocking transcriptional feedback loops [5,62]. The first loop to be described consists of TOC1, LHY (LATE ELONGATED HYPO-COTYL) and CCA1 (Figure 2) [63]. CCA1 and LHY encode MYB-like transcription factors that share considerable sequence similarity and have partly redundant functions in the clock [64,65]. Overexpression of either gene severely compromises detectable rhythms, indicating that they are components of the oscillator [63–65]. CCA1 and LHY bind to the promoter of TOC1, a PRR that is also required for the maintenance of circadian rhythms in constant light or constant dark [63,66–68]. TOC1 is predicted to promote both CCA1 and LHY expression by an unknown mechanism [63]. The CCA1–LHY–TOC1 feedback loop has been proposed to function as follows: CCA1 and LHY are expressed rhythmically with a peak in transcript abundance at subjective dawn. The cognate proteins are produced within 2–3 h and inhibit TOC1 expression by binding an evening element in the TOC1 promoter [63]. This causes a gradual decline in TOC1 levels, and consequently a decline in CCA1 and LHY production. As CCA1 and LHY levels decline, the repression of TOC1 expression is lifted, and the cycle is re-initiated (Figure 2) [15,63,69].
Figure 2. Proposed structure of the CCA/LHY–TOC1 feedback loop.
Light input, together with TOC1, induces the expression of CCA1 and LHY. CCA1 and LHY activate the expression of clock-controlled genes (CCGs, shown in mauve) and repress TOC1 expression [63] which in turn represses LHY/CCA1 expression. Light input is mediated by the CRYs and PHYs. Arrows represent positive regulatory steps, perpendicular lines indicate negative interactions. Genes are represented by rectangles and proteins as ovals.
Several lines of evidence suggest that this first model of the plant clock is incomplete. First, cca1/lhy double-null mutants are not completely arrhythmic [65,70]. Secondly, rhythms in ELF3 expression persist in mutants constitutively overexpressing LHY [71]. Thirdly, the model cannot account for why mutation and overexpression of TOC1 both lead to reductions in CCA1 and LHY expression [69]. Finally, it is unclear whether TOC1 is directly responsible for regulating CCA1 and LHY expression, or, if so, how this might be achieved [15,69]. Thus other components are required for the functioning of the core oscillator.
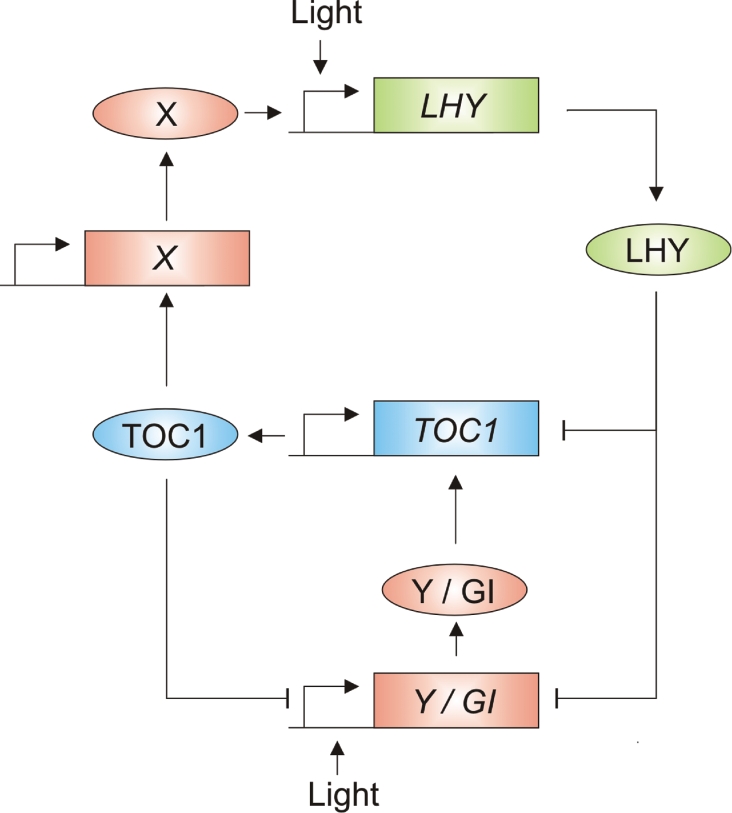
A mathematical model incorporating two additional components has recently been proposed [72,73]. In this model, light activates the expression of a hypothetical component, ‘Y’, which induces expression of TOC1. TOC1 acts via another hypothetical component, ‘X’, to induce LHY expression, and LHY and TOC1 both act to repress the expression of Y (Figure 3) [72,73]. Although the identity of Y has not been confirmed, both mathematical and experimental data suggest that its function might be fulfilled by GI (GIGANTEA) [72–74]. GI expression follows a circadian pattern, and has a broad peak in the late subjective afternoon, matching the predicted expression profile for Y [72,73]. GI is also rapidly but transiently induced by light, and its promoter regions contain several evening elements required for LHY-mediated repression [72,73]. Overexpression or mutation of GI affects circadian rhythms in both continuous light and continuous dark, implying that GI acts within the central clock mechanism [72–74].
Figure 3. Revised model of the LHY–TOC1 feedback loop [72,73].
Mathematical analysis indicates that the additional components X and Y are required in order to form a robust oscillator that matches the properties of the Arabidopsis clock. X is a gene that is required for the activation of LHY by TOC1. Y is a hypothetical gene that is activated by light and in turn activates TOC1. Y is repressed by both TOC1 and LHY. GI is a candidate for the identity of Y. X is currently unknown. Arrows represent positive regulatory steps, perpendicular lines indicate negative interactions. Genes are represented by rectangles and proteins as ovals.
Several members of the PRR gene family, of which TOC1 is a member, have a role in clock function and may have a dual role in light input [25,62,75,76]. The members of the family are expressed between dawn and dusk in the sequence PRR9–PRR7–PRR5–PRR3–TOC1 [25]. CCA1 regulates the expression of PRR7 and PRR9 directly by binding to a CBS (CCA1-binding site) in their promoters [62]. However, the prr7/prr9 double mutation results in a delay in the period of CCA1 and LHY expression of 4–5 h, indicating that the PRRs feed back to regulate CCA1 and LHY expression [62]. Additionally, the prr7/prr9 double mutant has extremely long periods in constant light and constant dark, which is consistent with a role in the central oscillator [62,75]. Nevertheless, the position of the PRRs in the core oscillator remains ambiguous because they have been implicated in both light and temperature entrainment [25,61,62,77].
