Abstract
Nuclear transport factor 2 (NTF2) is a soluble transport protein originally identified by its ability to stimulate nuclear localization signal (NLS)-dependent protein import in digitonin-permeabilized cells. NTF2 has been shown to bind nuclear pore complex proteins and the GDP form of Ran in vitro. Recently, it has been reported that NTF2 can stimulate the accumulation of Ran in digitonin-permeabilized cells. Evidence that NTF2 directly mediates Ran import or that NTF2 is required to maintain the nuclear concentration of Ran in living cells has not been obtained. Here we show that cytoplasmic injection of anti-NTF2 mAbs resulted in a dramatic relocalization of Ran to the cytoplasm. This provides the first evidence that NTF2 regulates the distribution of Ran in vivo. Moreover, anti-NTF2 mAbs inhibited nuclear import of both Ran and NLS-containing protein in vitro, suggesting that NTF2 stimulates NLS-dependent protein import by driving the nuclear accumulation of Ran. We also show that biotinylated NTF2-streptavidin microinjected into the cytoplasm accumulated at the nuclear envelope, indicating that NTF2 can target a binding partner to the nuclear pore complex. Taken together, our data show that NTF2 is an essential regulator of the Ran distribution in living cells and that NTF2-mediated Ran nuclear import is required for NLS-dependent protein import.
INTRODUCTION
Nucleocytoplasmic transport plays an important role in the regulation of diverse cellular processes, including transcription and translation, growth factor-mediated signaling, stress responses, and cell cycle control (for reviews, see Mattaj and Englmeier, 1998; Wilkinson and Millar, 1998). Proteins and RNAs are imported and exported through nuclear pore complexes (NPCs), supramolecular (125,000 kDa in vertebrates) channels that perforate the double bilayer of the nuclear envelope. NPCs mediate the active transport of most proteins and RNAs, as well as the passive diffusion of ions and small proteins less than ∼40 kDa (for review, see Nigg, 1997).
Proteins destined for the nucleus generally possess nuclear localization signals (NLSs) (for review, see Mattaj and Englmeier, 1998). NLSs were first identified in the SV40 large T antigen and the Xenopus protein nucleoplasmin and consist of one or two short stretches of basic amino acid residues, respectively. Other signals that are sufficient to mediate nuclear import include the M9 sequence from heterogeneous nuclear ribonucleoprotein A1 protein and the KNS sequence from heterogeneous nuclear ribonucleoprotein K protein (Siomi and Dreyfuss, 1995; Michael et al., 1997). The observation that nuclear import requires targeting signals and is saturable prompted the hypothesis that import is a receptor-mediated process (Goldfarb et al., 1986).
The use of cell-based assays has led to a general understanding of the proteins that mediate nuclear import. The import of NLS-containing reporter proteins into digitonin-permeabilized cell nuclei can be reconstituted with the addition of cytosol from various cells (Adam et al., 1990). Cell and molecular analyses of these soluble transport factors from Xenopus and mammalian cell cytosol established the identities of importin α and β (Adam and Gerace, 1991; Adam and Adam, 1994; Chi et al., 1995; Gorlich et al., 1994, 1995; Imamoto et al., 1995; Radu et al., 1995) and their paralogues (Pollard et al., 1996; Gorlich et al., 1997), the small GTPase Ran (Melchior et al., 1993a; Moore and Blobel, 1993), and nuclear transport factor 2 (NTF2) (Moore and Blobel, 1994; Paschal and Gerace, 1995). Orthologues of these soluble transport factors have also been identified in yeast, flies, and plants.
The soluble transport factors mediate recognition of NLS-containing proteins and their translocation through the NPC in a multistep process (for reviews, see Nigg, 1997; Mattaj and Englmeier, 1998). Import of proteins containing the SV40-type NLS is the best characterized pathway. In the cytoplasm, the import receptor heterodimer importin α/β forms an import complex with an NLS-containing protein and facilitates binding to the cytoplasmic surface of the NPC. Subsequent passage of the import complex through the central gated channel of the NPC is the least understood aspect of nuclear protein import; it probably involves transient interactions between the import complex and multiple NPC proteins. Upon reaching the nuclear side of the NPC, binding of RanGTP to importin β triggers disassembly of the import complex and release of the NLS-containing protein into the nucleoplasm. Importin α and β are then recycled to the cytoplasm for subsequent import reactions. Although Ran and NTF2 are required for efficient import in vitro, evidence that they are imported into the nucleus as stoichiometric components of the import complex has not been obtained.
Like other small GTPases of the Ras superfamily, Ran cycles between a GTP- and a GDP-bound form, adopting distinct structural conformations and interacting with different proteins depending on its nucleotide-bound state (Scheffzek et al., 1995; Vetter et al., 1999). Regulation of the nucleotide-bound state of Ran is controlled by a cytoplasmic GTPase-activating protein, termed RanGAP (Hopper et al., 1990; Melchior et al., 1993b), and a chromatin-associated guanine nucleotide exchange factor, termed RCC1 (Ohtsubo et al., 1987). Because of the mutually exclusive subcellular localizations of RanGAP and RCC1, a gradient of RanGTP is predicted to exist across the nuclear envelope, whereby the concentration of RanGTP is higher in the nucleus than in the cytoplasm (for review, see Cole and Hammell, 1998). Although Ran shuttles between nucleus and cytoplasm (Smith et al., 1998), it is a predominantly nuclear protein at steady state (Bischoff and Ponstingl, 1991; Ren et al., 1993), and this distribution is dependent on functional RCC1 (Ren et al., 1993). The high concentration of Ran in the nucleus and the RanGTP gradient are believed to confer compartment identity to the nucleus that favors either transport complex assembly or disassembly. Whereas export complexes are formed in the nucleus in the presence of RanGTP, import complexes are believed to dissociate in the nucleus in the presence of RanGTP (Gorlich et al., 1996; Fornerod et al., 1997). Perturbations of the Ran distribution have deleterious effects on nuclear transport (Tachibana et al., 1994; Carey et al., 1996; Izaurralde et al., 1997). Thus, cells are predicted to possess a mechanism that maintains the steady-state nuclear localization of Ran.
The properties of NTF2 suggest it may modulate the steady-state distribution of Ran (Ribbeck et al., 1998; Smith et al., 1998). NTF2 binds directly to RanGDP but not RanGTP (Nehrbass and Blobel, 1996; Paschal et al., 1996; Stewart et al., 1998), and mutant NTF2 proteins that cannot bind Ran do not stimulate nuclear protein import in vitro (Clarkson et al., 1997). NTF2 is an evolutionarily conserved protein that is essential for growth in yeast (Corbett and Silver, 1996; Paschal et al., 1997). Moreover, conditional alleles of yeast NTF2 (scNTF2) show defects in nuclear protein import (Corbett and Silver, 1996). scNTF2 and the yeast Ran gene (GSP1) exhibit informative genetic interactions. The null scntf2 allele can be suppressed by overexpression of GSP1 (Paschal et al., 1997). Also, temperature-sensitive alleles of gsp1 can be suppressed by overexpression of wild-type scNTF2 but not by mutants of scNtf2p that cannot bind to Gsp1p (Wong et al., 1997). Thus, the interaction of NTF2 and Ran is important in vivo, and NTF2 function can be bypassed by increased Ran dosage. Interestingly, the null gsp1 allele cannot be suppressed by overexpression of scNTF2 (Wong et al., 1997), indicating that NTF2 cannot function as a bypass suppressor of Ran. Together, these results provide strong evidence that NTF2 plays an important role in nuclear protein import and Ran regulation.
In addition to binding RanGDP, NTF2 also binds NPC proteins. Mutations in NTF2 that abolish Ran binding do not affect binding to NPC proteins (Clarkson et al., 1996), which suggests that Ran and NPC proteins bind to different domains of NTF2. NTF2 binds directly to p62, as well as to other NPC proteins containing multiple FxFG peptide repeats (Paschal and Gerace, 1995; Clarkson et al., 1996, 1997; Nehrbass and Blobel, 1996). These repeat-containing NPC proteins have been proposed to provide binding sites for import complexes during translocation through the NPC, although direct binding of NTF2 or other transport receptors to FxFG repeats has not been shown.
NTF2 has also been reported to have a negative effect on nuclear transport. Microinjected NTF2 has been shown to inhibit nuclear protein import in mammalian cells (Tachibana et al., 1996), and RNA import has been shown to be inversely related to the concentration of endogenous NTF2 in Xenopus oocytes (Feldherr et al., 1998). Furthermore, the addition of high concentrations of NTF2 to semi-intact cells has been reported to inhibit nuclear protein import (Hu and Jans, 1999; Lam et al., 1999). The significance and mechanisms of NTF2-related inhibition of protein and RNA import are not known.
NTF2 interactions with RanGDP and NPC proteins are properties consistent with NTF2 functioning in nuclear protein import as a transport receptor for Ran. Recently, NTF2 was shown to stimulate the accumulation of Ran into digitonin-permeabilized cell nuclei (Ribbeck et al., 1998; Smith et al., 1998). These results led to the proposal that NTF2 functions as a Ran import receptor. In the present study, we have used a panel of mAbs specific for NTF2 to characterize the function of NTF2 in nuclear protein import. We show that NTF2 regulates Ran distribution in living cells, and that NTF2-mediated Ran nuclear import is required for NLS-dependent protein import in vitro. We also provide the first evidence that NTF2 can target a binding partner to the NPC in vivo. Our data, together with previous studies on NTF2, indicate that NTF2 is an essential regulator of Ran function in the cell.
MATERIALS AND METHODS
Production of NTF2 Monoclonal Antibodies
Hybridoma cell lines secreting NTF2 mAbs were generated at the University of Virginia Lymphocyte Culture Center by standard methods, using spleen cells from immunized A/J mice and the Sp2/0 myeloma cell line. The initial screen was an ELISA using recombinant human NTF2 (Paschal and Gerace, 1995) and colorimetric detection with an alkaline phosphatase-coupled secondary antibody. Clones that were positive by ELISA were evaluted for specificity by immunoprecipitation and immunoblot analyses. Antibodies were purified by protein G chromatography, using either clarified ascites fluid or conditioned culture supernatant as the starting material. The antibody isotypes were determined using a kit from Sigma (St. Louis, MO).
