Abstract
When both genotype and environment are held constant, “chance” variation in the lifespan of individuals in a population is still quite large. Using isogenic populations of the nematode Caenorhabditis elegans we show that, on the first day of adult life, chance variation in the level of induction of a green fluorescent protein (GFP) reporter coupled to a promoter from the hsp-16.2 gene, predicts as much as a four-fold variation in subsequent survival. The same reporter is also a predictor of ability to withstand a subsequent lethal thermal stress. The level of induction of GFP is not heritable and GFP expression levels in other reporter constructs are not associated with differential longevity. HSP-16 alone is probably not responsible for the observed differences in survival but instead is likely reflective of a hidden, heterogeneous, but now quantifiable, physiological state that dictates the ability of the organism to deal with the rigors of living.
Chance plays a large and probably ineradicable role in determining variation among individuals in age at death1,2. In humans, as well as populations of laboratory animals, 60–90% of the variation in age at death is independent of genotype3. In isogenic populations (where genetic variance is essentially zero), under a uniform environment, some individuals die early in life and others live quite long1,4. Differences in individual life span of Caenorhabditis elegans can reach as much as 50-fold4,5 and still have almost as much variation in time of death as does the population of the United States1,2,6. Such observations make suspect the popular notion of a “genetic program that regulates longevity”7. Instead, geriatric, demographic and evolutionary evidence suggest an alternate paradigm of aging; one that encompasses a rich variety of often highly plastic processes, influenced by genetic, environmental, and stochastic phenomenon1,2,6. Here we demonstrate that the ability of individual isogenic worms to respond to stress on the first day of adult life has a large stochastic component and is a major predictor of their subsequent longevity.
The optical transparency of C. elegans allows non-invasive visual assessment of living worms without compromising subsequent measurement of longevity. We used a chromosomally-integrated transgenic strain (TJ375), containing the 400 bp hsp-16.2 promoter coupled to the gene encoding green fluorescent protein (GFP) and encoding no HSP-16.2 product itself (Fig. 1a). This reporter provides an accurate assessment of the total amount of native HSP-16.2 protein8 (see also Supplementary Fig. 1). No detectable GFP is observed in uninduced worms, but following a one or two-hour 35 °C pulse (Fig. 1b), GFP becomes readily apparent, peaking at 15–18 hours (Figs. 1c, d).
Figure 1.
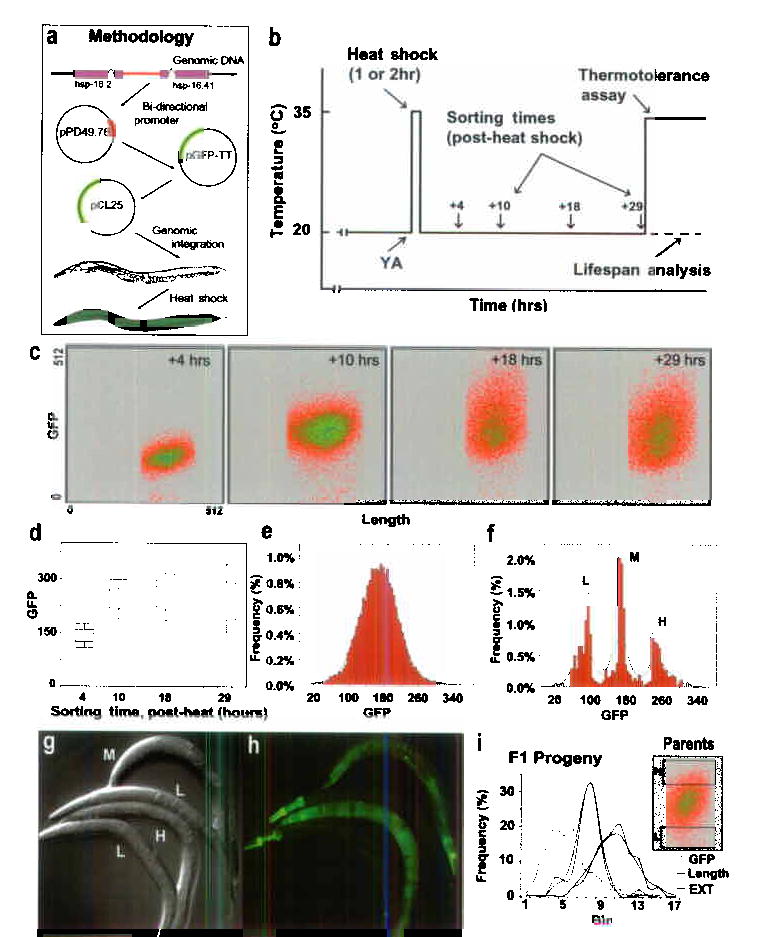
Overview. (a) Outline of experimental design and construction of TJ375. (b) Schematic of hsp-16::GFP induction, sorting, and analysis. (c) Individual fluorescence data from a representative experiment. Increasing density of events is color-coded (red to blue). (d) Box and whisker plots summarizing fluorescence distribution of worm populations at each sort time following the end of the heat shock. (e) Distribution of GFP levels in a typical population 19 hours after induction by a 2 hr 35 °C pulse (Mean ± SD is 168.0 ± 44.9 GFP units); the green line shows a normal distribution with the same mean and SD. (f) Distribution of individuals selected in a sort of low (L), median (M) and high (H) levels of expression. (g, h) Representative worms from each of the three sub-populations in (f). (i) Properties of progeny derived from parents with high or low GFP fluorescence demonstrating that level of GFP-expression is not heritable. Shown are the population distributions for three parameters: worm length (Length, black lines), optical absorption (EXT, blue lines) and green fluorescence (GFP, green lines). There were no significant differences between progeny derived from the original high or low sub-populations for any of the three parameters (t-test, p-values all >0.3).