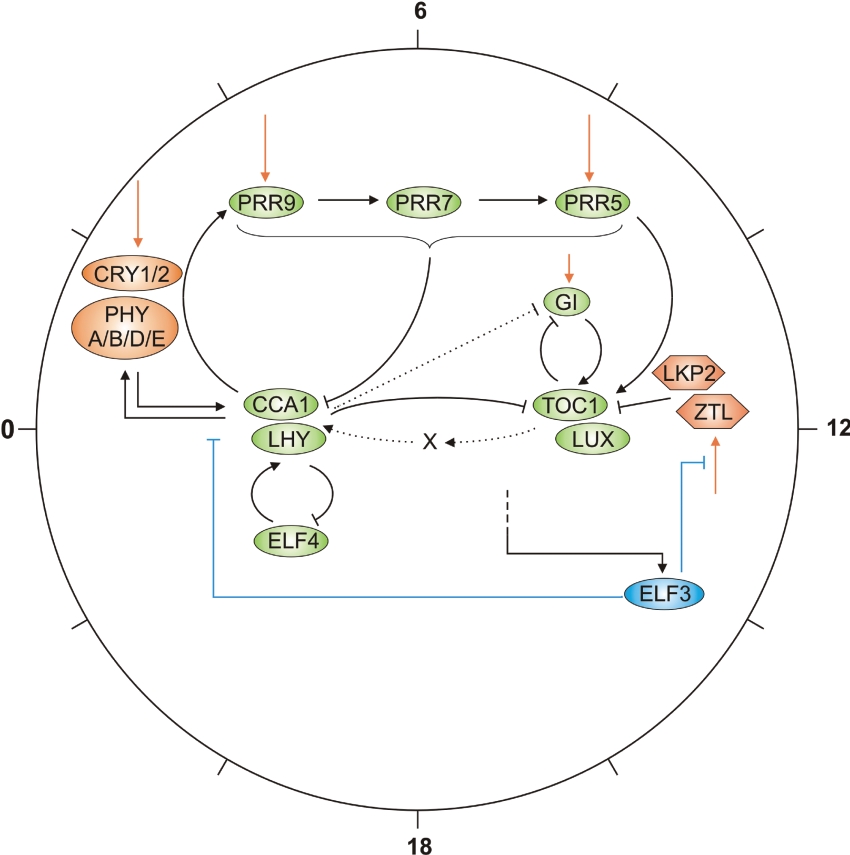
ELF4 and LUX (LUX ARRYTHMO, also called PHYTOCLOCK) have also recently been proposed to be components of the core oscillator [78–80]. LUX encodes a MYB transcription factor which is expressed co-ordinately with TOC1 in both different photoperiods and mutant backgrounds [78]. The LUX promoter contains an evening element which is bound by CCA1 and LHY, suggesting that these proteins repress LUX transcription [78]. Curiously, the MYB domain of LUX is also present in CCA1 and LHY. Mutation of the LUX gene leads to arrhythmia in constant light and constant dark for most outputs and CCA1/LHY are clamped low, while TOC1 is clamped high [78]. Overexpression of LUX reduced the levels of expression of both GI and LUX and damped oscillations of LUX as well as TOC1, CCA1 and LHY in continuous light [80]. ELF4 is required for maintenance of rhythmic outputs and for light-induced expression of CCA1 and LHY, but not TOC1 [79,81]. ELF4 expression is repressed by CCA1 and LHY, indicating the existence of an additional feedback loop in the central oscillator which includes CCA1, LHY and ELF4, but not TOC1 (Figure 4) [79]. Collectively, the data indicate that there are multiple interlocking feedback loops in the central oscillator of Arabidopsis. We propose one model of the arrangement these multiple regulatory loops in Figure 4. Positioning the many elements accurately requires further information and will be aided by mathematical analyses.
Figure 4. Model of the Arabidopsis circadian clock.
The model is based on recent analyses which indicate that multiple feedback loops exist within the Arabidopsis clock. The CCA1–LHY–TOC1–GI–X feedback loop is outlined in [72,73], the PRR loop in [62,25], and the ELF4 loop in [79]. PHYs, CRYs and ELF3 are under transcriptional control by the circadian clock, so essentially represent additional loops [48,53]. The circle around the model indicates time of day, with 0 representing dawn. Components are positioned within the circle according to their approximate maximal transcript abundance in continuous light, with the exception of LKP2 and ZTL which are not transcribed in a circadian-dependent manner [105,108] and thus are represented by hexagons. Only interactions demonstrated experimentally are shown; positive interactions are shown with arrows, and negative interactions are indicated by perpendicular lines. Dotted lines indicate interactions assumed from a mathematical model but not conclusively demonstrated experimentally [72,73]. Components and interactions associated with light perception are shown in orange, and components and interactions involved in gating are shown in blue. The circadian regulator of ELF3 is unknown and therefore control of ELF3 by the clock is indicated by a dotted line.
How many different types of oscillator are there?
The evidence suggests that every cell in plants has an oscillator and, at least in above-ground tissue, these oscillators are entrained by light and temperature independently of the oscillators in neighbouring cells. The circadian clocks of plants are thought to be cell autonomous because CAB2::LUC luminescence persists in cultured cells [82] and in isolated tissue explants [83]. Furthermore, CAB2::LUC luminescence rhythms can be entrained to opposite phases in the same leaf of a single plant and these oppositely phased rhythms are maintained in constant light, indicating that the oscillators driving the rhythms remained uncoupled [83].
Dissecting the molecular nature of the circadian oscillator is complicated by the possibility that tissue- or cell-specific oscillators may be present, yet most genetic dissection of oscillator structure and function is performed by measuring outputs at the whole-plant scale of observation which summates the outputs of many cell and tissue types [84]. Numerous studies have reported desynchronization of the periods of rhythmic outputs between distinct tissue types, raising the possibility that there are cell or tissue-specific oscillators (Figure 5a). The periods of PHYB or CHS (CHALCONE SYNTHASE) promoter activity, which are predominantly associated with epidermal cells, are 1–1.5 h longer than the period of CAB2 promoter activity, which is mainly associated with mesophyll tissue [85,86]. The periods of leaf movement and CAB2::LUC luminescence rhythms also sometimes differ markedly [87], as can the periods of leaf movement and stomatal conductance [88]. Finally, the phase of circadian rhythms in [Ca2+]cyt (cytosolic free Ca2+ concentration) differs depending on the promoter used to drive the expression of the aequorin reporter in different tissues [89]. However, it should be noted that, since the period of the clock in diurnal organisms lengthens with decreasing light intensity [43], some of these effects could be caused by attenuation of light input by outer cell layers [90]. In this scenario, the different period of rhythms of circadian markers may reflect the intensity of light input to the cell populations in which that marker is active. For example, in the ztl1 mutation, the period of the oscillator is particularly sensitive to light intensity, and stomatal and photosynthetic rhythms are desynchronized because these outputs occur in different cell layers [90]. This can be explained by the cell-autonomous nature of plant circadian oscillators rather than evoking cell-specific oscillator types. The period of the rhythms of stomatal conductance are longer than the period for photosynthesis in ztl1 because the majority of the stomata are on the lower epidermis and receive approx. 25% of the light received by the photosynthetic mesophyll cells, which are nearer the light source [90].
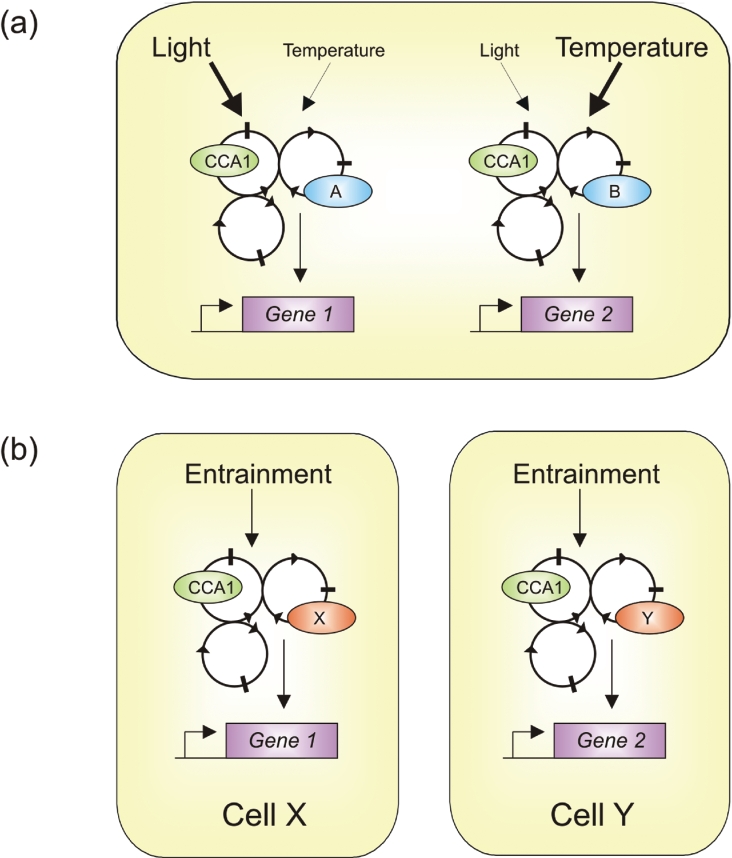
Figure 5. Model to explain how multiple rhythms may be generated from similar oscillators.