Production of Affinity-purified Ran Polyclonal Antibodies and Anti-NPC mAb QE5
The polyclonal antisera to Ran were generated in rabbits (Cocalico, Reamstown, PA) using the C-terminal peptide (C)MGDKPIWEQIGSSF coupled to keyhole limpet hemocyanin. The antisera were affinity purified using the Sepharose-immobilized peptide. The anti-NPC mAb QE5 was produced and purified by the University of Virginia Lymphocyte Culture Center. The QE5 hybridoma cell line was generously provided by Dr. Brian Burke (University of Calgary, Calgary, Alberta, Canada).
Cell Culture and Transfections
Adherent HeLa, BHK21, and temperature-sensitive BN2 (tsBN2) cells were grown on 10-cm plastic dishes in Dulbecco's modified Eagle's medium. HeLa and BHK21 cells were grown at 37°C in 5% CO2, and media were supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin, and 5% FBS or 10% newborn calf serum. tsBN2 cells were maintained at 33.5°C in 5% CO2, and media were supplemented with penicillin-streptomycin and 10% FBS. Temperature shift of tsBN2 cells was performed by rinsing dishes with 39.5°C medium and transferring the dishes to a 39.5°C incubator. All media, antibiotics and sera were purchased from Life Technologies (Gaithersburg, MD).
The FLAG-tagged NTF2 construct (pFLAG-NTF2) was made by fusing the NTF2 coding sequence in-frame downstream of the FLAG epitope sequence in a pcDNA3-based vector (Invitrogen, Carlsbad, CA). BHK21 cells were transfected with pFLAG-NTF2 using the calcium phosphate method. Approximately 48 h after tranfection, cells were processed for immunofluorescence and stained as described below with the M2 anti-FLAG mAb used at 1:5000 (Sigma).
Preparation of Biotinylated Proteins and Biotin–Streptavidin Complexes
BSA, recombinant myc-tagged NTF2 (in pET vector with N-terminal myc tag), and recombinant untagged NTF2 proteins (expressed and purified as described by Paschal and Gerace, 1995) were biotinylated using the Pierce (Rockford, IL) EZ-Link Sulfo-NHS-LC biotinylation kit according to the manufacturer's instructions. Biotinylation was confirmed by SDS-PAGE and Western blotting using FITC-neutravidin as probe and visualization by fluorimaging. Neutravidin, a carbohydrate-free derivative of streptavidin, was purchased from Pierce and labeled with FITC (Molecular Probes, Eugene, OR). Biotinylation products evaluated by laser desorption mass spectrometric analysis at the University of Virginia Biomolecular Research Facility indicated that biotinylation reactions resulted in the addition of one to three biotins per protein.
For preparation of biotin-streptavidin complexes, an excess of biotinylated protein was mixed with FITC-neutravidin on an end-over-end rotator at 4°C for several hours and then filtered for microinjection using 0.22-μm Millipore (Bedford, MA) centrifugal filtration units. Biotin-streptavidin complexes were microinjected as described below, and cells were processed for imaging by 3.7% formaldehyde fixation and mounting on glass slides.
SDS-PAGE and Western Blotting
Proteins were separated by SDS-PAGE (minigel system from Bio-Rad, Hercules, CA) and transferred to nitrocellulose using standard methods. Nitrocellulose was blocked overnight with 5% nonfat dry milk in PBS containing 0.1% Tween 20. Primary and secondary antibody incubations were performed at room temperature in 0.5% nonfat dry milk in PBS containing 0.1% Tween 20. NTF2 was detected with mAbs used at 1:1000 for ascites fluids, 1:5 for tissue culture supernatants, or 5 μg/ml for purified mAbs. Ran was detected with anti-Ran mAb diluted 1:5000 (Transduction Laboratories, Lexington, KY). Horseradish peroxidase-coupled goat-anti-mouse immunoglobulin G (IgG) secondary antibodies were diluted 1:5000 (Pierce). Detection was by chemiluminescence (New England Nuclear, Boston, MA).
Immunofluorescence Microscopy and Image Analysis
For immunofluorescence of intact cells, glass coverslips were seeded with cultured cells and incubated overnight at 37°C for BHK21 and HeLa cells or 33.5°C for tsBN2 cells. Coverslips were washed three times with PBS, fixed with 3.7% formaldehyde in PBS for 30 min, rinsed three times with PBS, permeabilized with 0.2% Triton X-100 in PBS for 5 min, rinsed three times with PBS, and incubated in a humid chamber with primary antibodies for 2 h and FITC-labeled donkey-anti-rabbit IgG or rhodamine-labeled goat-anti-mouse IgG secondary antibodies (Pierce) for 30 min. Primary antibodies were used at the following dilutions: anti-NTF2 ascites, 1:100; anti-Ran mAb (Transduction Laboratories), 1:200; affinity-purified anti-Ran polyclonal antibodies, 10 μg/ml; anti-NPC mAb RL1 ascites (a gift from Dr. Larry Gerace, The Scripps Research Institute, La Jolla, CA), 1:100; anti-biotin mAb (Sigma), 1:100; anti-myc mAb (Oncogene, Cambridge, MA), 1:600; and anti-histone polyclonal antibody, specific for the hyperacetylated H4 histone (a gift from Dr. David Allis, University of Virginia), 1:400. Coverslips were mounted onto glass slides using Vectashield mounting medium (Vector Laboratories, Burlingame, CA).
Fluorescence images were acquired using a 60× oil immersion lens on a Nikon (Tokyo, Japan) Microphot-SA microscope with Scanalytics (Fairfax, VA) CellScan, Photometrics (Tucson, AZ) PMIS, or Improvision (Coventry, England) Openlab software and a charge-coupled device camera (Photometrics CH250 or Hamamatsu [Bridgewater, NJ] ORCA). Within an experiment, images were captured with the same exposure times and transferred as 8-bit grayscale tagged image format files to Photoshop 4.0 (Adobe Systems, Mountain View, CA), and equivalent modifications were made to each image for the production of figures. All photomicrographs shown are representative of multiple experiments.
For deconvolution fluorescence microscopy, z stacks of 0.2-μm sections were acquired using a 60× water immersion lens and a stepper motor focus device on an Olympus (Tokyo, Japan) IX-70 microscope with a Photometrics PXL camera and Inovision (Raleigh, NC) Isee software. Captured z sections were deconvolved with Deltavision (Issaquah, WA) software version 2.0. Raw and deconvolved Isee images were converted to 8-bit grayscale tagged image format files and imported into Photoshop 4.0 for figure production.
Microinjection Assays
All microinjections were performed with an Eppendorf microinjector on an inverted Zeiss (Thornwood, NY) microscope. Cells were plated onto glass locator coverslips (Eppendorf, Hamburg, Germany) and grown overnight. Before injection, coverslips were rinsed and placed in 30-mm dishes containing phenol red-free Dulbecco's modified Eagle's medium supplemented with bovine serum and penicillin-streptomycin. For injections, proteins were diluted in PBS and clarified using 0.22-μm Millipore centrifugal filtration units. Anti-NTF2 mAbs were concentrated using Amicon/Centricon (Beverly, MA) 50-kDa cutoff centrifugal concentrators and clarified with 0.22-μm Millipore filtration units. Cells were injected using Eppendorf femtotips, placed in an incubator to recover, and processed for immunofluorescence microscopy as described above. The injection markers FITC-dextran (average molecular mass, 167 kDa) and TRITC-dextran (average molecular mass, 155 kDa), were purchased from Sigma.
In Vitro Nuclear Protein Import Assays
Import assays in digitonin-permeabilized cells were performed essentially as described (Adam et al., 1990). Adherent HeLa cells were plated onto glass coverslips and grown overnight. Coverslips were rinsed three times in cold transport buffer (TB; 20 mM HEPES, pH 7.4, 110 mM potassium acetate, 2 mM magnesium acetate, 0.5 mM EGTA), transferred to ice-cold complete TB (TB containing 1 mM DTT and protease inhibitors) with 0.005% digitonin, and incubated for 10 min. Coverslips were rinsed three times in ice-cold complete TB. Import reactions were assembled and incubated in a humid chamber for 15 min at room temperature. HeLa cytosol was prepared as described (Paschal and Gerace, 1995). Coverslips were rinsed in ice-cold complete TB, and cells were fixed and processed for immunofluorescence microscopy or fixed and visualized by direct fluorescence microscopy.
For Ran import and NLS-dependent protein import assays, cytosol was preincubated with anti-NPC mAb QE5 (1 mg/ml), wheat germ agglutinin (WGA; 0.5 mg/ml), or anti-NTF2 mAbs (1 mg/ml) on ice for 15 min. For WGA inhibition of import, permeabilized cells were preincubated with 0.5 mg/ml WGA for 5 min before addition of cytosol containing 0.5 mg/ml WGA.
For immunofluorescence microscopy of import assays, coverslips were placed in ice-cold 4% paraformaldehyde for 10 min, transferred to ice-cold methanol for 5 min, incubated at room temperature in Tris-buffered saline for 5 min, rinsed three times with PBS, and stained with primary and secondary antibodies at room temperature as described above. Coverslips were mounted onto glass slides with Vectashield mounting medium and imaged as described above.
Solid-Phase Binding Assays
Ran binding assays were performed as described (Black et al., 1999). Recombinant myc-NTF2 or untagged NTF2 was adsorbed to microtiter wells of 96-well plates (5 μg/well) in TB overnight at 4°C. The wells were blocked with BSA (30 mg/ml) in TB overnight at 4°C. Before the addition of Ran, the wells were preincubated with 50 μg of the indicated antibodies for 2 h in 0.5× TB containing BSA (5 mg/ml). Ran (preloaded with [α-32P]GDP) was added directly to these wells and incubated for an additional 2 h. The wells were washed three times, and the bound Ran was eluted with 5% SDS and analyzed by scintillation counting. Ran binding to wells containing BSA alone (background) was subtracted from NTF2 wells. The assays were performed in duplicate, and nonspecific mouse IgG was used as a negative control.