Heat-shocked populations displayed a wide and normally-distrubuted variation in individual GFP fluorescence (Figs. 1c–g), even though individuals were isogenic and grown in an environment designed to minimize environmental heterogeneity. Such heterogeneity was observed from the earliest times at which GFP expression was detectable and continued until the time GFP had completely dissipated (several days, data not shown). The degree of heterogeneity increased markedly with time (Figs. 1 c, d), and was both replicable and quantifiable (Figs. 1 g, h).
We asked if HSP-16::GFP expression might predict longevity. Our initial findings (Supplementary Figure 2), on individual, isogenic worms measured manually, suggested a significant correlation between GFP-expression level and subsequent longevity (r = 0.48; P = 0.002); so we extended our studies to large populations. Worms were sorted into high, intermediate, and low GFP-expression classes at various times after heat induction (Figs. 1 b, f) and were subsequently tested for resistance to a lethal thermal stress or kept for longevity analysis. We routinely observed significant differences in subsequent longevity and thermotolerance among worms that expressed GFP at high, average or low levels (Figs. 2, 3). When sorted after a two-hour induction at 35°C, worms that differentially expressed GFP showed large differences in life expectancy and thermotolerance. In a typical experiment, we found life expectancies of 16.4 days in the brightest worms, while the worms expressing the lowest levels of GFP after heat shock lived only about 3.2 days (Fig. 2a, Supplementary Table I). Similarly, worms with the highest GFP levels also showed higher thermotolerance (9.5 hours) than the average (6.7 hours) or than worms showing the lowest levels of thermotolerance (4.0 hours; Fig. 2d; P < .001).
Figure 2.
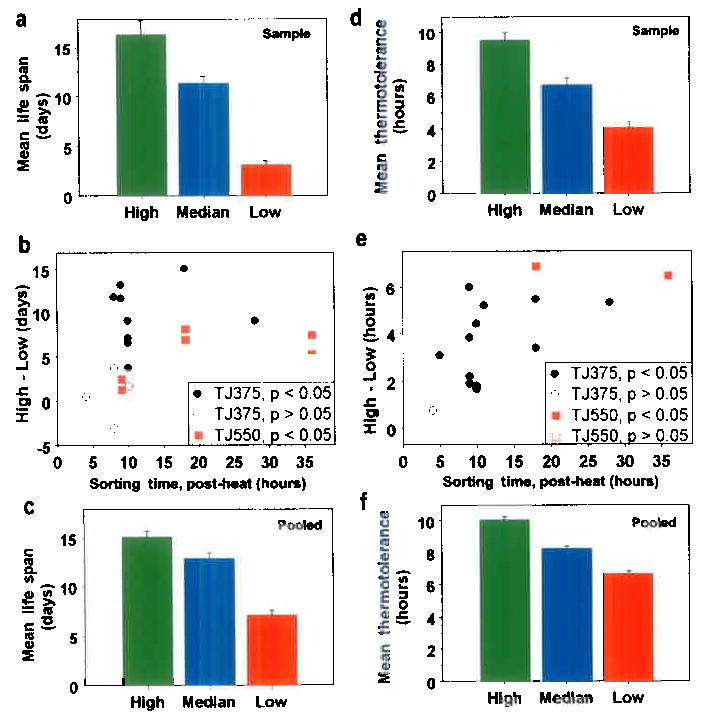
Survival and thermotolerance of worms previously sorted on differential hsp-16-2::GFP expression following a two-hour heat shock. (a) A representative longevity assessment showing adult life expectancy following heat shock (mean life span and SEM; High: 16.4 ± 1.5 days; Median: 11.3 ± 0.7 days and Low: 3.2 ± 0.4 days, N = 30 and P < .001, for each). (b) The difference between the average longevity of high and low sub-populations for every study is shown as a point. Significant differences in survival are shown as filled symbols, non-significant as open symbols; TJ375 symbols are black and TJ550 are red. (For details see Supplementary Table II). (c) Combined data for all 13 longevity experiments using TJ375 (High: 15.13 ± 0.61 days, N = 530; Median: 12.92 ± 0.56 days, N = 545; Low: 7.13 ± 0.49 days, N = 550). (For TJ550 see Supplementary Table II). (d) Thermotolerance of worms derived from the same populations that were sampled to generate the longevity data in (a), (mean survival at 35 °C and SE; High: 9.5 ± 0.5 hours; Median: 6.7 ± 0.4 hours; Low: 4.0 ± 0.4 hours, N = 31 to 33, and P < .00001, for each comparison). (e) Each point represents the difference in thermotolerance of the high and low sub-populations for each experiment performed. (Symbols as in Fig. 2b.) (f) Combined data for all thermotolerance experiments (High: 10.07 ± 0.20 hours, N = 394; Median: 8.27 ± 0.15 hours, N = 401; Low: 6.68 ± 0.18 hours, N = 404; all P’s < 10−10.