Gene 1 and Gene 2 are expressed in similar tissues, but have different free running periods and sensitivity to entrainment signals. (a) This could be due to multiple oscillators present within each cell. Each oscillator is based on the CCA1/LHY model shown in Figure 4, with multiple interlocking loops. Gene 1 expression is preferentially regulated by light which is mediated by an oscillator containing component A. Gene 2 expression is entrained primarily by temperature cycles, and is controlled via an oscillator containing component B. (b) Alternatively, different cells within the same tissue contain different oscillators. Circadian expression of Gene 1 and Gene 2 occur in different cells (X and Y), and the expression data obtained experimentally are an average of all cell types. Gene 1 expression is controlled by an oscillator with the cell-specific factor X, and Gene 2 expression is controlled by an oscillator containing cell-specific factor Y.
The most compelling evidence for the existence of oscillators with different properties comes from studies of temperature and light entrainment. In plants transformed with a CAB2::LUC reporter, the rhythm of luminescence is set preferentially by light/dark cycles relative to temperature cycles, and is responsive to differences in the entrainment photoperiod [60]. In contrast, luminescence rhythms due to a CAT3 (CATALASE 3)::LUC reporter are set preferentially by temperature cycles rather than light/dark cycles, and are insensitive to the entrainment photoperiod [60]. Additionally, the luminescence rhythms in the CAB2::LUC and CAT3::LUC lines have distinct phase responses to low temperature pulses [60]. Since the two promoters drive LUC expression in similar tissues, the results indicate the presence of two circadian oscillators with different sensitivities to entraining signals [60]. To conclusively determine whether the different circadian regulation of the promoters of the CAB2 and CAT3 genes is due to differences in oscillator function between cell types or is due to the presence of multiple oscillators within cells requires simultaneous measurement of both promoter activities over a circadian time course within a single cell (Figure 5) [4,60,82].
If multiple oscillator types are present in different cells or tissues, it is likely that they have a broadly similar molecular architecture, since lesions in core oscillator components tend to have similar effects on rhythmic outputs in all tissues. For example, although the periods of the luminescence rhythms in CHS::LUC and CAB2::LUC plants differ, the toc1-1 mutation shortens both [86]. The toc1-1 mutation also shortens the periods of leaf movement [87] and stomatal conductance [67]. elf3-1 and mutants, and LHY- and CCA1-overexpressing lines all have arrhythmic PHYB and CAB2 promoter activity, despite the fact that PHYB and CAB2 promoter activities are associated with different tissues [85]. Consequently, specialization of the circadian clock in different cell types could be dependent on factors that affect the entrainment of the core oscillator, impinge on its functioning, or modify components of the output pathways [21].
MOLECULAR BASIS OF CLOCK GENE REGULATION
Most clock genes have rhythms in transcript abundance that are followed by rhythms in the abundance of the cognate proteins [5,6]. These proteins bind to promoter elements in other clock genes and initiate their expression. The secondary proteins in turn bind to the promoter elements in the initial gene, and negatively regulate its expression. This process generates rhythms in the steady-state transcript and protein abundance of both components [4,5,29]. However, at least one component must show a significant delay between the expression of the transcript and the expression of the active protein to maintain 24 h periodicity and prevent attenuation of the rhythms [5,6,37]. Several levels of control can contribute to the imposition of appropriate delays and the maintenance of rhythms in transcript and/or protein abundance.
Transcriptional control of clock genes
Cis-acting elements that confer circadian expression have been characterized in various organisms [13,91–94]. In Arabidopsis, these include the evening element (AAAATATCT) [13] in genes which encode transcripts expressed towards the end of the subjective afternoon, and the CBS (AAAAATCT) [95] and morning element [94] in genes which encode transcripts expressed around subjective dawn. However, rhythmic transcription is not always necessarily a prerequisite for the generation of rhythms in steady-state transcript abundance. In Arabidopsis, there are variations in degradation rates for the circadian transcripts COLD AND CLOCK REGULATED-LIKE and SENESCENCE-ASSOCIATED GENE 1, suggesting there are rhythmic changes in the half-life of the mRNA [96]. Rhythmic production of natural aRNA (antisense RNA) may underlie the rhythms in RNA degradation rates. aRNAs complementary to clock transcripts have been found in N. crassa, Antheraea pernyi and mammals [97,98]. In N. crassa, an aRNA complementary to the core clock transcript FRQ has circadian rhythms in abundance, with the peak abundance 180° out of phase with the peak in FRQ transcript abundance. The antisense PER (PERIOD) transcript shows a similar cycle inversion relative to PER abundance in Antheraea pernyi [98].
Alternative splicing of transcripts may also play a role in generating rhythmic phenomena. In Arabidopsis, alternative splicing is involved in the GRP7/8 (GLYCINE-RICH PROTEIN 7/8) slave oscillator feedback loop. GRP7 accumulates during the subjective day, binds to the 3′-untranslated region of the GRP7/8 transcripts and shifts the splice site selection in favour of a truncated version of both transcripts. These transcripts are degraded rapidly, ultimately leading to a decrease in the abundance of GRP7 [99].
Translational and post-translational control of clock genes
Translational control may be important in introducing a delay between the production of certain clock transcripts and the synthesis of the corresponding proteins. Control of protein degradation is integral to the maintenance of robust circadian oscillations in protein levels [6,100]. Targeted degradation of at least one key clock protein occurs in cyanobacteria, N. crassa, D. melanogaster, Arabidopsis and mammals [5]. Progressive phosphorylation before degradation appears to be a key feature in all these systems, and in many instances, CK1 and CK2 may be the primary protein kinases responsible [20,101].
The proteins phosphorylated by CK1, CK2 and other kinases are targeted for degradation via the ubiquitin–proteasome pathway [58]. This involves the addition of a polyubiquitin chain to the protein, a process mediated by the SCF [Skp1 (S-phase kinase-associated protein 1)/cullin/F-box] ubiquitin ligase-mediated proteasomal pathway [102,103]. Different F-box proteins can be assembled on to the SCF to target core clock components for degradation, including TOC1 in Arabidopsis [58]. Since yeast-two hybrid assays indicate that LKP2 interacts with both Skp1-like proteins and TOC1 [104], LKP2 may have a similar function to ZTL in mediating TOC1 degradation. The potential redundancy of ZTL and LKP2 is underscored by the fact that neither ztl nor lkp2 mutants are arrhythmic, whereas mutants overexpressing either ZTL or LKP2 are, presumably due to increased degradation of TOC1 [56,59,105]. The ztl/lkp2 double mutant has a phenotype identical with that of ztl, suggesting that the effects of lkp2 on circadian function are subtle [41]. A third member of the ZTL family, FKF1 (FLAVIN-BINDING KELCH REPEAT F-BOX 1), is structurally quite similar to ZTL and LKP2 but appears to function in the regulation of flowering time by mediating the degradation of a repressor of CO (CONSTANS) [55,106,107]. Protein degradation may also be important in the control of the ZTL protein itself because these oscillate, whereas the abundance of ZTL transcripts do not [108,109].
Targeted degradation of clock elements also appears to be a feature of light input to the clock. DET1 (DE-ETIOLATED 1) and the E3 ubiquitin ligase, COP1 (CONSTITUTIVELY PHOTOMORPHOGENIC 1) act as negative regulators of light input into the clock [110,111]. DET1 functions by targeting the core clock component LHY for degradation [110] and it is likely that COP1 acts in a similar manner. Both det1 and cop1 mutations lead to period shortening in constant light [87,110]. However, DET1 may also regulate light-independent processes, and therefore might function close to, or within, the core oscillator [110].