RESULTS
Characterization of mAbs Specific for NTF2
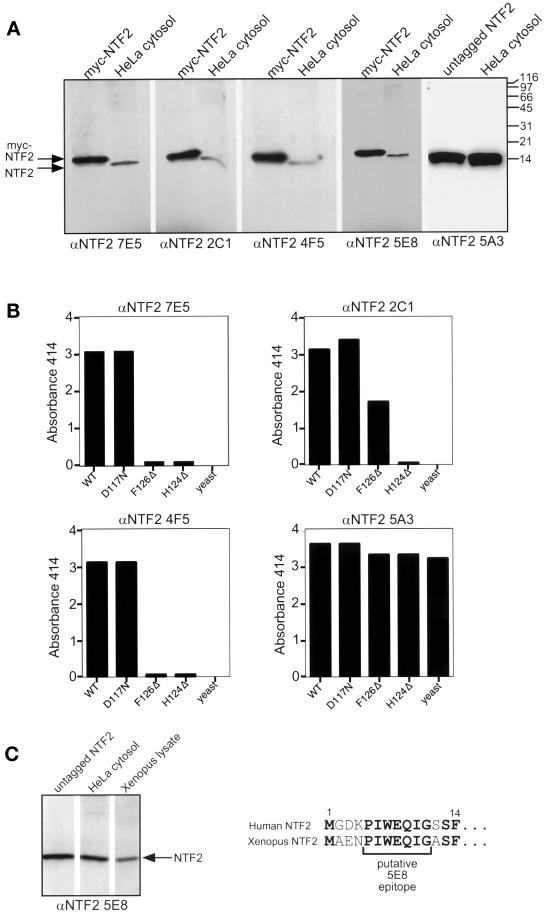
We generated a panel of mAbs to NTF2 to characterize its role in nucleocytoplasmic transport. Mice were injected with full-length (127 amino acids) recombinant human NTF2 or a synthetic peptide corresponding to the N-terminal 14 amino acids (MGDKPIWEQIGSSF) of NTF2. Hybridomas were initially screened for antibody binding to native NTF2 immobilized in microtiter wells. Clones that scored positive by ELISA were further screened by Western blotting and immunoprecipitation using rat liver cytosol. Using this approach, we were able to generate five mAbs specific for NTF2 (Table 1). Four mAbs were produced by injection of the full-length protein and are designated 7E5, 2C1, 4F5, and 5A3. The mAb 5E8 was produced by injection of the N-terminal peptide. Western blotting of recombinant NTF2 and HeLa cytosol showed that each of these five mAbs was monospecific for NTF2 (Figure 1A). The myc tag at the N terminus of recombinant NTF2 results in a slightly slower migrating protein compared with the endogenous NTF2 in HeLa cytosol (14 kDa). In addition to HeLa cytosol, each mAb recognized a single 14-kDa polypeptide in Xenopus lysate, BHK21 cell lysate, and C2C12 cell lysate (our unpublished data). We also confirmed that each of the five mAbs was able to recognize native protein by immunoprecipitating NTF2 from HeLa cell lysate (our unpublished data).
Table 1.
Properties of NTF2 mAbs
| Name | Subtype | Epitope | Binds
|
Inhibits
|
NTF2-Ran binding (% inhibition)a | ||
|---|---|---|---|---|---|---|---|
| Native NTF2 | Denatured NTF2 | NLS protein import | Ran import | ||||
| 2C1 | IgG 2b | C terminus | + | + | + | + | 29 ± 6 |
| 4F5 | IgG 2b | C terminus | + | + | + | + | 44 ± 17 |
| 7E5 | IgG 2b | C terminus | + | + | + | + | 38 ± 7 |
| 5E8 | IgG 1 | N terminus | + | + | − | +/− | 58 ± 1 |
| 5A3 | IgG 2a | Not defined | + | + | − | − | 91 ± 2 |
Experiments were performed in duplicate and values shown represent mean values ± SD.
Percent inhibition of NTF2-Ran binding by NTF2 mAbs was normalized to control IgG levels.
Figure 1.
Characterization of anti-NTF2 mAbs. (A) Anti-NTF2 mAbs are monospecific for NTF2. Recombinant human untagged (15 ng) or myc-tagged (85 ng) NTF2 protein and HeLa cytosol were separated on 15% SDS-PAGE gels, transferred to nitrocellulose, and probed with anti-NTF2 mAbs. 5A3, 7E5, 2C1, and 4F5 were used at 5 μg/ml purified antibody. 5E8 was used at 1:5 dilution of tissue culture supernatant. (B) Epitope mapping of anti-NTF2 mAbs. ELISAs were performed using recombinant wild-type human NTF2, recombinant mutant human NTF2 (D117N, H124Δ, and F126Δ), and recombinant wild-type yeast NTF2. (C) Assignment of the epitope of anti-NTF2 5E8. Western blot of recombinant untagged human NTF2 (15 ng), HeLa cytosol, and Xenopus lysate probed with 5E8 (1:1000 of ascites fluid). The synthetic N-terminal peptide of human NTF2 used to generate 5E8 is shown above the corresponding N-terminal 14 amino acids from Xenopus NTF2. Residues conserved between human and Xenopus NTF2 are in bold. The bracket indicates residues that likely encompass the 5E8 epitope.
Epitope mapping of the four anti-NTF2 mAbs produced against the full-length protein was performed using wild-type and mutant NTF2 proteins. The mutant proteins used in this analysis included the C-terminal point mutant D117N and two truncation mutants that lack either the final two residues (F126Δ) or the final four residues (H124Δ) of the C terminus (Clarkson et al., 1997). NTF2 protein was adsorbed to microtiter wells, and antibody binding was measured by ELISA (Figure 1B). mAb 4F5 recognized wild-type NTF2 and D117N but detected neither of the C-terminal truncation mutants (Figure 1B, lower left). Therefore, the 4F5 epitope lies within the extreme C terminus of NTF2. The ELISA profile for 7E5 was nearly identical to the 4F5 profile, indicating that the 7E5 epitope may be identical to the 4F5 epitope (Figure 1B, upper left). Like 4F5 and 7E5, 2C1 did not detect the H124Δ deletion mutant. mAb 2C1 did detect the F126Δ mutant, indicating that its epitope is slightly more N-terminal than the 4F5 and 7E5 epitopes (Figure 1B, upper right). mAb 5A3 reacts with wild-type NTF2 and all of the mutant proteins (Figure 1B, lower right), and it detects both human and Xenopus NTF2 by Western blotting (our unpublished data). These results indicate that the 5A3 epitope is outside of the C terminus in a domain that is conserved between these two orthologues. mAb 5E8, produced by injection of the N-terminal peptide, detects both human and Xenopus NTF2 by Western blotting (Figure 1C, left). We deduce that the 5E8 epitope lies within amino acids 5–11 of the N terminus, residues that are conserved between human and Xenopus NTF2 (Figure 1C, right). In summary, the panel of anti-NTF2 mAbs consists of the anti-N-terminal peptide antibody (5E8), the anticonserved region antibody (5A3), and the three anti-C terminus antibodies (4F5, 2C1, and 7E5) (Table 1).
NTF2 Localizes to the Nucleus at Steady State
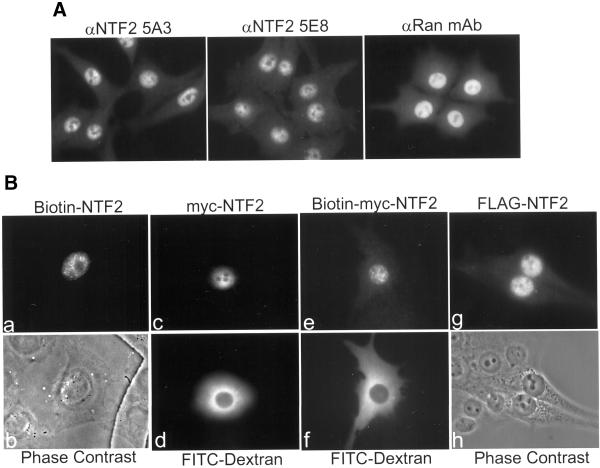
To fully understand the function of NTF2 in vivo, it is necessary to determine its subcellular location. Gold-labeled NTF2 injected into Xenopus oocytes has been shown to localize to the NPC (Feldherr et al., 1998), and fluorescently labeled NTF2 has been shown to localize to the nuclear envelope of permeabilized cells (Ribbeck et al., 1998); however, NTF2 added to protein import assays localizes to the nuclear interior (Moroianu et al., 1995). The localization of endogenous NTF2 in mammalian cells has not been shown. To address this, multiple lines of cultured cells were processed for immunofluorescence microscopy with our panel of anti-NTF2 mAbs. We found that NTF2 (Figure 2A, left and middle panels), like Ran (Figure 2A, right panel; Ren et al., 1993), is a nuclear protein at steady state that is excluded from nucleoli. None of the mAbs reveals localization of NTF2 at the NPC. NTF2 nuclear staining was also observed in HeLa, COS, C2C12, and primary chicken embryo fibroblast cells (our unpublished data). Given that multiple anti-NTF2 mAbs stain the nuclei of five different cell types, and that preincubation of 5E8 with its antigenic peptide blocks nuclear staining (our unpublished data), we conclude that the steady-state subcellular localization of endogenous NTF2 is nuclear.
Figure 2.
Localization of endogenous and microinjected NTF2 protein. (A) Endogenous NTF2 is a nuclear protein at steady state. BHK21 cells grown on glass coverslips were fixed, permeabilized, and stained with 5A3, 5E8, or anti-Ran mAb. (B) Microinjected and transfected NTF2 protein localizes to the nucleus. Recombinant biotinylated NTF2 protein (a and b), myc-NTF2 protein (c and d), or biotinylated myc-NTF2 protein (e and f) were coinjected with FITC-dextran (as an injection marker) into the cytoplasm of HeLa (a–d) or BHK21 (e and f) cells. Cells were incubated at 37°C for 30 min after injection, fixed, permeabilized, and stained with anti-biotin (a and e) or anti-myc (c) mAbs and rhodamine anti-mouse IgG. (b) Phase-contrast image of the injected cell in a. (d and f) Injection sites of the cells in c and e, respectively. BHK21 cells were transfected with FLAG-tagged NTF2 and stained with an anti-FLAG mAb (g and h).
To confirm and extend our results showing that NTF2 is a nuclear protein, the localization of microinjected NTF2 was analyzed by immunofluorescence microscopy. Untagged recombinant NTF2 purified from bacteria (Paschal and Gerace, 1995) was biotinylated and injected into the cytoplasm of HeLa cells (Figure 2B, a and b). Anti-biotin immunofluorescence microscopy showed that the injected protein localized exclusively to the nucleoplasm 30 min after injection. Likewise, myc-tagged NTF2 (Figure 2B, c and d) injected into HeLa cells and biotinylated myc-tagged NTF2 (Figure 2B, e and f) injected into BHK21 cells also localized to the nucleus. Importantly, both myc-NTF2 and biotinylated myc-NTF2, when used in in vitro import assays, stimulated import of a fluorescent NLS-containing ligand (Holaska and Paschal, unpublished data), indicating that these epitope-tagged versions of NTF2 are functional. In addition to microinjected NTF2, transfected NTF2 also localized to the nucleoplasm (Figure 2B, g and h). Nuclear localization of cytoplasmically injected and transfected NTF2 confirms that NTF2 is a nuclear protein at steady state and shows that either NTF2 itself contains information sufficient to specify its nuclear import or NTF2 associates with binding partners that mediate its nuclear localization.