Figure 3.
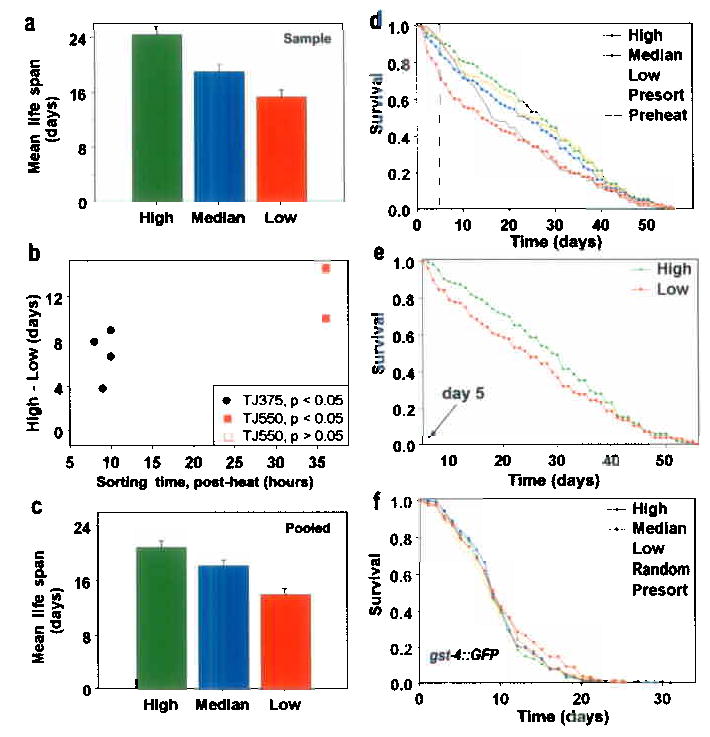
Survival of worms previously sorted on differential HSP-16-2::GFP expression after 1 hour of induction at 35 °C. Sorting and other conditions were as in Fig. 2. (a) Data from a typical longevity analysis shown as per Fig. 2; (mean life span and SE; High: 24.4 ± 1.1 days; Median: 18.7 ± 1.1 days and Low: 15.35 ± 1.0 days, N = 40 and P < .025, for each). (b) The difference between the average longevity of the high and low sub-populations for each of nine experiments is shown by a point as in Fig. 2. (For details see Supplementary Table II.) (c) Combined data for all longevity experiments (mean resistance and SE; High: 20.86 ± 0.93 days; Median: 18.20 ± 0.80 days; Low: 14.03 ± 0.85 days, N = 149–150 for each, P = .03 for High vs Median and P < .001, for others). (d) Survival trajectories of worms used to generate the data in Fig. 3a. (See also Supplementary Tables I and IV). (e) Survival trajectories of High vs Low subpopulations (as in Fig. 3d) plotted from days 5 onward. The curves are significantly different (P < .05). (f) Not all GFP reporter constructs are biomarkers for longevity. Worms expressing the oxidative stress reporter gst-4::GFP were sorted into constitutively High, Intermediate or Low GFP-expressing populations then their survival was assessed. Shown is the combined data (N = 291 total for each subpopulation) from five independent experiments. Controls include an unsorted population (Presort) and a sorted but unselected population (Random). The curves are not significantly different (P all >0.1). (See Supplementary Table I and also Supplementary Fig. 3).