Other post-translational mechanisms that affect the functioning of the circadian clock include rhythmic activation by phosphorylation [112], modification of protein–protein interactions, alteration of subcellular localization and poly(ADP-ribosyl)ation [113,114]. Only the latter has been studied extensively in plants. In Arabidopsis, the tej (‘Tej’ means ‘bright’ in Sanskrit) mutation indicates a role for poly(ADP-ribosyl)ation in the control of clock period [115]. TEJ encodes a functional poly(ADP-ribosyl) glycohydrolase, which degrades ADP-ribosyl polymers [115]. The tej mutation lengthens the period of the expression of all known circadian transcripts, suggesting that a core clock component may be the target of poly(ADP-ribosyl)ation [115].
THE CIRCADIAN CLOCK OF ARABIDOPSIS: CLOCK OUTPUTS
The outputs of the central oscillator are pathways that lead to physiological and biochemical rhythms such as photosynthesis, leaf movement, hypocotyl elongation, stomatal movement and circumnutation [4,5,21,26,116]. It is these rhythms in clock outputs that ultimately provide the advantages in growth and competition conferred by the clock [12]. In general, the pathways by which temporal information from the core oscillator regulates cell physiology have not been extensively described. Rhythms in steady-state transcript abundance are almost certainly an initial output of the circadian oscillator, since several transcription factors that make up the core clock regulate directly the expression of downstream components. CCA1, for example, initiates the expression of light-harvesting-complex genes [65] and represses the expression of several other genes by binding the evening element in their promoter regions [13,63]. The oscillator also regulates the expression of key transcription factors, which in turn regulate the circadian expression of distal elements [13,94,117]. Approx. 2–16% of transcripts encoded in the Arabidopsis genome have rhythms in steady-state abundance [9,13,117]. These include transcripts encoding proteins involved in flowering, flavonoid synthesis, lignin synthesis, cell elongation, nitrogen fixation, carbon metabolism, mineral assimilation and photosynthesis [9,13,117]. Transcripts that encode proteins involved in related pathways tend to be co-regulated, with the peak in transcript abundances occurring at specific phases of the light/dark cycle [13,15]. For example, transcripts encoding components of the light-harvesting complexes tend to be co-expressed during the middle part of the day when light intensity is greatest [13,15]. Similarly, transcripts that encode proteins involved in auxin transport and cell elongation tend to be maximally expressed in the subjective afternoon, which is coincident with the peak in cell-elongation rates [4,13]. Cell elongation is responsible for rhythms in both leaf movement [118] and hypocotyl elongation [119]. Diurnal changes in sugar content of the plants appear interact with output pathways of the clock to reinforce circadian signals and contribute to the control of transcript abundance in light and dark cycles [120]. Collectively, the data suggest that rhythmic transcript abundance underlies many of the known physiological rhythms in Arabidopsis.
Mechanisms other than rhythmic transcript or protein abundance may also couple the central oscillator to rhythms in cellular physiology. At least one output pathway is likely to be based on rhythms in [Ca2+]cyt [21,26,28,84]. In Arabidopsis, [Ca2+]cyt oscillates with a circadian rhythm in constant light and light/dark cycles. The levels of Ca2+ peak in the middle or late subjective afternoon at concentrations between 350 and 700 nmol·l−1 [3,121]. The phase and shape of the [Ca2+]cyt oscillations depend on the photoperiod during entrainment, with the peak in [Ca2+]cyt occurring later in plants entrained to long photoperiods compared with those entrained to short photoperiods [121]. Ca2+ is a potent second messenger and therefore circadian modulation of [Ca2+]cyt could have profound effects on physiology. Although the functions of circadian [Ca2+]cyt oscillations are unknown, roles in light input, photoperiodism, stomatal and leaf movements and Crassulacean acid metabolism have been proposed [21,26,28,84,121]. Identifying a role for circadian oscillations of [Ca2+]cyt has been hampered by the technical difficulties of measuring this output at the single-cell level, resulting in problems in determining the precise nature of the Ca2+ signal and identifying downstream targets [28,84].
One well-defined output pathway from the Arabidopsis clock is the photoperiodic response pathway that controls the transition from vegetative growth to flowering. Arabidopsis is sensitive to day length and flowers earlier in long days than in short days [69]. This response is partly regulated by the circadian oscillator [69], and mutant analyses have identified GI as a clock component that regulates the induction of flowering [74]. GI interacts with SPY (SPINDLY), a protein implicated in gibberellin signalling [122]. Together, GI and SPY induce the circadian expression of CO. CO encodes a zinc-finger protein which is activated by light. Under short day conditions, CO expression peaks during the night phase. In long days, however, the peak in the abundance of CO occurs during the light period [123]. This leads to the production of active CO which is then capable of inducing the expression of a putative kinase inhibitor, FT (FLOWERING LOCUS T) [123]. Like CO, FT is expressed predominantly in the leaves, but FT mRNAs relocate to the shoot apex [124]. In the shoot apex, FT interacts with a transcription factor, FD, to induce the expression of additional genes that mediate the transition to flowering [125,126]. It is possible that circadian oscillations of [Ca2+]cyt may participate in the photoperiodic control of flowering. Circadian oscillations of [Ca2+]cyt and oscillations of [Ca2+]cyt in day/night cycles encode photoperiodic relevant information [121] and a cell-surface sensor of Ca2+ has a role in regulating the transition from vegetative growth to flowering [127].
CONCLUSIONS
Tremendous progress has been made in identifying the genetic components of the circadian oscillator in plants, the photoreceptors governing the light-dependent resetting of the oscillator and understanding how these interact. It is a tribute to genomic technologies that this progress has been made in just over ten years. This contrasts sharply with physiological investigations, which, in the nearly three centuries since de Mairan's initial identification of circadian rhythms, yielded little information about the players in the circadian network of plants. However, large gaps remain: the processes of phototransduction from the photoreceptors to the core oscillator(s) lack detail; only slowly is the role of post-transcriptional events in the clock work being revealed; the emergent properties of the oscillator that underlie robust 24 h rhythms are a matter of speculation; and the mechanisms by which outputs are controlled by the oscillator are virtually unknown. Ironically, investigation of the signalling pathways by which clock outputs are regulated requires a return to physiological approaches, because it is very probable that fluxes of ions, particularly Ca2+ and K+, and protein modifications will be central to these processes. Another exciting challenge for circadian biologists has emerged with the recent demonstrations that an important role of the circadian clock in plants [12] and mammals [18] is to enhance the performance of metabolic pathways. Our current understanding of metabolism in plants does not easily explain how the circadian clock enhances the operation of the biochemical pathways leading to chlorophyll accumulation and carbon assimilation [12]. It is likely that progress in understanding these mechanisms will not only inform circadian biology, but also provide insight into more general aspects of the control of biochemistry and metabolism.
Acknowledgments
A.A.R.W. is grateful to the Royal Society for the award of University Research Fellowship and the BBSRC (Biotechnology and Biological Sciences Research Council) for supporting research in his laboratory. M.J.G. is grateful to the Gates Cambridge Trust and Corpus Christi College, Cambridge, for support. C.T.H. is supported by a CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior) Brazil studentship. A.N.D. and K.E.H. acknowledge the financial support of the BBSRC.