NTF2 Localizes to Numerous Small Foci within the Nucleus
Although NTF2 binds directly to NPC proteins in vitro (Paschal and Gerace, 1995; Clarkson et al., 1996, 1997; Nehrbass and Blobel, 1996), our immunofluorescence microscopy did not reveal the localization of NTF2 at the nuclear envelope (see Figure 2). We considered that NTF2 may be located at the NPC but that its detection may be complicated by the high nucleoplasmic concentration of NTF2. To address this issue, we performed immunofluorescence and deconvolution microscopy and compared cells stained with an anti-NPC mAb to cells stained with an anti-NTF2 mAb.
To visualize NPCs, a stack of 0.2-μm optical sections of a HeLa cell nucleus stained with the anti-NPC mAb RL1 (Snow et al., 1987) was acquired and deconvolved. Comparison of the raw and deconvolved z sections (Figure 3A) showed that RL1 stained the surface of the nucleus in a punctate pattern (Snow et al., 1987) and that the apparent nucleoplasmic signal in the raw sections resulted from out-of-focus fluorescence. This result is consistent with the fact that NPCs are in the same plane as the nuclear envelope. Deconvolved images of a nucleus stained with anti-NTF2 mAb 5A3 (Figure 3B) showed no detectable signal at the nuclear envelope, confirming that NTF2 does not colocalize with NPCs at steady state. Instead, NTF2 localized to numerous foci within the nucleoplasm. Comparison of the raw and deconvolved z sections showed that most of the diffuse nucleoplasmic NTF2 staining was reassigned as out-of-focus fluorescence to bright foci. Our high-resolution microscopic analysis indicates that NTF2 localizes to nucleoplasmic foci but not to NPCs at steady state. We speculate that these NTF2-containing foci may represent intranuclear structures involved in nuclear transport.
Figure 3.
Deconvolution immunofluorescence microscopy of anti-NTF2 5A3- and anti-NPC RL1-stained cells. Serial sections through one nucleus of a HeLa cell stained with RL1 (A) and one nucleus of a BHK21 cell stained with 5A3 (B) are shown.
Nuclear Localization of NTF2 Is Independent of Nuclear Ran, Functional RCC1, and NLS-dependent Protein Import
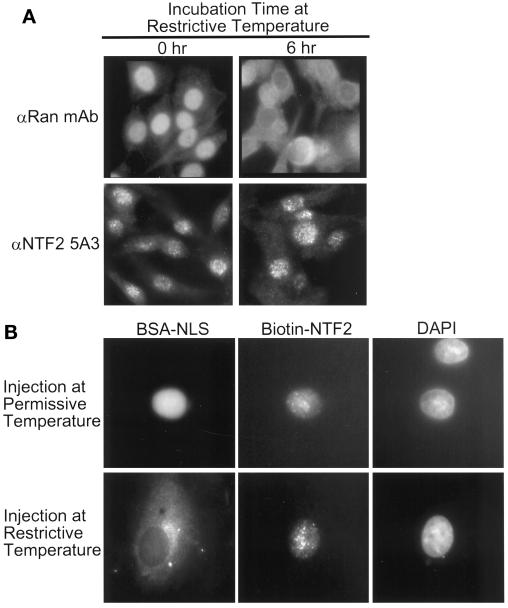
Nuclear localization of Ran has been shown to be dependent on functional RCC1 in vivo (Ren et al., 1993) and dependent on binding to NTF2 in vitro (Ribbeck et al., 1998; Smith et al., 1998). We used the BHK21-derived tsBN2 cell line (Ohtsubo et al., 1987) to test whether NTF2 nuclear localization was dependent on functional RCC1, nuclear Ran, and NLS-dependent protein import. When tsBN2 cells are shifted to the restrictive temperature of 39.5°C, RCC1 is degraded, Ran accumulates in the cytoplasm, and NLS-dependent protein import is inhibited (Ohtsubo et al., 1987; Ren et al., 1993; Tachibana et al., 1994).
As is well established, 6 h after shifting tsBN2 cells to the restrictive temperature, most Ran has been lost from the nucleus and has accumulated in the cytoplasm (Figure 4A, upper panels). Surprisingly, NTF2 remained in nuclei of tsBN2 cells under the condition in which Ran redistributed to the cytoplasm (Figure 4A, lower panels). We also note that NTF2 has persisted in nuclear foci. This result shows that NTF2 remains localized to the nucleus in the absence of functional RCC1, nuclear Ran, and NLS-dependent import. To address whether transport of NTF2 into the nucleus depends on functional RCC1, nuclear Ran, and NLS-dependent import, biotinylated NTF2 (bNTF2) and a fluorescent import ligand (FITC-BSA-NLS) were coinjected into the cytoplasm of tsBN2 cells preincubated at the permissive and restrictive temperatures. At the permissive temperature, both FITC-BSA-NLS and bNTF2 accumulated in the nucleus (Figure 4B, upper panels). At the restrictive temperature, FITC-BSA-NLS was excluded from the nucleus, indicating that NLS-dependent protein import was inhibited (Figure 4B, lower left panel). In contrast, bNTF2 efficiently accumulated in the nucleus of the same cell (Figure 4B, lower center panel), indicating that import of NTF2 is mechanistically different from import of NLS ligand.
Figure 4.
Nuclear localization of NTF2 is independent of functional RCC1, nuclear Ran, and NLS-dependent protein import. (A) tsBN2 cells were incubated for 0 h (left panels) or 6 h (right panels) at the restrictive temperature (39.5°C). Cells were fixed, permeabilized, and stained with either anti-Ran mAb (upper panels) or 5A3 (lower panels). (B) tsBN2 cells were preincubated at the permissive temperature (33.5°C; upper panels) or the restrictive temperature (39.5°C; lower panels) and cytoplasmically coinjected with bNTF2 and FITC-BSA coupled to NLS peptide. Cells were further incubated at the respective temperatures for 30 min after injection, fixed, permeabilized, and stained with anti-biotin mAb, rhodamine anti-mouse IgG, and DAPI. Left, center, and right panels, FITC, rhodamine, and DAPI images, respectively, of the same cells.
Ran-independent Nuclear Import of NTF2
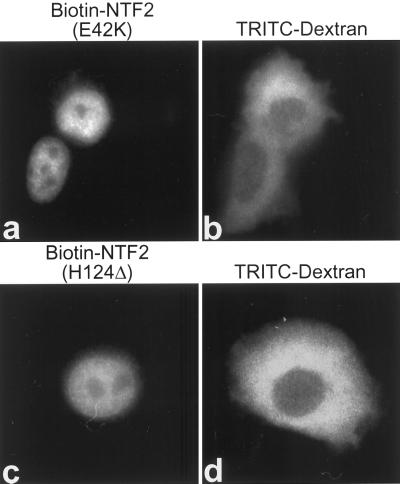
To directly address whether an interaction with Ran is necessary for NTF2 nuclear entry and accumulation in living cells, the localization of NTF2 mutants E42K and H124Δ was determined by microinjection analysis. The E42K mutation in NTF2 abolishes binding to Ran, whereas the H124Δ mutation significantly reduces binding to Ran (Clarkson et al., 1997). Each mutant protein was biotinylated and injected into the cytoplasm of HeLa cells, which were then stained with anti-biotin mAb. Wild-type bNTF2 injected into the cytoplasm localized to the nucleus (Figure 2B, a). Surprisingly, both NTF2 mutants also localized to the nucleus (Figure 5, a and c). These results show that nuclear import of NTF2 is independent of direct binding to Ran.
Figure 5.
Nuclear localization of NTF2 is independent of Ran binding. Ran binding mutants of NTF2, E42K (a) and H124Δ (c), which eliminate or significantly reduce binding to Ran, respectively, were biotinylated and coinjected with FITC-dextran as an injection marker (b and d) into the cytoplasm of HeLa cells. Cells were incubated at 37°C for 30 min aftet injection, fixed, permeabilized, and stained with antibiotin mAb and rhodamine anti-mouse IgG.
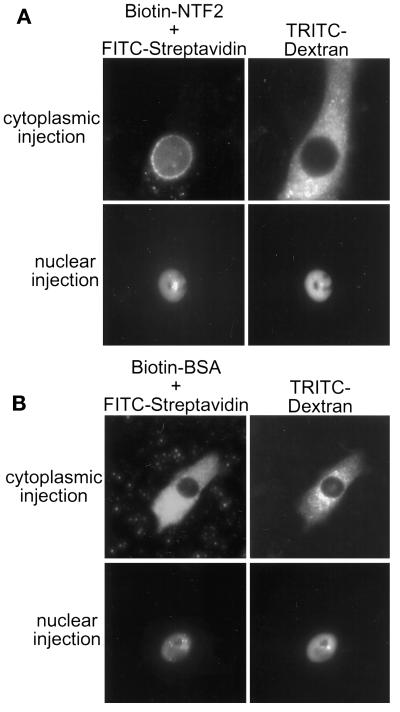
NTF2-Streptavidin Injected into the Cytoplasm Accumulates at the Nuclear Envelope
To investigate whether NTF2 can mediate the nuclear entry of a binding partner, we tested whether bNTF2 could import FITC-streptavidin to the nucleus in vivo. Complexes of bNTF2-streptavidin were injected into BHK21 cells, and fluorescence microscopy was performed to determine the localization of the injected complexes. bNTF2-streptavidin injected into the cytoplasm exhibited a striking accumulation at the nuclear envelope, in apparent association with NPCs (Figure 6A, upper panels), and with little or no localization in the cytoplasm or nucleoplasm. In contrast to the result obtained with the cytoplasmic injection, bNTF2-streptavidin injected into the nucleus remained in the nucleoplasm, with no evidence of export to the cytoplasm or accumulation at the nuclear envelope (Figure 6A, lower panels). In the control experiments, biotinylated BSA-streptavidin remained diffusely distributed in the compartment into which it was injected, with no localization at the nuclear envelope (Figure 6B). This experiment shows that NTF2 can target a binding partner from the cytoplasm to the NPC in vivo. Because NTF2 can bind to NPC proteins (and streptavidin does not), NTF2-streptavidin localized at the nuclear envelope may represent an intermediate step in NTF2-mediated import. Furthermore, our observation that nuclear-injected NTF2-streptavidin does not accumulate at the nuclear envelope suggests a difference in apparent affinities of NTF2 for the cytoplasmic and nuclear sides of the NPC.