We asked whether sorting at different times after the heat shock affected the observation of differential survival. We sorted worms at various times following induction and found differential life expectancy to be as much as 10 to 15 days (Fig. 2b, Supplementary Table II), averaging 8.0 days over all 19 experiments (Fig. 2c). Following two-hours of induction, lifespan averaged 15.1 days for the high and only 7.1 days for the low, more than a two-fold difference (Figs. 2 c, Supplementary Table I; P = 8.0 x 10−29). Sorting earlier than 9 hours led to non-significant results. Worms expressing different levels of GFP, 9 to 36 hours after induction, also differed significantly with respect to subsequent thermotolerance (Figs. 2d, e and Supplementary Tables I, III). We found statistically significant differences in all but two of sixteen replicates. This differential thermotolerance was very robust, averaging about 3.4 hours (Fig. 2f, Supplementary Table I; P = 1.7 x 10−28). Differential survival and thermotolerance were highest at about 18 hours after induction, about the time when variance is maximal.
In prior studies8, incubation of the TJ375 reporter strain for two hours at 35°C resulted in subsequent thermotolerance and increased longevity in a process termed hormesis. Under the two-hour heat induction used in the current experiments we also saw hormesis for both longevity and thermotolerance, but only in a subpopulation of worms. Our calculations indicate that 27% of the worms were damaged by this treatment regimen (D.W., S.L.R., T.E.J., J.W.V., manuscript in preparation). These results are different because we used new induction conditions to maintain a more uniform environment (abrupt vs slow temperature shift, see Methods). When we decreased induction time to one hour, we again found significant differences in survival between the brightest and dimmest worms in 7 of 9 experiments (Figs. 3a–e and Supplementary Tables I, II and IV). Average life span differed by as much as 14 days in one study (details in Fig. 3b legend). Furthermore, after one hour of heat we now observed a robust hormesis effect, consistent with previous observations9,10.
To correct for possible interrelationships between the effect of heat on survival and its effect on fertility11we crossed two temperature-sensitive (ts) fertility mutations, fer-15(b26) and spe-9(hc88), into the TJ375 reporter strain to form a new strain: TJ550. At the non-permissive temperature, the combination of both mutations completely blocked reproduction, but not germ cell formation or proliferation5. Differential survival was, however, still observed in this background with all six replicates showing significant differences in longevity between bright and dim worms (Fig. 2b, Supplementary Table II).
Individual differences in hsp-16.2::GFP reporter expression may result from genetic variation – i.e. epigenetic changes may occur in isogenic individuals during propagation leading to differential inactivation or expression of one or more of the large number of repeats in the transgenic array present in the reporter strains12. To address this question, we asked whether differences in levels of GFP expression were heritable. We sorted a population at 11 hours post heat shock into sub-populations containing a few hundred of the total initial population of 60,000 worms (Fig. 1i). Progeny were collected, allowed to grow to maturity, induced by heat shock and assessed for level of GFP expression using the identical protocol. We found that progeny of both the high- and low-expressing parents showed almost identical average levels of GFP expression (Fig. 1i). Progeny of both high- and low-expression parental classes had almost the same mean expression level (298.0 vs 288.6 GFP units) and both displayed almost identical variation in levels of GFP expression (p = 0.5, χ2 test for distributional difference), essentially recapitulating that of the parental population. Thus, the precise level of GFP expression is not heritable. While it is possible that further experimentation may reveal discrete causal factors determining variance of GFP expression, the results shown here were obtained from an isogenic population, maintained in a uniform environment during their propagation. Non-heritability of GFP expression level suggests the presence of a large underlying stochastic component specifying level of GFP expression in individual worms, similar to that observed in bacteria13,14.
Finally, we also asked whether level of GFP fluorescence is a predictor of longevity when GFP is tagged to promoters of non stress-inducible genes (myo-2 and mtl-2). It is not. Since GFP fluorescence is dependent upon redox activation we also utilized a promoter tag of a gene normally activated in response to oxidative stress (gst-4) and again found no relationship between GFP levels and subsequent longevity (Fig. 3f, Supplementary Fig. 3 and Supplementary Table I).
HSP-16.2 expression level in young adults is a robust predictor of remaining life expectancy. This variation is not heritable.
For C. elegans, mutational analysis has long been the preferred approach for understanding gene action and biological function15, no less so for aging and life span. Despite the success of the genetic approach in explaining life-span extension between distinct genotypes in C. elegans, most life-span variation is not under genetic control. Even under rigidly controlled laboratory conditions, 60% of the variation in longevity in F2 intercrosses in nematodes is not genetic16. Similar findings are true in all species that have been studied1,3; in humans, only about 25% of the variation in life span (even after excluding early deaths due to childhood disease and accident), is due to measurable genetic effects1,2,17leaving the vast majority of variation in life span as unexplained or “environmental”, some of which results from chance or stochastic events within individuals1,2,18.