References
- 1.Pittendrigh C. S. Temporal organization: reflections of a Darwinian clock-watcher. Annu. Rev. Physiol. 1993;55:17–54. doi: 10.1146/annurev.ph.55.030193.000313. [DOI] [PubMed] [Google Scholar]
- 2.Barak S., Tobin E. M., Andronis C., Sugano S., Green R. M. All in good time: the Arabidopsis circadian clock. Trends Plant Sci. 2000;5:517–522. doi: 10.1016/s1360-1385(00)01785-4. [DOI] [PubMed] [Google Scholar]
- 3.Johnson C. H., Knight M. R., Kondo T., Masson P., Sedbook J., Haley A., Trewavas A. Circadian oscillations of cytosolic and chloroplastic free calcium in plants. Science. 1995;269:1863–1865. doi: 10.1126/science.7569925. [DOI] [PubMed] [Google Scholar]
- 4.McClung C. R., Salomé S. A., Michael T. P. The Arabidopsis circadian system. In: Somerville C. R., Meyerowitz E. M., editors. The Arabidopsis Book. Rockville: American Society of Plant Biologists; 2002. 10.1199/tab.0044, http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harmer S. L., Panda S., Kay S. A. Molecular bases of circadian rhythms. Annu. Rev. Cell Dev. Biol. 2001;17:215–253. doi: 10.1146/annurev.cellbio.17.1.215. [DOI] [PubMed] [Google Scholar]
- 6.Dunlap J. C. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 7.Lakin-Thomas P. L. Circadian rhythms: new functions for old clock genes? Trends Genet. 2000;16:135–142. doi: 10.1016/s0168-9525(99)01945-9. [DOI] [PubMed] [Google Scholar]
- 8.Roenneberg T., Merrow M. Molecular circadian oscillators: an alternative hypothesis. J. Biol. Rhythms. 1998;13:167–179. doi: 10.1177/074873098129000011. [DOI] [PubMed] [Google Scholar]
- 9.Edwards K. D., Anderson P. E., Hall A., Salathia N. S., Locke J. C. W., Lynn J. R., Straume M., Smith J. Q., Millar A. J. FLOWERING LOCUS C mediates natural variation in the high-temperature response of the Arabidopsis circadian clock. Plant Cell. 2006;18:639–650. doi: 10.1105/tpc.105.038315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson C. H. Endogenous timekeepers in photosynthetic organisms. Annu. Rev. Physiol. 2001;63:695–728. doi: 10.1146/annurev.physiol.63.1.695. [DOI] [PubMed] [Google Scholar]
- 11.Ouyang Y., Andersson C. R., Kondo T., Golden S. S., Johnson C. H. Resonating circadian clocks enhance fitness in cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 1998;95:8660–8664. doi: 10.1073/pnas.95.15.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dodd A. N., Salathia N., Hall A., Tóth E.-K., Nagy F., Hibberd J. M., Millar A. J., Webb A. A. R. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309:630–633. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- 13.Harmer S. L., Hogenesch J. B., Straume M., Chang H. S., Han B., Zhu T., Wang X., Kreps J. A., Kay S. A. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- 14.Kreps J. A., Wu Y., Chang H. S., Zhu T., Wang X., Harper J. F. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 2002;130:2129–2141. doi: 10.1104/pp.008532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Millar A. J. Input signals to the plant circadian clock. J. Exp. Bot. 2004;55:277–283. doi: 10.1093/jxb/erh034. [DOI] [PubMed] [Google Scholar]
- 16.Pittendrigh C. S., Bruce V. G. Daily rhythms as coupled to oscillator systems and their relationship to thermoperiodism and photoperiodism. In: Withrow R. B., editor. Photoperiodism and Related Phenomena in Plants and Animals. Washington D.C.: American Association for the Advancement of Science; 1959. pp. 475–505. [Google Scholar]
- 17.Green R. M., Tingay S., Wang Z. Y., Tobin E. M. Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol. 2002;129:576–584. doi: 10.1104/pp.004374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turek F. W., Joshu C., Kohsaka A., Lin E., Ivanova G., McDearmon E., Laposky A., Losee-Olson S., Easton A., Jensen D. R., et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Young M. W., Kay S. A. Time zones: a comparative genetics of circadian clocks. Nat. Rev. Genet. 2001;2:702–715. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- 20.Sugano S., Andronis C., Green R. M., Wang Z. Y., Tobin E. M. Protein kinase CK2 interacts with and phosphorylates the Arabidopsis circadian clock-associated 1 protein. Proc. Natl. Acad. Sci. U.S.A. 1998;95:11020–11025. doi: 10.1073/pnas.95.18.11020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Webb A. A. R. The physiology of circadian rhythms in plants. New Phytol. 2003;160:281–303. doi: 10.1046/j.1469-8137.2003.00895.x. [DOI] [PubMed] [Google Scholar]
- 22.Merrow M., Brunner M., Roenneberg T. Assignment of circadian function for the Neurospora clock gene frequency. Nature (London) 1999;399:584–586. doi: 10.1038/21190. [DOI] [PubMed] [Google Scholar]
- 23.Devlin P. F., Kay S. A. Circadian photoperception. Annu. Rev. Physiol. 2001;63:677–694. doi: 10.1146/annurev.physiol.63.1.677. [DOI] [PubMed] [Google Scholar]
- 24.Bell-Pedersen D., Cassone V. M., Earnest D. J., Golden S. S., Hardin P. E., Thomas T. L., Zoran M. J. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat. Rev. Genet. 2005;6:544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mizuno T., Nakamichi N. Pseudo response regulators (PRRs) or true oscillator components (TOCs) Plant Cell Physiol. 2005;46:677–685. doi: 10.1093/pcp/pci087. [DOI] [PubMed] [Google Scholar]
- 26.Webb A. A. R. Stomatal rhythms. In: Lumsden P. J., Millar A. J., editors. Biological Rhythms and Photoperiodism in Plants. Oxford: Bios Scientific Publications; 1998. pp. 69–79. [Google Scholar]
- 27.Ikeda M. Calcium dynamics and circadian rhythms in suprachiasmatic nucleus neurons. Neuroscientist. 2004;10:315–324. doi: 10.1177/10738584031262149. [DOI] [PubMed] [Google Scholar]
- 28.Gardner M. J., Dodd A. N., Hotta C. T., Sanders D., Webb A. A. R. Circadian regulation of Ca2+ signalling. Annu. Plant Rev. 2005;21:191–210. [Google Scholar]
- 29.Roenneberg T., Merrow M. Circadian light input: omnes viae Romam ducunt. Curr. Biol. 2000;10:742–745. doi: 10.1016/s0960-9822(00)00739-9. [DOI] [PubMed] [Google Scholar]
- 30.Lillo C., Meyer C., Ruoff P. The nitrate reductase circadian system: the central clock dogma contra multiple oscillatory feedback loops. Plant Physiol. 2001;125:1554–1557. doi: 10.1104/pp.125.4.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roenneberg T., Morse D. Two circadian oscillators in one cell. Nature (London) 1993;362:362–364. doi: 10.1038/362362a0. [DOI] [PubMed] [Google Scholar]
- 32.Morse D., Hastings J. W., Roenneberg T. Different phase responses of the two circadian oscillators in Gonyaulax. J. Biol. Rhythms. 1994;9:263–274. doi: 10.1177/074873049400900307. [DOI] [PubMed] [Google Scholar]
- 33.Ramsdale M., Lakin-Thomas P. L. sn-1,2-diacylglycerol levels in the fungus Neurospora crassa display circadian rhythmicity. J. Biol. Chem. 2000;275:41–50. doi: 10.1074/jbc.M002911200. [DOI] [PubMed] [Google Scholar]
- 34.Christensen M. K., Falkeid G., Loros J. J., Dunlap J. C., Lillo C., Ruoff P. A nitrate induced frq-less oscillator in Neurospora crassa. J. Biol. Rhythms. 2004;19:280–286. doi: 10.1177/0748730404265532. [DOI] [PubMed] [Google Scholar]