Figure 6.
Localization of injected bNTF2-streptavidin complexes. bNTF2 (A) or biotinylated BSA (B) was preincubated with FITC-streptavidin and coinjected with TRITC-dextran as an injection marker into the cytoplasm or nucleus of BHK21 cells. Cells were incubated at 37°C for 1 h after injection and fixed, and fluorescent proteins were visualized directly.
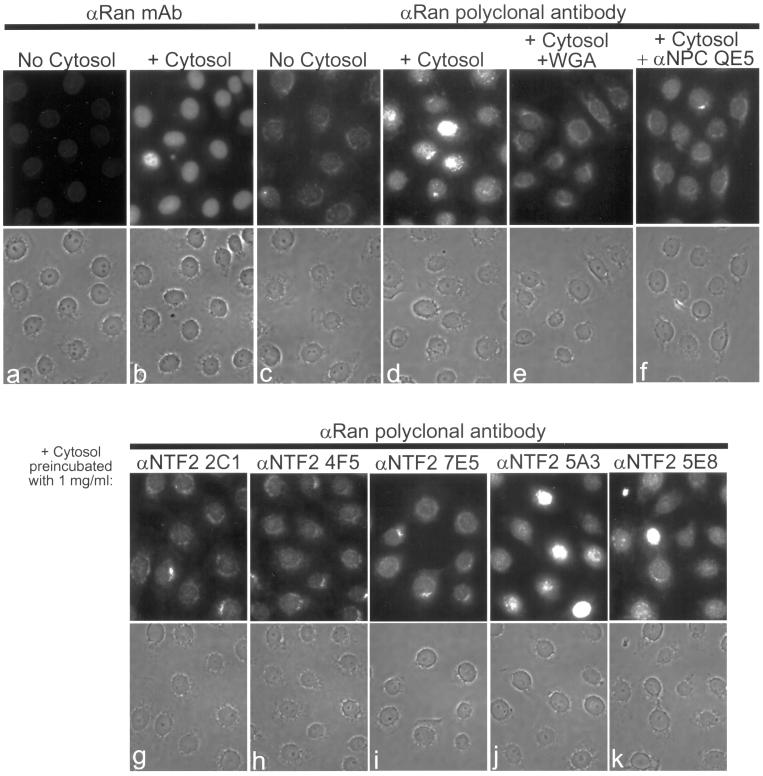
Preincubation of Cytosol with mAbs to the C Terminus of NTF2 Blocks Nuclear Import of Ran
We examined whether cytosol preincubated with our panel of anti-NTF2 mAbs could support nuclear import of Ran in digitonin-permeabilized cells. To carefully assess the effects of the mAbs on Ran import, we first examined 1) the level of Ran remaining in permeabilized cells, 2) the level of Ran that can be imported from cytosol, and 3) the level of Ran import that can be inhibited by nuclear transport inhibitors. To assay for Ran import, we added exogenous HeLa cytosol to permeabilized cells and performed immunofluorescence microscopy with anti-Ran antibodies. As previously shown, HeLa cells permeabilized with digitonin are mostly depleted of Ran (Figure 7, a and c; Melchior et al., 1995). Addition of cytosol and an energy-regenerating system to permeabilized cells resulted in the accumulation of Ran in the nuclear interior (Figure 7, b and d; Melchior et al., 1995). Addition of cytosol in the presence of WGA or anti-NPC mAb QE5 inhibited nuclear accumulation of Ran (Figure 7, e and f). These control experiments suggest that Ran import does not occur by passive diffusion, although it is formally possible that Ran passively diffuses through the NPC and the active import of another protein is requisite for nuclear accumulation of Ran.
Figure 7.
Preincubation of cytosol with anti-NTF2 mAbs inhibits Ran nuclear import in permeabilized cells. (a–k) Adherent HeLa cells grown on glass coverslips were permeabilized with digitonin and incubated with an energy-regenerating system for 15 min at room temperature. Cells were incubated without exogenous HeLa cytosol (a and c), with exogenous HeLa cytosol (b and d), or with exogenous HeLa cytosol that had been preincubated on ice for 15 min with 0.5 mg/ml WGA (e), 1 mg/ml anti-NPC mAb QE5 (f), or 1 mg/ml anti-NTF2 mAb (g–k). Cells were stained for Ran with either anti-Ran mAb and rhodamine anti-mouse IgG (a and b) or anti-Ran polyclonal antibodies and FITC anti-rabbit IgG (c–k).
Anti-NTF2 mAbs were analyzed individually for the ability to inhibit Ran accumulation in permeabilized cell nuclei (Figure 7, g–k). Each anti-C terminus mAb (1 mg/ml) inhibited Ran import (Figure 7, g–i) to an extent comparable with the inhibition observed with 0.5 mg/ml WGA or 1 mg/ml QE5 (Figure 7, e and f). Ran nuclear accumulation was inhibited to the greatest extent by the anti-C terminus mAb 2C1. In contrast, Ran import was not inhibited by preincubation of cytosol with either 5A3 or 5E8 (Figure 7, j and k). These data show that Ran import in vitro can be inhibited by mAbs to the NTF2 C terminus but not by mAbs to other regions of NTF2.
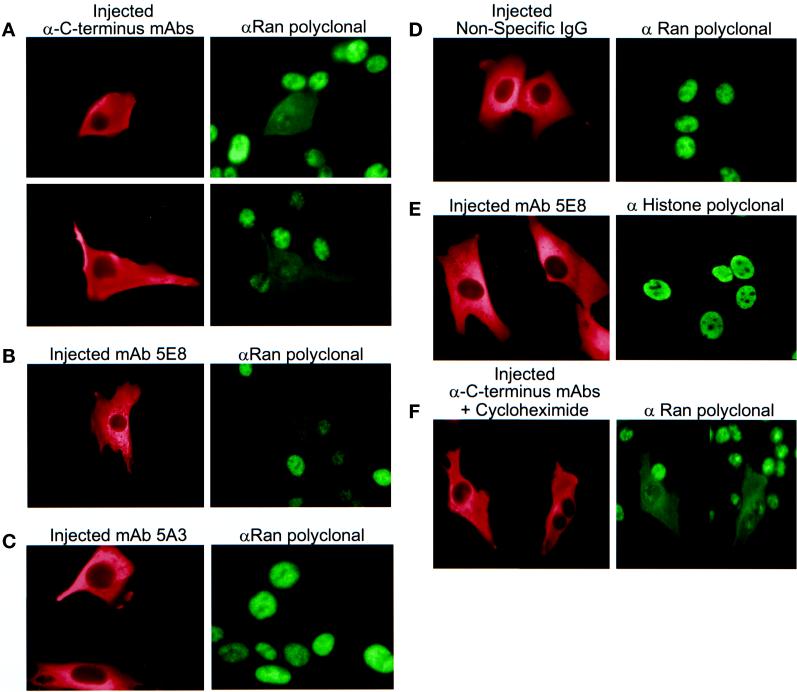
Injection of mAbs to NTF2 Results in the Redistribution of Ran in Living Cells
We investigated whether injection of mAbs to NTF2 would affect Ran distribution in living cells. Because the anti-C terminus mAbs inhibited Ran import in vitro, we predicted that these antibodies would block Ran import in vivo. Both the cocrystal structure of RanGDP-NTF2 (Stewart et al., 1998) and binding assays (Clarkson et al., 1997) show that the NTF2 C terminus is crucial for stabilization of the interaction with Ran. Therefore, we predicted that the anti-C terminus mAbs (2C1, 4F5, and 7E5) would act by specifically blocking NTF2 binding to Ran.
Because the anti-C terminus mAbs recognize the same or overlapping epitopes, we injected these antibodies as a mixture. Cytoplasmic injection of the anti-C terminus mAb mixture (4F5–7E5–2C1) resulted in a striking redistribution of Ran to the cytoplasm (Figure 8A). Ran is predominantly nuclear at steady state, but in the presence of the 4F5–7E5–2C1 mixture, Ran equilibrated between nucleus and cytoplasm and was absent from the nuclear envelope. Injection of 5E8 also induced Ran redistribution (Figure 8B). Injection of 5A3 had no detectable effect (Figure 8C), and Ran distribution was unaffected when NTF2 mAbs were injected into the nucleus (our unpublished data). Experiments were performed to address the specificity of the effect of the mAbs and to demonstrate that microinjection of NTF2 mAbs does not induce a general redistribution of nuclear proteins. Neither cytoplasmic (Figure 8D) nor nuclear injection (our unpublished data) of nonspecific mouse IgG caused a change in Ran distribution. Also, microinjection of mAb 5E8 caused no change in the nuclear localization of histone H4 (Figure 8E). Because injection of the NTF2 mAbs into the cytoplasm (but not the nucleus) induces Ran redistribution to the cytoplasm, we conclude that these antibodies are affecting a cytoplasmic NTF2-Ran interaction that is important for Ran nuclear import.
Figure 8.
Injection of mAbs to NTF2 results in the redistribution of endogenous Ran in living cells. A mixture of anti-NTF2 mAbs (2C1, 4F5, and 7E5; ∼25 mg/ml total concentration; A and F), ∼12 mg/ml 5E8 (B and E), ∼12 mg/ml 5A3 (C), or 12 mg/ml nonspecific mouse IgG (D) was injected into the cytoplasm of BHK21 cells. Two hours after injection the cells were fixed, permeabilized, and stained with anti-Ran polyclonal antibodies and FITC anti-rabbit IgG to visualize endogenous Ran (A–D and F) or anti-histone H4 polyclonal antibody and FITC anti-rabbit IgG to visualize endogenous histone H4 (E). All cells were also stained with rhodamine anti-mouse IgG to visualize the injected mAbs. In F, cells were incubated before injection in the presence of 50 μg/ml or 200 μg/ml cycloheximide, injected, and incubated after injection with cycloheximide to inhibit protein synthesis. Each image in C, E, and F is merged from two separate fields of view.
The mAbs could be affecting Ran distribution by trapping preexisting NTF2 and/or preexisting Ran in the cytoplasm as they shuttle between compartments or by trapping newly synthesized NTF2 and/or Ran in the cytoplasm before their initial nuclear import. To address whether the effect was due to anti-NTF2 mAbs binding to newly synthesized NTF2 (or trapping of newly synthesized Ran), the 2C1–4F5–7E5 cocktail was injected into BHK21 cells that had been preincubated with cycloheximide to inhibit protein synthesis. Ran redistributed to the cytoplasm in the presence and absence of cycloheximide (Figure 8, compare F and A). Therefore, mislocalization of Ran is not a consequence of trapping newly synthesized NTF2 or Ran in the cytoplasm. Rather, the redistribution of Ran likely results from the inhibition of the reimport of shuttling Ran.