Stochastic variation arises from fundamental thermodynamic and statistical mechanical considerations. A large fraction of individual variation in life span must stem from the fundamental fact that life results from an integrated series of metabolic reactions which themselves are under fundamental physical constraints on the specificity and rigidity with which they, too, can be regulated19. At the molecular level, two points are germane to the present study. First, when the number of molecules regulating a biological process becomes countably small, “chance” distributions come into play such that some regulatory molecules can vary several-fold between individual cells20. Second, the Maxwell-Boltzmann (M-W) equation specifies the distribution of kinetic energies among molecules and requires kinetic energy to be a distributed function. Strehler and Mildivan21 utilized this equation to develop a general theory explaining mortality kinetics. Several sources of variation at the molecular level could conceivably alter GFP (HSP-16.2) expression level while simultaneously affecting more global processes. These include intracellular differences and fluctuations in the rates of molecular processes such as transcription, ribosome loading and translation (as postulated by Kirkwood et al22). Chance variation in the number of HSF effecter molecules present within each cell at the time of heat shock also could have dramatic phenotypic consequences. Variation in the frequency of mitochondrial genomic rearrangements, as previously observed in isogenic populations of C. elegans 23–25, could have an effect. There is an increasing literature describing variation among isogenic individuals at the molecular level, typically in microbial or yeast cultures where such effects can be visualized13,14,26. Clearly, significant variation among genetically identical individuals is a fact of nature and inherent molecular variability implies that biochemical and molecular genetic processes must exhibit inherent variability.
From the earliest studies of C. elegans aging populations27, it has been apparent that individual age at death varies greatly in isogenic populations - spanning several weeks between those dying on the first day of adult life (excluding larval and embryonic death) and individuals who died last. In the case of long-lived Age mutants, this span can be several months. Stochastic variation provides a means by which one can start to understand this huge variation in lifespan. Genetic regulatory systems can be viewed in terms of robustness or sensitivity toward chance environmental fluctuations, maintaining expression of either a single phenotype or leading to the expression of a distributed phenotype. When multiple phenotypes are useful, such as for sampling changing environments, genetic systems that have a built in capacity to reveal variance, or indeed amplify it, could be selected28. Such systems might be of particular use to self-fertilizing organisms like C. elegans.
A Biomarker of Aging (BoA) is defined as “a biological parameter of an organism that either alone or in some multivariate composite will, in the absence of disease, better predict functional capability at some later age than will chronological age”29. Our present studies suggest that level of HSP-16 production may now also be such a BoA and that it is a robust predictor of subsequent individual longevity. Note, however, that the HSP-16::GFP reporter construct used in this study provided no functional HSP-16 protein. It is unlikely that GFP was conferring the longevity effect since other GFP reporters we tested showed no longevity differential. Also, it is unlikely that endogenous HSP-16 proteins alone were responsible for the differential longevity of bright and dim worms, since overexpression of HSP-16 increases longevity by only a few days30. Instead, it seems likely that the hsp-16.2::GFP reporter is conveying information about the general physiological state of the cell and/or organism with respect to its ability to withstand stress and its subsequent likelihood of survival. Future studies are likely to reveal additional biomarkers for longevity in C. elegans that also reveal something about the “Physiologic State” of the organism.
METHODS
Strains and Construction
TJ375 [hsp-16.2::GFP(gpIs1)]
The 431 bp HinDIII/BamH1 fragment from pPD49.78 (A. Fire, Carnegie Institution of Washington), containing the 393 bp bi-directional promoter from hsp-16.2/hsp16.41, was cloned into pGFP-TT (S65T, I176T) to form pCL25. GFP in pCL25 is in the direction of hsp-16.2. pCL25 was integrated into N2 (wild-type) using standard injection and γ-irradiation techniques. One integrant was back-crossed into N2 ten times (subsequently referred to as TJ375), and it behaved indistinguishably from N2 except that it uniformly fluoresced green (excluding gonad region) when heat-shocked. Expression lasted several days depending on the length of the heat pulse (unpublished results). No GFP expression was observed in the absence of a heat pulse when cultured at 16–25ºC. (Constructed by S. Henderson and C. Link, University of Colorado).