- 35.Cheng P., Yang Y. H., Liu Y. Interlocked feedback loops contribute to the robustness of the Neurospora circadian clock. Proc. Natl. Acad. Sci. U.S.A. 2001;98:7408–7413. doi: 10.1073/pnas.121170298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smolen P., Baxter D. A., Byrne J. H. Modeling circadian oscillations with interlocking positive and negative feedback loops. J. Neurosci. 2001;21:6644–6656. doi: 10.1523/JNEUROSCI.21-17-06644.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roenneberg T., Merrow M. The network of time: understanding the molecular circadian system. Curr. Biol. 2003;13:198–207. doi: 10.1016/s0960-9822(03)00124-6. [DOI] [PubMed] [Google Scholar]
- 38.Rand D. A., Shulgin B. V., Salazar J. D., Millar A. J. Uncovering the design principles of circadian clocks: mathematical analysis of flexibility and evolutionary goals. J. Theor. Biol. 2006;238:616–635. doi: 10.1016/j.jtbi.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 39.Michael T. P., Salomé P. A., Yu H. J., Spencer T. R., Sharp E. L., McPeek M. A., Alonso J. M., Ecker J. R., McClung C. R. Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science. 2003;302:1049–1053. doi: 10.1126/science.1082971. [DOI] [PubMed] [Google Scholar]
- 40.Devlin P. F. Signs of the time: environmental input to the circadian clock. J. Exp. Bot. 2002;53:1535–1550. doi: 10.1093/jxb/erf024. [DOI] [PubMed] [Google Scholar]
- 41.Somers D. E. Entrainment of the circadian clock. Annu. Plant Rev. 2005;21:85–105. [Google Scholar]
- 42.Nelson D. E., Takahashi J. S. Sensitivity and integration in a visual pathway for circadian entrainment in the hamster (Mesocricetus auratus) J. Physiol. 1991;439:115–145. doi: 10.1113/jphysiol.1991.sp018660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aschoff J. Circadian rhythms: influences of internal and external factors on the period measured in constant conditions. Z. Tierpsychol. 1979;49:225–249. doi: 10.1111/j.1439-0310.1979.tb00290.x. [DOI] [PubMed] [Google Scholar]
- 44.Somers D. E., Devlin P., Kay S. A. Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science. 1998;282:1488–1490. doi: 10.1126/science.282.5393.1488. [DOI] [PubMed] [Google Scholar]
- 45.Devlin P. F., Kay S. A. Cryptochromes and phytochromes are required for phytochrome signalling to the circadian clock but not for rhythmicity. Plant Cell. 2000;12:2499–2509. doi: 10.1105/tpc.12.12.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kozma-Bognár L., Hall A., Adam E., Thain S. C., Nagy F., Millar A. J. The circadian clock controls the expression pattern of the circadian input photoreceptor, phytochrome B. Proc. Natl. Acad. Sci. U.S.A. 1999;96:14652–14657. doi: 10.1073/pnas.96.25.14652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hall A., Kozma-Bognár L., Toth R., Nagy F., Millar A. J. Conditional circadian regulation of phytochrome A gene expression. Plant Physiol. 2001;127:1808–1818. [PMC free article] [PubMed] [Google Scholar]
- 48.Tóth R., Kevei É., Hall A., Millar A. J., Nagy F., Kozma-Bognár L. Circadian clock-regulated expression of phytochrome and cryptochrome genes in Arabidopsis. Plant Physiol. 2001;127:1607–1616. doi: 10.1104/pp.010467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sharrock R. A., Clack T. Patterns of expression and normalized levels of the five Arabidopsis phytochromes. Plant Physiol. 2002;130:442–456. doi: 10.1104/pp.005389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Millar A. J., Kay S. A. Integration of circadian and phototransduction pathways in the network controlling CAB gene expression in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 1996;93:15491–15496. doi: 10.1073/pnas.93.26.15491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hicks K. A., Millar A. J., Carré I. A., Somers D. E., Straume M., Meeks-Wagner R., Kay S. A. Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science. 1996;274:790–792. doi: 10.1126/science.274.5288.790. [DOI] [PubMed] [Google Scholar]
- 52.McWatters H. G., Bastow R. M., Hall A., Millar A. J. The ELF3 zeitnehmer regulates light signalling to the circadian clock. Nature (London) 2000;408:716–720. doi: 10.1038/35047079. [DOI] [PubMed] [Google Scholar]
- 53.Covington M. F., Panda S., Liu X. L., Strayer C. A., Wagner D. R., Kay S. A. ELF3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell. 2001;13:1305–1315. doi: 10.1105/tpc.13.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mizoguchi T., Coupland G. ZEITLUPE and FKF1: novel connections between flowering time and circadian clock control. Trends Plant Sci. 2000;5:409–411. doi: 10.1016/s1360-1385(00)01747-7. [DOI] [PubMed] [Google Scholar]
- 55.Imaizumi T., Tran H. G., Swartz T. E., Briggs W. R., Kay S. A. FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature (London) 2003;426:302–306. doi: 10.1038/nature02090. [DOI] [PubMed] [Google Scholar]
- 56.Salomé P. A., McClung C. R. What makes Arabidopsis tick: light and temperature entrainment of the circadian clock. Plant Cell Environ. 2005;28:21–38. [Google Scholar]
- 57.Kevei E., Gyula P., Hall A., Kozma-Bognár L., Kim W. Y., Eriksson M. E., Toth R., Hanano S., Feher B., Southern M. M., et al. Forward genetic analysis of the circadian clock separates the multiple functions of ZEITLUPE. Plant Physiol. 2006;140:933–945. doi: 10.1104/pp.105.074864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Más P., Kim W.-Y., Somers D. E., Kay S. A. Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature (London) 2003;426:567–570. doi: 10.1038/nature02163. [DOI] [PubMed] [Google Scholar]
- 59.Somers D. E., Kim W. Y., Geng R. The F-box protein ZEITLUPE confers dosage-dependent control on the circadian clock, photomorphogenesis, and flowering time. Plant Cell. 2004;16:769–782. doi: 10.1105/tpc.016808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Michael T. P., Salomé P. A., McClung C. R. Two Arabidopsis circadian oscillators can be distinguished by differential temperature sensitivity. Proc. Natl. Acad. Sci. U.S.A. 2003;100:6878–6883. doi: 10.1073/pnas.1131995100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Salomé P. A., McClung C. R. PRR7 and PRR9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell. 2005;17:791–803. doi: 10.1105/tpc.104.029504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Farré E. M., Harmer S. L., Harmon F. G., Yanovsky M. J., Kay S. A. Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr. Biol. 2005;15:47–54. doi: 10.1016/j.cub.2004.12.067. [DOI] [PubMed] [Google Scholar]
- 63.Alabadí D., Oyama T., Yanovsky M. J., Harmon F. G., Más P., Kay S. A. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science. 2001;293:880–883. doi: 10.1126/science.1061320. [DOI] [PubMed] [Google Scholar]
- 64.Schaffer R., Ramsay N., Samach A., Corden S., Putterill J., Carré I. A., Coupland G. LATE ELONGATED HYPOCOTYL, an Arabidopsis gene encoding a MYB transcription factor, regulates circadian rhythmicity and photoperiodic responses. Cell. 1998;93:1219–1229. doi: 10.1016/s0092-8674(00)81465-8. [DOI] [PubMed] [Google Scholar]
- 65.Mizoguchi T., Wheatley K., Hanzawa Y., Wright L., Mizoguchi M., Song H.-R., Carré I. A., Coupland G. LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev. Cell. 2002;2:629–641. doi: 10.1016/s1534-5807(02)00170-3. [DOI] [PubMed] [Google Scholar]
- 66.Millar A. J., Carré I. A., Strayer C. A., Chua N. H., Kay S. A. Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science. 1995;267:1161–1163. doi: 10.1126/science.7855595. [DOI] [PubMed] [Google Scholar]