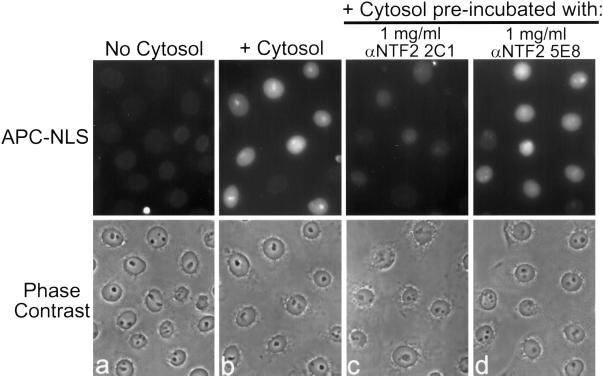
mAbs to the C Terminus of NTF2 Inhibit NLS-dependent Nuclear Protein Import
The high concentration of Ran in the nucleus is apparently essential for nuclear transport. Nuclear RanGTP is thought to be required for the dissociation of import complexes upon reaching the nucleoplasm (Gorlich et al., 1996), and RanGTP is required for the assembly of export complexes (Fornerod et al., 1997). Therefore, we predicted that blocking the nuclear accumulation of Ran by anti-NTF2 mAbs would inhibit NLS-dependent protein import. To test this prediction, we preincubated cytosol with our panel of mAbs and assayed the import of an NLS ligand in permeabilized cells by fluorescence microscopy. In these experiments, the nuclear accumulation of fluorescent NLS ligand requires the addition of exogenous cytosol (Figure 9, a and b).
Figure 9.
Preincubation of cytosol with anti-NTF2 mAbs inhibits nuclear import of a fluorescent NLS ligand. (a–d) HeLa cells were permeabilized, and protein import was assayed using the naturally fluorescent allophycocyanin (APC) protein coupled to NLS peptides. Transport reactions were preincubated on ice in the absence (a and b) or presence of 1 mg/ml purified anti-NTF2 mAb (c and d). Cells were fixed, and APC-NLS protein was visualized directly.
Preincubation of cytosol with each anti-C terminus mAb (1 mg/ml) inhibited import of NLS ligand to an extent comparable with the level of inhibition by 0.5 mg/ml WGA (Figure 9c; our unpublished data). In contrast, cytosol preincubated with either 5E8 (Figure 9d) or 5A3 (our unpublished data) was fully competent for NLS import. These data show that mAbs to NTF2 that block Ran import also block NLS-dependent protein import.
NTF2-Ran Binding Can Be Partially Blocked by mAbs to NTF2
There are at least two different mechanisms by which the NTF2 mAbs could inhibit Ran import. The antibodies could block the interaction between NTF2 and Ran. The epitopes for several of the mAbs include residues that directly participate in Ran binding. Alternatively, the antibodies could bind to NTF2-Ran complexes. In this case, antibody binding to the NTF2-Ran complex would prevent nuclear import of Ran. To address the first possibility, we tested whether the mAbs could block NTF2-Ran binding in vitro (Table 1). Microtiter plate binding assays were performed in which NTF2 was adhered to the bottom of microtiter wells, an excess of antibodies was added, and radiolabeled Ran was added to the NTF2-mAb complexes. Binding of Ran to NTF2 in the presence and absence of the antibodies was quantitated by scintillation counting of bound, radiolabeled Ran. This type of assay has been used successfully to show that NTF2 bound to a microtiter well can discriminate between the GDP and GTP forms of Ran (Black et al., 1999).
The anti-C terminus mAbs, 2C1, 4F5, and 7E5, inhibited Ran binding to NTF2 by 29, 44, and 38%, respectively. 5E8 inhibited Ran binding to NTF2 by 58%. 5A3 inhibited Ran binding to NTF2 by 91% (Table 1). These data show that 5A3, and to a lesser extent 5E8, is a more potent inhibitor of NTF2-Ran binding in vitro than each of the anti-C terminus mAbs. This is somewhat surprising given that the anti-C terminus antibodies were effective at redistributing Ran in vivo. It suggests, therefore, that the redistribution of Ran in vivo could result from cytoplasmic tethering of Ran-NTF2-mAb complexes.
DISCUSSION
NTF2 Regulates the Subcellular Distribution of Ran
Many lines of evidence indicate that the concentration of Ran in the nucleus is essential for nuclear transport. Moreover, nuclear RanGTP is predicted to be continually depleted during the export of both import receptors and export complexes. Therefore, a mechanism should exist to ensure the steady-state nuclear accumulation of Ran. Studies using permeabilized cells indicated that Ran is imported into the nucleus in the GDP-bound form (Ribbeck et al., 1998; Smith et al., 1998). In microinjection experiments, the nuclear accumulation of Ran was shown to be saturable (Smith et al., 1998), suggesting that Ran import is receptor mediated. NTF2 is an attractive candidate for a Ran import receptor because it binds specifically to RanGDP and to multiple NPC proteins.
In this study, we have presented evidence that NTF2 is necessary for the nuclear accumulation of Ran in living cells and in permeabilized cells. We found that cytosol preincubated with anti-NTF2 mAbs does not support Ran import or NLS-dependent protein import in vitro, which indicates that the function of NTF2 in NLS protein import may be to stimulate the nuclear accumulation of Ran. In addition, we provide the first evidence that NTF2 can target a binding partner to the nuclear envelope in vivo. Taken together, these data strongly suggest that NTF2 mediates interactions between NPC proteins and RanGDP to drive the steady-state nuclear accumulation of Ran that is required for nuclear protein import.
Model of NTF2 and Ran Transport
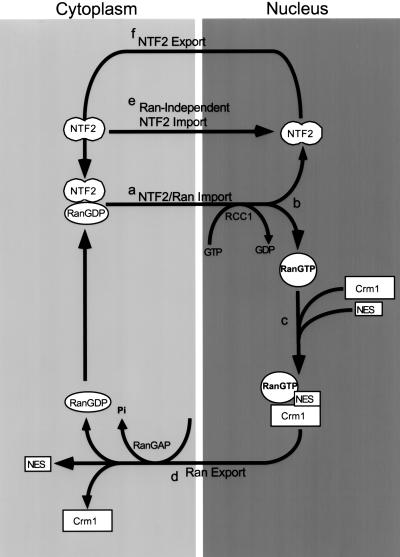
Ran import is likely to be a multistep process, similar to the process of NLS-dependent protein import, which can be divided into four steps: receptor-ligand binding in the cytoplasm, receptor-mediated interactions with NPC proteins and translocation of the import complex through the NPC, import complex dissociation in the nucleus, and receptor recycling to the cytoplasm. Immunofluorescence microscopy of endogenous factors, microinjection analysis, and in vitro assays have provided insights into this process by establishing many of the protein–protein interactions involved in each step. Our results, together with other studies, suggest a model that describes the import and export pathways of Ran and NTF2 (Figure 10). In this model, NTF2 binds to RanGDP in the cytoplasm and mediates translocation through the NPC by interacting with FxFG-containing NPC proteins. Dissociation of the NTF2-RanGDP complex in the nucleus occurs by RCC1-catalyzed nucleotide exchange on Ran and is followed by NTF2 export to the cytoplasm. RanGTP is exported to the cytoplasm bound to importin β-type receptors, and dissociation of cargo and Ran from the receptors is triggered by RanGAP-catalyzed GTP hydrolysis. Thus, NTF2 and Ran are both shuttling proteins that enter the nucleus together but exit the nucleus separately.
Figure 10.
Model of NTF2 and Ran trafficking. (a) NTF2 homodimers bind to RanGDP in the cytoplasm, and NTF2-RanGDP complexes are translocated through NPCs by the interaction of NTF2 with FxFG-containing NPC proteins, in a manner analogous to translocation of transport complexes via importin β-type receptors. (b) Dissociation of the NTF2-RanGDP complex occurs by RCC1-catalyzed nucleotide exchange on Ran. (c) RanGTP is exported to the cytoplasm complexed to importin β-type receptors (e.g., Crm1). (d) RanGAP-catalyzed GTP hydrolysis on Ran triggers dissociation of Ran from the receptors in the cytoplasm. (e) NTF2 can also enter the nucleus independently of Ran. (f) After dissociation of NTF2 and Ran in the nucleus, NTF2 recycles to the cytoplasm. RanGDP then binds to NTF2 in the cytoplasm for a new round of nuclear import and export.
For the first and second steps of import (Figure 10a), NTF2 and RanGDP bind in the cytoplasm and translocate through the NPC as a complex, mediated by NTF2–NPC interactions. This is analogous to the first and second steps of NLS-dependent protein import, whereby shuttling receptors bind NLS-containing proteins in the cytoplasm and mediate translocation by receptor–NPC interactions. Although it is still possible that NTF2 and Ran translocate through the NPC separately, our data indicate that a functional interaction between these proteins is required on the cytoplasmic side of the nuclear envelope (see Figure 8). Moreover, we have shown that NTF2 can target a binding partner (streptavidin) to the cytoplasmic side of the nuclear envelope. Taken together, these data suggest that NTF2 binds Ran in the cytoplasm and targets it to the NPC (Figure 10a).
Receptor-mediated nuclear import is predicted to require dissociation of the cargo protein from the receptor upon reaching the nucleus. Therefore, NTF2-mediated Ran import requires dissociation of these proteins after translocation to the nuclear side of the NPC (Figure 10b). Conversion of RanGDP to RanGTP by RCC1 is a plausible mechanism for triggering NTF2-Ran dissociation in the nucleus, because NTF2 has no affinity for RanGTP. RanGTP could then bind directly to nucleoplasmic importin β and transportin, contributing to the NTF2-mediated nuclear accumulation of Ran observed in vitro (Ribbeck et al., 1998; Smith et al., 1998). Interestingly, however, NTF2 has recently been shown to inhibit RCC1-catalyzed GDP dissociation from Ran (Yamada et al., 1998). This observation suggests that the NTF2-Ran dissociation step in the nucleus may be regulated by additional factors.
After dissociation in the nucleus, the transport pathways of NTF2 and Ran diverge. Ran is exported to the cytoplasm bound to importin β-type receptors, e.g., Crm1 (Figure 10c). Upon reaching the cytoplasmic side of the NPC, RanGAP-catalyzed GTP hydrolysis would result in conversion of RanGTP to RanGDP and disassembly of the export complex (Figure 10d). RanGDP would then be able to bind NTF2 in the cytoplasm for a new cycle of import and export.