TJ550 spe-9(hc88ts)I; fer-15(b26ts)II; [hsp-16.2::GFP(gpIs73)]
TJ375 males were mated into TJ1060 spe-9(hc88ts)I; fer-15(b26ts)II and F2 progeny doubly homozygous for spe-9(hc88ts) and fer-15(b26ts) were identified based on reduced fertility at 23ºC. Double mutants homozygous for hsp-16.2::GFP also, were identified by a sub-lethal heat shock (30 mins 35 ºC). The TJ550 line was selected. Fertility assessment of F1 progeny from TJ550 crossed with BA671 spe-9(hc88ts)I and BA713 fer-15(b26ts)II independently confirmed homozygosity at the spe-9 and fer-15 loci.
Other strains constitutively expressing GFP
CL2122 dvIs15 [mtl-2::GFP, pPD30.38 (unc-54 expression vector)] expresses GFP under the control of the intestinal-specific metallothionine-2 (mtl-2) promoter. PD4788 mIs13 [myo-2::GFP, pes-10::GFP, gut::GFP] expresses GFP under the control of the pharynx-specific myosin-2 (myo-2) promoter, the germline-specific pes-10 promoter as well as a gut specific enhancer. In the adult, expression appears restricted to the pharynx only. CL2166 dvIs19 [K08F4.7::GFP] expresses GFP under the direction of the glutathione-S-transferase 4 (gst-4) promoter. All three strains, are transgenic derivatives of Bristol N2 and were the gift of C. Link.
Nematode Maintenance and Mass Cultures
Standard techniques were used for maintenance of nematode strains15,16. For large-scale production of synchronized populations (>120,000 worms), sixteen 10 cm NGM/E. coli (OP50) plates were each seeded with 1200 arrested first stage larvae (L1). After 76 hrs at 20 ºC, gravid adults (GAs) were collected (2 x 4 mls S-Basal media), transferred into a single tube and allowed to pellet under gravity (~5 mins, room temperature, RT). After washing to remove eggs and hatched L1 progeny (5 x 50mls, S-Basal), GAs were resuspended into ~1 ml S-Basal then spread onto 4 x 10 cm NGM/OP50 plates. Food was suffucient so cultures starved and stopped laying after 4 hrs. After ~ 24 hrs, hatched L1s and GAs were washed from plates (8 mls S-Basal, total) and GAs were separated and discarded by allowing adults to differentially pellet under gravity (~5 mins). After determining L1 concentration, animals were seeded onto 16 new starter NGM/OP50 plates (1200 L1/plate) or twelve 10 cm NGM/RW2 (wild-type E. coli) plates supplemented with 2% peptone (RNGM, 12,000 L1/plate). Plates were incubated at 20ºC (TJ375) or 25ºC (TJ550), allowed to reach adulthood (65 hrs or 50 hrs, respectively), harvested (2 x 4 mls S-Basal/plate), pooled, washed (2 x 50 ml S-Basal), and finally resuspended in 3 mls S-Basal ready for subsequent assays. Populations were supplemented with 1 x 1011 OP50 per RNGM plate at +52 hrs to prevent starvation.
Heat-Shock
Pooled GAs were heat-shocked (35ºC) for 1 or 2 hrs (with rotation, 100rpm) following transfer into pre-heated S-Basal (35ºC) containing 1 x 109 OP50/ml and 10 μg cholesterol/ml (liquid food, 300 worms/ml). After heat-shock worms were gravity pelleted (~ 5 mins) then quickly transferred to pre-chilled (20ºC) liquid food. Cultures were maintained at 20ºC with rotation (120 rpm) until ready for sorting. Cultures were re-fed every 12–16 hours with fresh liquid food. For each experiment, a sub-sample of worms not exposed to the heat treatment (Pre-heat) were retained in 4 mls of liquid food (20ºC) in a 6 cm dish until later use.
Sorting Procedure
Recovering worms were collected (gravity), washed (2 x 50 mL S-Basal), then resuspended (1000 worms/mL) in S-Basal + 1 x 109 OP50 (this concentration of bacteria did not interfere with subsequent sorting). Worms were sorted using a COPAS Biosort 250 Worm Sorter (Union Biometrica Inc., Harvard Biosciences), using three criteria – length (Length), optical absorbance (extinction, EXT) and integrated fluorescence intensity at 488 nm (GFP). Values for Length and EXT (gating) were defined prior to the beginning of each sort experiment and typically 60–75% of the entire population was included. Four sub-populations were then sorted, each expressing differing amounts of GFP - highest 2% (High), lowest 2% (Low), a median sample (2%, Median) and a gated only sample (All). Percentages represent fraction of the total starting population. Worms that were not subjected to the COPAS were also kept as controls in each experiment (Pre-sort); we found negligible changes in this population when compared to the “All” group (data not shown). Worms were not recycled through the sorter; so a population comprised of 30,000 individuals yielded ~150 worms per group. Worms were sorted directly into 6 cm dishes containing 4 mls of liquid food and incubated at 20ºC until further use. All sorting was performed at RT.