- 67.Somers D. E., Webb A. A. R., Pearson M., Kay S. A. The short-period mutant, toc1–1, alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development. 1998;125:485–494. doi: 10.1242/dev.125.3.485. [DOI] [PubMed] [Google Scholar]
- 68.Strayer C., Oyama T., Schultz T. F., Raman R., Somers D. E., Más P., Panda S., Kreps J. A., Kay S. A. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science. 2000;289:768–771. doi: 10.1126/science.289.5480.768. [DOI] [PubMed] [Google Scholar]
- 69.Hayama R., Coupland G. Shedding light on the circadian clock and the photoperiodic control of flowering. Curr. Opin. Plant Biol. 2003;6:13–19. doi: 10.1016/s1369-5266(02)00011-0. [DOI] [PubMed] [Google Scholar]
- 70.Alabadí D., Yanovsky M. J., Más P., Harmer S. L., Kay S. A. Critical role for CCA1 and LHY in maintaining circadian rhythmicity in Arabidopsis. Curr. Biol. 2002;12:757–761. doi: 10.1016/s0960-9822(02)00815-1. [DOI] [PubMed] [Google Scholar]
- 71.Hicks K. A., Albertson T. M., Wagner D. R. EARLY FLOWERING 3 encodes a novel protein that regulates circadian clock function and flowering in Arabidopsis. Plant Cell. 2001;13:1281–1292. doi: 10.1105/tpc.13.6.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Locke J. C. W., Southern M. M., Kozma-Bognár L., Hibberd V., Brown P. E., Turner M. S., Millar A. J. Extension of a genetic network model by iterative experimentation and mathematical analysis. Mol. Syst. Biol. 2005 doi: 10.1038/msb4100018. 10.1038/msb4100018. [DOI] [PMC free article] [PubMed]
- 73.Locke J. C. W., Millar A. J., Turner M. S. Modelling genetic networks with noisy and varied experimental data: the circadian clock in Arabidopsis thaliana. J. Theor. Biol. 2005;234:383–393. doi: 10.1016/j.jtbi.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 74.Mizoguchi T., Wright L., Fujiwara S., Cremer F., Lee K., Onouchi H., Mouradov A., Fowler S., Kamada H., Putterill J., Coupland G. Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell. 2005;17:2255–2270. doi: 10.1105/tpc.105.033464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nakamichi N., Kita M., Ito S., Sato E., Yamashino T., Mizuno T. PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol. 2005;46:686–698. doi: 10.1093/pcp/pci086. [DOI] [PubMed] [Google Scholar]
- 76.Hanano S., Davies S. J. Pseudo-response regulator genes ‘tell’ the time of day: multiple feedbacks in the circadian system of higher plants. Annu. Plant Rev. 2005;21:25–56. [Google Scholar]
- 77.Kaczorowski K. A., Quail P. H. Arabidopsis PSEUDO-RESPONSE REGULATOR 7 is a signaling intermediate in phytochrome-regulated seedling deetiolation and phasing of the circadian clock. Plant Cell. 2003;15:2654–2665. doi: 10.1105/tpc.015065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hazen S. P., Schultz T. F., Pruneda-Paz J. L., Borevitz J. O., Ecker J. R., Kay S. A. LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc. Natl. Acad. Sci. U.S.A. 2005;102:10387–10392. doi: 10.1073/pnas.0503029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kikis E. A., Khanna R., Quail P. H. ELF4 is a phytochrome-regulated component of a negative-feedback loop involving the central oscillator components CCA1 and LHY. Plant J. 2005;44:300–313. doi: 10.1111/j.1365-313X.2005.02531.x. [DOI] [PubMed] [Google Scholar]
- 80.Onai K., Ishiura M. PHYTOCLOCK 1 encoding a novel GARP protein essential for the Arabidopsis circadian clock. Genes Cells. 2005;10:963–972. doi: 10.1111/j.1365-2443.2005.00892.x. [DOI] [PubMed] [Google Scholar]
- 81.Doyle M. R., Davis S. J., Bastow R. M., McWatters H. G., Kozma-Bognár L., Nagy F., Millar A. J., Amasino R. M. The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature (London) 2002;419:74–77. doi: 10.1038/nature00954. [DOI] [PubMed] [Google Scholar]
- 82.Sai J., Johnson C. H. Different circadian oscillators control Ca2+ fluxes and LHCB gene expression. Proc. Natl. Acad. Sci. U.S.A. 1999;96:11659–11663. doi: 10.1073/pnas.96.20.11659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thain S. C., Hall A., Millar A. J. Functional independence of circadian clocks that regulate plant gene expression. Curr. Biol. 2000;10:951–956. doi: 10.1016/s0960-9822(00)00630-8. [DOI] [PubMed] [Google Scholar]
- 84.Dodd A. N., Love J., Webb A. A. R. The plant clock shows its metal: the circadian regulation of cytosolic free Ca2+ Trends Plant Sci. 2005;10:15–21. doi: 10.1016/j.tplants.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 85.Hall A., Kozma-Bognár L., Bastow R. M., Nagy F., Millar A. J. Distinct regulation of CAB and PHYB gene expression by similar circadian clocks. Plant J. 2002;32:529–537. doi: 10.1046/j.1365-313x.2002.01441.x. [DOI] [PubMed] [Google Scholar]
- 86.Thain S. C., Murtas G., Lynn J. R., McGrath R. B., Millar A. J. The circadian clock that controls gene expression in Arabidopsis is tissue specific. Plant Physiol. 2002;130:102–110. doi: 10.1104/pp.005405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Millar A. J., Straume M., Chory J., Chua N. H., Kay S. A. The regulation of circadian period by phototransduction pathways in Arabidopsis. Science. 1995;267:1163–1166. doi: 10.1126/science.7855596. [DOI] [PubMed] [Google Scholar]
- 88.Hennessy T. L., Field C. B. Evidence of multiple circadian oscillators in bean plants. J. Biol. Rhythms. 1992;7:105–113. doi: 10.1177/074873049200700202. [DOI] [PubMed] [Google Scholar]
- 89.Wood N. T., Haley A., Viry-Moussaïd M., Johnson C. H., van der Luit A. H., Trewavas A. J. The calcium rhythms of different cell types oscillate with different circadian phases. Plant Physiol. 2001;125:787–796. doi: 10.1104/pp.125.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dodd A. N., Parkinson K., Webb A. A. R. Independent circadian regulation of assimilation and stomatal conductance in the ztl-1 mutant of Arabidopsis. New Phytol. 2004;162:63–70. [Google Scholar]
- 91.Millar A. J., Kay S. A. Circadian control of cab gene transcription and mRNA accumulation in Arabidopsis. Plant Cell. 1991;3:541–550. doi: 10.1105/tpc.3.5.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Darlington T. K., Wager-Smith K., Ceriani M. F., Staknis D., Gekakis N., Steeves T. D. L., Weitz C. J., Takahashi J. S., Kay S. A. Closing the circadian loop: Clock-induced transcription of its own inhibitors per and tim. Science. 1998;280:1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- 93.Ueda H. R., Matsumoto A., Kawamura M., Iino M., Tanimura T., Hashimoto S. Genome-wide transcriptional orchestration of circadian rhythms in Drosophila. J. Biol. Chem. 2002;277:14048–14052. doi: 10.1074/jbc.C100765200. [DOI] [PubMed] [Google Scholar]
- 94.Harmer S. L., Kay S. A. Positive and negative factors confer phasespecific circadian regulation of transcription in Arabidopsis. Plant Cell. 2005;17:1926–1940. doi: 10.1105/tpc.105.033035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Michael T. P., McClung C. R. Phase-specific circadian clock regulatory elements in Arabidopsis thaliana. Plant Physiol. 2002;130:627–638. doi: 10.1104/pp.004929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lidder P., Gutiérrez R. A., Salomé P. A., McClung C. R., Green P. J. Circadian control of mRNA stability: association with DST-mediated mRNA decay. Plant Physiol. 2005;138:2374–2385. doi: 10.1104/pp.105.060368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kramer C., Loros J. J., Dunlap J. C., Crosthwaite S. K. Role for antisense RNA in regulating circadian clock function in Neurospora crassa. Nature (London) 2003;421:948–952. doi: 10.1038/nature01427. [DOI] [PubMed] [Google Scholar]