As a shuttling import receptor, NTF2 needs to be recycled to the cytoplasm for further rounds of RanGDP import (Figure 10f). The mechanism of NTF2 export has not been addressed, but it could involve 1) passive diffusion, 2) export mediated by an NTF2 export receptor, or 3) export involving direct interactions of NTF2 with NPC proteins. We favor a direct interaction with NPC proteins as a mechanism for NTF2 export, because like importin β-type proteins (Kose et al., 1997, 1999; Nakielny and Dreyfuss, 1997), NTF2 can translocate across the NPC without its cargo, Ran (see below).
The import and export cycles of NTF2 and Ran clearly gives rise to a high nuclear steady-state concentration of both proteins. This could be due to faster import rates compared with export rates for NTF2 and Ran or binding interactions in the nucleus. In the case of Ran, nuclear RCC1 could provide a stable binding site, or Ran could associate with importin β-type receptors, as has been previously suggested (Ribbeck et al., 1998; Smith et al., 1998). In addition, the steady-state nuclear distribution of both NTF2 and Ran could reflect a stable interaction between these proteins in the nucleus. We speculate that the localization of NTF2 to small foci might reflect an association with other nuclear proteins in addition to Ran.
Import of NTF2 Can Occur by a Ran-independent Mechanism
We found that nuclear import and localization of NTF2 are independent of the nuclear localization of Ran, independent of NTF2 binding to Ran, independent of functional RCC1, and independent of ongoing NLS-protein import. Ran-independent transport of the E42K mutant of NTF2 clearly demonstrates that NTF2 can enter the nucleus without Ran.
Nucleocytoplasmic trafficking of NTF2 may be analogous to the transport of importin β-type receptors, which can occur independent of Ran and the RanGTP gradient (Kose et al., 1997, 1999; Nakielny and Dreyfuss, 1997). Both importin β and transportin contain import modules, which can translocate into the nucleus independent of cargo binding and independent of Ran (Kose et al., 1997; Nakielny and Dreyfuss, 1997). Similarly, we find that nuclear import of NTF2 can occur independent of binding to Ran (its apparent cargo) and independent of the RanGTP gradient. Because importin β transport in the absence of cargo and Ran requires binding to NPC proteins (Kose et al., 1997, 1999), we predict that NTF2 nuclear import and export in the absence of Ran likewise requires NTF2 interactions with NPC proteins.
NTF2 Is Not Stably Associated with the NPC
Fluorescein-labeled NTF2 has been shown to localize exclusively to the nuclear envelope in vitro (Ribbeck et al., 1998), and colloidal gold-labeled NTF2 has been shown to accumulate at NPCs in Xenopus oocytes (Feldherr et al., 1998). Also, it was reported that microinjected FITC-NTF2 is enriched at the nuclear envelope in intact somatic cells (Yamada et al., 1998), and endogenous NTF2 is concentrated at the nuclear envelope in yeast (Corbett and Silver, 1996). In contrast to these reports, we find that multiple mAbs show that NTF2 is a nuclear protein at steady state. Moreover, deconvolution immunofluorescence microscopy reveals that NTF2 does not localize to the NPC but does localize to numerous small nucleoplasmic foci. In addition, biochemically active, epitope-tagged NTF2 localizes to the nucleus after microinjection into the cytoplasm. We also found that after microinjection, fluorescently labeled NTF2 localizes to the nucleus, showing no accumulation at the NPC (our unpublished observations). Interestingly, addition of the same preparation of fluorescent NTF2 to digitonin-permeabilized cells results in marked accumulation at the NPC, with little or no accumulation in the nucleoplasm (our unpublished observations). The latter result has been reported previously (Ribbeck et al., 1998). In light of the distribution of endogenous, microinjected, and transfected NTF2, the accumulation of exogenous NTF2 at the NPC in vitro may be a consequence of the permeabilized cell system. The absence of NTF2 at the NPC under more physiological conditions probably reflects the highly transient nature of NTF2–NPC interactions, similar to the transient interactions of other soluble transport factors such as Ran with the NPC.
Basis for mAb Inhibition of Ran Import
Inhibition of Ran import by the anti-C terminus mAbs and 5E8 can be explained by at least two different mechanisms: 1) the mAbs could block NTF2 binding to Ran in the cytoplasm, thereby preventing formation of a Ran import complex, or 2) the mAbs could bind to the cytoplasmic Ran-NTF2 complex, creating a trimeric complex that is sterically inhibited from undergoing import.
Because the anti-C terminus mAbs recognize residues that are known to be involved in Ran binding (Clarkson et al., 1997; Stewart et al., 1998), we predicted that the anti-C terminus antibodies would inhibit Ran import by the first mechanism, namely blockage of NTF2-Ran binding. The cocrystal structure of NTF2 and Ran demonstrates that there are several interactions between Ran and NTF2 (Stewart et al., 1998). The most important interaction is the insertion of a conserved phenylalanine in Ran (F72) into the hydrophobic cavity of the NTF2 monomer. Stabilization of NTF2-Ran binding is provided in part by the interaction of the switch I domain of Ran with the C-terminal residues L123, H124, and N125 of NTF2. Studies with truncation mutants of NTF2 (Clarkson et al., 1997) showed that removal of the last two residues from the C terminus (126 and 127) did not influence NTF2-Ran binding but that further removal of residues 124 and 125 abolished NTF2-Ran binding. This underscores the importance of H124 and N125 in stabilizing NTF2 binding to Ran. Epitope mapping of the three anti-C terminus mAbs indicates that the epitopes for these antibodies include residues H124 and/or N125 (see Figure 1). Thus, the epitope mapping, truncation mutant studies, and the cocrystal structure suggest that the anti-C terminus mAbs should inhibit Ran import by blocking the NTF2–Ran interaction. However, using quantitative microtiter plate binding assays, we have found that the NTF2-Ran interaction is inhibited 29–44% by the anti-C terminus mAbs. It is unclear why these mAbs do not completely block the NTF2–Ran interaction in vitro, given their capacity to inhibit Ran import in cells. It may be related to a reduced ability to bind NTF2 adsorbed in a microtiter well versus NTF2 in the cytoplasm of a living cell. mAb 5A3 displayed the highest level of inhibition of Ran binding to NTF2 in vitro. Surprisingly, this antibody does not induce Ran redistribution in vivo. It is conceivable that mAb 5A3 fails to interact with NTF2 in vivo because of epitope masking by another protein. Alternatively, it is formally possible that Ran import in the presence of 5A3 may be due to an NTF2-independent mechanism.
In summary, we have presented a cellular characterization of NTF2 and Ran transport pathways. Our studies have revealed that NTF2 is a regulator of nuclear transport that controls Ran distribution in the cell. The continued analysis of NTF2 is expected to provide additional insights into the functional regulation of the nuclear transport machinery.
ACKNOWLEDGMENTS
We thank Dr. Ammasi Periasamy (Center for Cellular Imaging, University of Virginia) for assistance with deconvolution microscopy, Dr. Murray Stewart for the mutant NTF2 cDNAs, Sally Adams for assistance with monoclonal antibody production, and Dr. Lizabeth Allison for preliminary experiments analyzing the distribution of NTF2 in the Xenopus oocyte. We also thank James Holaska for assistance with in vitro import assays and Dr. Amy Brownawell for reading the manuscript. This research was supported by American Cancer Society grant RPG98-048-01-CSM and by funds from the Lucille P. Markey charitable trust.