Thermotolerance Assay
After sorting, and at defined times following the original two-hour heat shock, groups of 30 worms from each sample were transferred from liquid food onto 6 cm NGM/OP50 plates and then immediately placed into a 35 ºC incubator. Survivorship until 100% death was scored at periodic intervals (typically over a span of 16 hours).
Longevity Assay
All lifespan analyses were performed in liquid food, as described previously 16. Briefly, 60–80 worms per sub-population were cultured in 4 mls liquid food, transferred every day to fresh food while egg-laying, then transferred every third day until none remained alive.
Microscopy
Fluorescent images were collected using a Zeiss Axioskop. Nematodes were immoblized with 1mM sodium azide, mounted on 2% agarose cushions, then images captured using a PCO SensiCam charge-coupled diode camera.
Heritability of Fluorescence Intensity
Sixty thousand arrested L1 were grown to adulthood (72 hrs, 20ºC), heat-shocked for 1 hr at 35ºC, allowed to recover for 11 hrs at 20ºC, then sorted into the highest (2%) and lowest (2%) GFP expressing worms (~350 worms each) exactly as described above. Each sub-population was transferred sequentially onto two NGM/OP50 6 cm plates and allowed to lay for 20 hrs at a time (this strategy was adopted in order to increase the number of F1 progeny for re-sorting). Each set of F1 progeny was pooled, heat shocked for 1 hr at 35ºC, allowed to recover for 16 hrs at 20ºC then sequentially sorted. Statistics for individual F1 worms were collected and analyzed.
Statistical Analyses
Longevity and thermotolerance information for 5836 and 2389 individuals, respectively, was derived from a combination of thirty TJ375 data sets (nine were completely independent and sourced from populations of 120,000 worms each). Fourteen longevity and thermotolerance data sets were collected for TJ550 from a single, 120,000 individual population. In total, close to 2,000,000 worms were employed for this study.
Mean life span was estimated using the formula:
where dj is the number of worms that died in the age interval (xj,xj+1) and N is total number of worms. The standard error of the mean life span estimate was calculated using:
Mean life span estimates for two populations were compared by calculating the normalized statistic:
where MLS0 and MLS1 are mean life span estimates, and SE0 and SE1 are standard error estimates, for populations 1 and 2, respectively.
A log-rank test was used for assessing differences in survival curves. A chi-square test was used for assessing the difference in GFP fluorescence distributions for the progeny experiments. Use of the discrete two-population frailty model mentioned in this paper will be described elsewhere (contacted T.E.J. for details). All additional significance testing was undertaken using Statistica (’99 edition) software (StatSoft, Inc, OK) and Gauss (Aptech Systems, Inc., WA).
Sequence Information
The Genbank accession number for hsp-16.2 is AC006774.
Supplementary Material
Acknowledgments
We wish to thank members of the Johnson lab for comments and support, especially C. Link and S. Henderson, as well as G. Amdam for her insight into control theory. We wish to acknowledge the help of A. Smith, Support for this work was provided by the National Institutes of Health, P01 AG08761 to JWV, RO1 AG16219 and KO2 AA00195 to TEJ, and a Polis Foundation Grant (SLR).
Footnotes
FEDEX: Institute for Behavioral Genetics, University of Colorado at Boulder, 1480 30th Street, Boulder, Colorado 80303, USA
References
- 1.Finch, C.E., & Kirkwood, T.B. Chance, Development and Aging, (Oxford University Press, London, 2000).