- 98.Crosthwaite S. Circadian clocks and natural antisense RNA. FEBS Lett. 2004;567:49–54. doi: 10.1016/j.febslet.2004.04.073. [DOI] [PubMed] [Google Scholar]
- 99.Staiger D., Zecca L., Kirk D. A. W., Apel K., Eckstein L. The circadian clock regulated RNA-binding protein AtGRP7 autoregulates its expression by influencing alternative splicing of its own pre-mRNA. Plant J. 2003;33:361–371. doi: 10.1046/j.1365-313x.2003.01629.x. [DOI] [PubMed] [Google Scholar]
- 100.Daniel X., Sugano S., Tobin E. M. CK2 phosphorylation of CCA1 is necessary for its circadian oscillator function in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2004;101:3292–3297. doi: 10.1073/pnas.0400163101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Millar A. J. Clock proteins: turned over after hours? Curr. Biol. 2000;10:529–531. doi: 10.1016/s0960-9822(00)00586-8. [DOI] [PubMed] [Google Scholar]
- 102.Deshaies R. J. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 1999;15:435–467. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- 103.del Pozo J. C., Estelle M. F-box proteins and protein degradation: an emerging theme in cellular regulation. Plant Mol. Biol. 2000;44:123–128. doi: 10.1023/a:1006413007456. [DOI] [PubMed] [Google Scholar]
- 104.Yasuhara M., Mitsui S., Hirano H., Takanabe R., Tokioka Y., Ihara N., Komatsu A., Seki M., Shinozaki K., Kiyosue T. Identification of ASK and clock-associated proteins as molecular partners of LKP2 (LOV kelch protein 2) in Arabidopsis. J. Exp. Bot. 2004;55:2015–2027. doi: 10.1093/jxb/erh226. [DOI] [PubMed] [Google Scholar]
- 105.Schultz T. F., Kiyosue T., Yanovsky M., Wada M., Kay S. A. A role for LKP2 in the circadian clock of Arabidopsis. Plant Cell. 2001;13:2659–2670. doi: 10.1105/tpc.010332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nelson D. C., Lasswell J., Rogg L. E., Cohen M. A., Bartel B. FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell. 2000;101:331–340. doi: 10.1016/s0092-8674(00)80842-9. [DOI] [PubMed] [Google Scholar]
- 107.Imaizumi T., Schultz T. F., Harmon F. G., Ho L. A., Kay S. A. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science. 2005;309:293–297. doi: 10.1126/science.1110586. [DOI] [PubMed] [Google Scholar]
- 108.Somers D. E., Schultz T. F., Milnamow M., Kay S. A. ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell. 2000;101:319–329. doi: 10.1016/s0092-8674(00)80841-7. [DOI] [PubMed] [Google Scholar]
- 109.Kim W.-Y., Geng R., Somers D. E. Circadian phase-specific degradation of the F-box protein ZTL is mediated by the proteasome. Proc. Natl. Acad. Sci. U.S.A. 2003;100:4933–4938. doi: 10.1073/pnas.0736949100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Song H.-R., Carré I. A. DET1 regulates the proteasomal degradation of LHY, a component of the Arabidopsis circadian clock. Plant Mol. Biol. 2005;57:761–771. doi: 10.1007/s11103-005-3096-z. [DOI] [PubMed] [Google Scholar]
- 111.Yang J., Lin R., Sullivan J., Hoecker U., Liu B., Xu L., Deng X. W., Wang H. Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell. 2005;17:804–821. doi: 10.1105/tpc.104.030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Iwasaki H., Nishiwaki T., Kitayama Y., Nakajima M., Kondo T. KaiA-stimulated KaiC phosphorylation in circadian timing loops in cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 2002;99:15788–15793. doi: 10.1073/pnas.222467299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Harms E., Kivimae S., Young M. W., Saez L. Posttranscriptional and posttranslational regulation of clock genes. J. Biol. Rhythms. 2004;19:361–373. doi: 10.1177/0748730404268111. [DOI] [PubMed] [Google Scholar]
- 114.Meyer P., Saez L., Young M. W. PER-TIM interactions in living Drosophila cells: an interval timer for the circadian clock. Science. 2006;311:226–229. doi: 10.1126/science.1118126. [DOI] [PubMed] [Google Scholar]
- 115.Panda S., Poirier G. G., Kay S. A. tej defines a role for poly(ADP-ribosyl)ation in establishing period length of the Arabidopsis circadian oscillator. Dev. Cell. 2002;3:51–61. doi: 10.1016/s1534-5807(02)00200-9. [DOI] [PubMed] [Google Scholar]
- 116.Niinuma K., Someya N., Kimura M., Yamaguchi I., Hamamoto H. Circadian rhythm of circumnutation in inflorescence stems of Arabidopsis. Plant Cell Physiol. 2005;46:1423–1427. doi: 10.1093/pcp/pci127. [DOI] [PubMed] [Google Scholar]
- 117.Schaffer R., Landgraf J., Accerbi M., Simon V., Larson M., Wisman E. Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. Plant Cell. 2001;13:113–123. doi: 10.1105/tpc.13.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Englemann W., Johnsson A. Rhythms in organ movement. In: Lumsden P. J., Millar A. J., editors. Biological Rhythms and Photoperiodism in Plants. Oxford: BIOS Scientific Publishers; 1998. pp. 35–50. [Google Scholar]
- 119.Dowson-Day M. J., Millar A. J. Circadian dysfunction causes aberrant hypocotyl elongation patterns in Arabidopsis. Plant J. 1999;17:63–71. doi: 10.1046/j.1365-313x.1999.00353.x. [DOI] [PubMed] [Google Scholar]
- 120.Bläsing O. E., Gibon Y., Günther M., Höhne M., Morcuende R., Osuna D., Thimm O., Usadel B., Scheible W.-R., Stitt M. Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell. 2005;17:3257–3281. doi: 10.1105/tpc.105.035261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Love J., Dodd A. N., Webb A. A. R. Circadian and diurnal calcium oscillations encode photoperiodic information in Arabidopsis. Plant Cell. 2004;16:956–966. doi: 10.1105/tpc.020214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tseng T.-S., Salomé P. A., McClung C. R., Olszewski N. E. SPINDLY and GIGANTEA interact and act in Arabidopsis thaliana pathways involved in light responses, flowering, and rhythms in cotyledon movements. Plant Cell. 2004;16:1550–1563. doi: 10.1105/tpc.019224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Valverde F., Mouradov A., Soppe W., Ravenscroft D., Samach A., Coupland G. Photoreceptor regulation of CONSTANS protein and the mechanism of photoperiodic flowering. Science. 2004;303:1003–1006. doi: 10.1126/science.1091761. [DOI] [PubMed] [Google Scholar]
- 124.Huang T., Bohlenius H., Eriksson S., Parcy F., Nilsson O. The mRNA of the Arabidopsis gene FT moves from leaf to shoot apex and induces flowering. Science. 2005;309:1694–1696. doi: 10.1126/science.1117768. [DOI] [PubMed] [Google Scholar]
- 125.Abe M., Kobayashi Y., Yamamoto S., Daimon Y., Yamaguchi A., Ikeda Y., Ichinoki H., Notaguchi M., Goto K., Araki T. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science. 2005;309:1052–1056. doi: 10.1126/science.1115983. [DOI] [PubMed] [Google Scholar]
- 126.Wigge P. A., Kim M. C., Jaeger K. E., Busch W., Schmid M., Lohmann J. U., Weigel D. Integration of spatial and temporal information during floral induction in Arabidopsis. Science. 2005;309:1056–1059. doi: 10.1126/science.1114358. [DOI] [PubMed] [Google Scholar]
- 127.Han S., Tang R., Anderson L. K., Woerner T. E., Pei Z.-M. A cell surface receptor mediates extracellular Ca2+ sensing in guard cells. Nature (London) 2003;425:196–200. doi: 10.1038/nature01932. [DOI] [PubMed] [Google Scholar]