Abbreviations used:
- bNTF2
biotinylated NTF2
- IgG
immunoglobulin G
- NLS
nuclear localization signal
- NPC
nuclear pore complex
- NTF2
nuclear transport factor 2
- TB
transport buffer
- ts
temperature-sensitive
- WGA
wheat germ agglutinin
REFERENCES
- Adam EJ, Adam SA. Identification of cytosolic factors required for nuclear location sequence-mediated binding to the nuclear envelope. J Cell Biol. 1994;125:547–555. doi: 10.1083/jcb.125.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam SA, Gerace L. Cytosolic proteins that specifically bind nuclear location signals are receptors for nuclear import. Cell. 1991;66:837–847. doi: 10.1016/0092-8674(91)90431-w. [DOI] [PubMed] [Google Scholar]
- Adam SA, Sterne-Marr RE, Gerace L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff FR, Ponstingl H. Mitotic regulator protein RCC1 is complexed with a nuclear ras-related polypeptide. Proc Natl Acad Sci USA. 1991;88:10830–10834. doi: 10.1073/pnas.88.23.10830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Lévesque L, Holaska JM, Wood TC, Paschal BM. Identification of an NTF2-related factor that binds Ran-GTP and regulates nuclear protein export. Mol Cell Biol. 1999;19:8616–8624. doi: 10.1128/mcb.19.12.8616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey KL, Richards SA, Lounsbury KM, Macara IG. Evidence using a green fluorescent protein-glucocorticoid receptor chimera that the Ran/TC4 GTPase mediates an essential function independent of nuclear protein import. J Cell Biol. 1996;133:985–996. doi: 10.1083/jcb.133.5.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi NC, Adam EJ, Adam SA. Sequence and characterization of cytoplasmic nuclear protein import factor p97. J Cell Biol. 1995;130:265–274. doi: 10.1083/jcb.130.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson WD, Corbett AH, Paschal BM, Kent HM, McCoy A, Gerace L, Silver P, Stewart M. Nuclear protein import is decreased by engineered mutants of nuclear transport factor 2 (NTF2) that do not bind GDP-Ran. J Mol Biol. 1997;272:716–730. doi: 10.1006/jmbi.1997.1255. [DOI] [PubMed] [Google Scholar]
- Clarkson WD, Kent HM, Stewart M. Separate binding sites on nuclear transport factor 2 (NTF2) for GDP-Ran and the phenylalanine-rich repeat regions of nucleoporins p62 and Nsp1p. J Mol Biol. 1996;263:517–524. doi: 10.1006/jmbi.1996.0594. [DOI] [PubMed] [Google Scholar]
- Cole CN, Hammell CM. Nucleocytoplasmic transport—driving and directing transport. Curr Biol. 1998;8:R368–R372. doi: 10.1016/s0960-9822(98)70239-8. [DOI] [PubMed] [Google Scholar]
- Corbett AH, Silver PA. The NTF2 gene encodes an essential, highly conserved protein that functions in nuclear transport in vivo. J Biol Chem. 1996;271:18477–18484. doi: 10.1074/jbc.271.31.18477. [DOI] [PubMed] [Google Scholar]
- Feldherr C, Akin D, Moore MS. The nuclear import factor p10 regulates the functional size of the nuclear pore complex during oogenesis. J Cell Sci. 1998;111:1889–1896. doi: 10.1242/jcs.111.13.1889. [DOI] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. Crm1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Goldfarb DS, Gariepy J, Schoolnik G, Kornberg RD. Synthetic peptides as nuclear location signals. Nature. 1986;326:641–644. doi: 10.1038/322641a0. [DOI] [PubMed] [Google Scholar]
- Gorlich D, Dabrowski M, Bischoff FR, Kutay U, Bork P, Hartmann E, Prehn S, Izaurralde E. A novel class of RanGTP binding proteins. J Cell Biol. 1997;138:65–80. doi: 10.1083/jcb.138.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich D, Kostka S, Kraft R, Dingwall C, Laskey RA, Hartmann E, Prehn S. Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr Biol. 1995;5:383–392. doi: 10.1016/s0960-9822(95)00079-0. [DOI] [PubMed] [Google Scholar]
- Gorlich D, Pante N, Kutay U, Aebi U, Bischoff FR. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- Gorlich D, Prehn S, Laskey RA, Hartmann E. Isolation of a protein that is essential for the first step of nuclear protein import. Cell. 1994;79:767–778. doi: 10.1016/0092-8674(94)90067-1. [DOI] [PubMed] [Google Scholar]
- Hopper AK, Traglia HM, Dunst RW. The yeast RNA1 gene product necessary for RNA processing is located in the cytosol and apparently excluded from the nucleus. J Cell Biol. 1990;111:309–321. doi: 10.1083/jcb.111.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Jans DA. Efficiency of importin α/β-mediated nuclear localization sequence recognition and nuclear import. J Biol Chem. 1999;274:15820–15827. doi: 10.1074/jbc.274.22.15820. [DOI] [PubMed] [Google Scholar]
- Imamoto N, Shimamoto T, Takao T, Tachibana T, Kose S, Matsubae M, Sekimoto T, Shimonishi Y, Yoneda Y. In vivo evidence for involvement of a 58 kDa component of nuclear pore-targeting complex in nuclear protein import. EMBO J. 1995;14:3617–3626. doi: 10.1002/j.1460-2075.1995.tb00031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E, Kutay U, Vonkobbe C, Mattaj IW, Gorlich D. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 1997;16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kose S, Imamoto N, Tachibana T, Shimamoto T, Yoneda Y. Ran-unassisted nuclear migration of a 97-kDa component of the nuclear pore-targeting complex. J Cell Biol. 1997;139:841–849. doi: 10.1083/jcb.139.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kose S, Imamoto N, Tachibana T, Yoshida M, Yoneda Y. Beta-subunit of nuclear pore-targeting complex (importin-beta) can be exported from the nucleus in a Ran-independent manner. J Biol Chem. 1999;274:3946–3952. doi: 10.1074/jbc.274.7.3946. [DOI] [PubMed] [Google Scholar]
- Lam MHC, Briggs LJ, Hu W, Martin TJ, Gillespie MT, Jans DA. Importin β recognizes parathyroid hormone-related protein with high affinity and mediates its nuclear import in the absence of importin α. J Biol Chem. 1999;274:7391–7398. doi: 10.1074/jbc.274.11.7391. [DOI] [PubMed] [Google Scholar]
- Mattaj IW, Englmeier L. Nucleocytoplasmic transport—the soluble phase. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- Melchior F, Guan T, Yokoyama N, Nishimoto T, Gerace L. GTP hydrolysis by Ran occurs at the nuclear pore complex in an early step of protein import. J Cell Biol. 1995;131:571–581. doi: 10.1083/jcb.131.3.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior F, Paschal B, Evans J, Gerace L. Inhibition of nuclear protein import by nonhydrolyzable analogues of GTP and identification of the small GTPase Ran/TC4 as an essential transport factor. J Cell Biol. 1993a;123:1649–1659. doi: 10.1083/jcb.123.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior F, Weber K, Gerke V. A functional homologue of the RNA1 gene product in Schizosaccharomyces pombe: purification, biochemical characterization, and identification of a leucine-rich repeat motif. Mol Biol Cell. 1993b;4:569–581. doi: 10.1091/mbc.4.6.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael WM, Eder PS, Dreyfuss G. The K nuclear shuttling domain: a novel signal for nuclear import and nuclear export in the hnRNP K protein. EMBO J. 1997;16:3587–3598. doi: 10.1093/emboj/16.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MS, Blobel G. The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature. 1993;365:661–663. doi: 10.1038/365661a0. [DOI] [PubMed] [Google Scholar]
- Moore MS, Blobel G. Purification of a Ran-interacting protein that is required for protein import into the nucleus. Proc Natl Acad Sci, USA. 1994;91:10212–10216. doi: 10.1073/pnas.91.21.10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroianu J, Hijikata M, Blobel G, Radu A. Mammalian karyopherin alpha 1 beta and alpha 2 beta heterodimers: alpha 1 or alpha 2 subunit binds nuclear localization signal and beta subunit interacts with peptide repeat-containing nucleoporins. Proc Natl Acad Sci USA. 1995;92:6532–6536. doi: 10.1073/pnas.92.14.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny S, Dreyfuss G. Import and export of the nuclear protein import receptor transportin by a mechanism independent of GTP hydrolysis. Curr Biol. 1997;8:89–95. doi: 10.1016/s0960-9822(98)70039-9. [DOI] [PubMed] [Google Scholar]
- Nehrbass U, Blobel G. Role of the nuclear transport factor p10 in nuclear import. Science. 1996;272:120–122. doi: 10.1126/science.272.5258.120. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- Ohtsubo M, et al. Isolation and characterization of the active cDNA of the human cell cycle gene (RCC1) involved in the regulation of onset of chromosome condensation. Genes Dev. 1987;1:585–593. doi: 10.1101/gad.1.6.585. [DOI] [PubMed] [Google Scholar]
- Paschal BM, Delphin C, Gerace L. Nucleotide-specific interaction of Ran/TC4 with nuclear transport factors NTF2 and p97. Proc Natl Acad Sci USA. 1996;93:7679–7683. doi: 10.1073/pnas.93.15.7679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschal BM, Fritze C, Guan T, Gerace L. High levels of the GTPase Ran/TC4 relieve the requirement for nuclear protein transport factor 2. J Biol Chem. 1997;272:21534–21539. doi: 10.1074/jbc.272.34.21534. [DOI] [PubMed] [Google Scholar]
- Paschal BM, Gerace L. Identification of NTF2, a cytosolic factor for nuclear import that interacts with nuclear pore complex protein p62. J Cell Biol. 1995;129:925–937. doi: 10.1083/jcb.129.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard VW, Michael WM, Nakielny S, Siomi MC, Wang F, Dreyfuss G. A novel receptor-mediated nuclear protein import pathway. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- Radu A, Blobel G, Moore MS. Identification of a protein complex that is required for nuclear protein import and mediates docking of import substrate to distinct nucleoporins. Proc Natl Acad Sci USA. 1995;92:1769–1773. doi: 10.1073/pnas.92.5.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren M, Drivas G, D'Eustachio P, Rush MG. Ran/TC4: a small nuclear GTP-binding protein that regulates DNA synthesis. J Cell Biol. 1993;120:313–323. doi: 10.1083/jcb.120.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K, Lipowsky G, Kent HM, Stewart M, Gorlich D. NTF2 mediates nuclear import of Ran. EMBO J. 1998;17:6587–6598. doi: 10.1093/emboj/17.22.6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffzek K, Klebe C, Fritz-Wolf K, Kabsch W, Wittinghofer A. Crystal structure of the nuclear Ras-related protein Ran in its GDP-bound form. Nature. 1995;374:378–381. doi: 10.1038/374378a0. [DOI] [PubMed] [Google Scholar]
- Siomi H, Dreyfuss G. A nuclear localization domain in the hnRNP A1 protein. J Cell Biol. 1995;129:551–560. doi: 10.1083/jcb.129.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A, Brownawell A, Macara IG. Nuclear import of Ran is mediated by the transport factor NTF2. Curr Biol. 1998;8:1403–1406. doi: 10.1016/s0960-9822(98)00023-2. [DOI] [PubMed] [Google Scholar]
- Snow CM, Senior A, Gerace L. Monoclonal antibodies identify a group of nuclear pore complex glycoproteins. J Cell Biol. 1987;104:1143–1156. doi: 10.1083/jcb.104.5.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M, Kent HM, McCoy AJ. Structural basis for molecular recognition between nuclear transport factor 2 (NTF2) and the GDP-bound form of the Ras-family GTPase Ran. J Mol Biol. 1998;277:635–646. doi: 10.1006/jmbi.1997.1602. [DOI] [PubMed] [Google Scholar]
- Tachibana T, Hieda M, Sekimoto T, Yoneda Y. Exogenously injected nuclear import factor p10/NTF2 inhibits signal-mediated nuclear import and export of proteins in living cells. FEBS Lett. 1996;397:177–182. doi: 10.1016/s0014-5793(96)01180-5. [DOI] [PubMed] [Google Scholar]
- Tachibana T, Imamoto N, Seino H, Nishimoto T, Yoneda Y. Loss of RCC1 leads to suppression of nuclear protein import in living cells. J Biol Chem. 1994;269:24542–24545. [PubMed] [Google Scholar]
- Vetter IR, Nowak C, Nishimoto T, Kuhlmann J, Wittinghofer A. Structure of a Ran-binding domain complexed with Ran bound to a GTP analogue: implications for nuclear transport. Nature. 1999;398:39–46. doi: 10.1038/17969. [DOI] [PubMed] [Google Scholar]
- Wilkinson MG, Millar JB. SAPKs and transcription factors do the nucleocytoplasmic tango. Genes Dev. 1998;12:1391–1397. doi: 10.1101/gad.12.10.1391. [DOI] [PubMed] [Google Scholar]
- Wong DH, Corbett AH, Kent HM, Stewart M, Silver PA. Interaction between the small GTPase Ran/Gsp1p and Ntf2p is required for nuclear transport. Mol Cell Biol. 1997;17:3755–3767. doi: 10.1128/mcb.17.7.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Tachibana T, Imamoto N, Yoneda Y. Nuclear transport factor p10/NTF2 functions as a Ran-GDP dissociation inhibitor (Ran-GDI) Curr Biol. 1998;8:1339–1342. doi: 10.1016/s0960-9822(07)00566-0. [DOI] [PubMed] [Google Scholar]