- 2.Kirkwood TB, Finch CE. Ageing: the old worm turns more slowly. Nature. 2002;419:794–5. doi: 10.1038/419794a. [DOI] [PubMed] [Google Scholar]
- 3.Finch CE, Tanzi RE. Genetics of Aging. Science. 1997;278:407–411. doi: 10.1126/science.278.5337.407. [DOI] [PubMed] [Google Scholar]
- 4.Johnson TE. Increased life-span of age-1 mutants in Caenorhabditis elegans and lower Gompertz rate of aging. Science. 1990;249:908–12. doi: 10.1126/science.2392681. [DOI] [PubMed] [Google Scholar]
- 5.Johnson TE, et al. Gerontogenes mediate health and longevity in nematodes through increasing resistance to environmental toxins and stressors. Exp Gerontol. 2000;35:687–94. doi: 10.1016/s0531-5565(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 6.Vaupel JW, et al. Biodemographic trajectories of longevity. Science. 1998;280:855–60. doi: 10.1126/science.280.5365.855. [DOI] [PubMed] [Google Scholar]
- 7.Kenyon C. Ponce d’elegans: genetic quest for the fountain of youth. Cell. 1996;84:501–4. doi: 10.1016/s0092-8674(00)81024-7. [DOI] [PubMed] [Google Scholar]
- 8.Link CD, Cypser JR, Johnson CJ, Johnson TE. Direct observation of stress response in Caenorhabditis elegans using a reporter transgene. Cell Stress Chaperones. 1999;4:235–42. doi: 10.1379/1466-1268(1999)004<0235:doosri>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cypser JR, Johnson TE. Multiple stressors in Caenorhabditis elegans induce stress hormesis and extended longevity. J Gerontol A Biol Sci Med Sci. 2002;57:B109–14. doi: 10.1093/gerona/57.3.b109. [DOI] [PubMed] [Google Scholar]
- 10.Calabrese EJ, Baldwin LA. Defining hormesis. Hum Exp Toxicol. 2002;21:91–7. doi: 10.1191/0960327102ht217oa. [DOI] [PubMed] [Google Scholar]
- 11.Lithgow GJ, White TM, Melov S, Johnson TE. Thermotolerance and extended life-span conferred by single-gene mutations and induced by thermal stress. Proc Natl Acad Sci U S A. 1995;92:7540–4. doi: 10.1073/pnas.92.16.7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelly WG, Xu S, Montgomery MK, Fire A. Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics. 1997;146:227–38. doi: 10.1093/genetics/146.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–6. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 14.Raser JM, O’Shea EK. Control of stochasticity in eukaryotic gene expression. Science. 2004;304:1811–4. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson TE, Wood WB. Genetic analysis of life-span in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1982;79:6603–7. doi: 10.1073/pnas.79.21.6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herskind AM, et al. The heritability of human longevity: a population-based study of 2872 Danish twin pairs born 1870–1900. Hum Genet. 1996;97:319–23. doi: 10.1007/BF02185763. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, T.E., & Vaupel, J.W. Stochastic Effects in Aging. Science In preparation(2005).
- 19.Gillooly JF, Brown JH, West GB, Savage VM, Charnov EL. Effects of size and temperature on metabolic rate. Science. 2001;293:2248–51. doi: 10.1126/science.1061967. [DOI] [PubMed] [Google Scholar]
- 20.McAdams HH, Arkin A. It’s a noisy business! Genetic regulation at the nanomolar scale. Trends Genet. 1999;15:65–9. doi: 10.1016/s0168-9525(98)01659-x. [DOI] [PubMed] [Google Scholar]
- 21.Strehler BL, Mildivan AS. General theory of mortality and aging. Science. 1960;132:14–21. doi: 10.1126/science.132.3418.14. [DOI] [PubMed] [Google Scholar]
- 22.Kirkwood TB, et al. What accounts for the wide variation in life span of genetically identical organisms reared in a constant environment? Mech Ageing Dev. 2005;126:439–43. doi: 10.1016/j.mad.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 23.Melov S, Lithgow GJ, Fischer DR, Tedesco PM, Johnson TE. Increased frequency of deletions in the mitochondrial genome with age of Caenorhabditis elegans. Nucleic Acids Res. 1995;23:1419–25. doi: 10.1093/nar/23.8.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Golden TR, Melov S. Microarray analysis of gene expression with age in individual nematodes. Aging Cell. 2004;3:111–24. doi: 10.1111/j.1474-9728.2004.00095.x. [DOI] [PubMed] [Google Scholar]
- 25.Herndon LA, et al. Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans. Nature. 2002;419:808–14. doi: 10.1038/nature01135. [DOI] [PubMed] [Google Scholar]
- 26.Fedoroff N, Fontana W. Genetic networks. Small numbers of big molecules. Science. 2002;297:1129–31. doi: 10.1126/science.1075988. [DOI] [PubMed] [Google Scholar]
- 27.Klass MR. Aging in the nematode Caenorhabditis elegans: major biological and environmental factors influencing life span. Mech Ageing Dev. 1977;6:413–29. doi: 10.1016/0047-6374(77)90043-4. [DOI] [PubMed] [Google Scholar]
- 28.Rao CV, Wolf DM, Arkin AP. Control, exploitation and tolerance of intracellular noise. Nature. 2002;420:231–237. doi: 10.1038/nature01258. [DOI] [PubMed] [Google Scholar]
- 29.Baker GT, 3rd, Sprott RL. Biomarkers of aging. Exp Gerontol. 1988;23:223–39. doi: 10.1016/0531-5565(88)90025-3. [DOI] [PubMed] [Google Scholar]
- 30.Walker GA, Lithgow GJ. Lifespan extension in C. elegans by a molecular chaperone dependent upon insulin-like signals. Aging Cell. 2003;2:131–9. doi: 10.1046/j.1474-9728.2003.00045.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


